Abstract
Epigenetic regulation of histones mediates neurotrophin actions with histone acetylation enhancing cAMP response element-binding (CREB)-associated transcription elicited by brain-derived neurotrophic factor (BDNF) and nerve-growth factor (NGF). Roles for histone methylation in CREB's transcriptional activity have not been well characterized. We show that depletion of the histone methyltransferase suppressor of variegation 3–9 homolog 1 (SUV39H1) selectively augments BDNF- and NGF-mediated neurite outgrowth. SUV39H1 is the principal enzyme responsible for trimethylation of histone H3 at lysine 9, a molecular mark associated with transcriptional silencing. BDNF and NGF act via a signaling cascade wherein degradation of SUV39H1 down-regulates trimethylation of H3K9 in a nitric oxide-dependent pathway. BDNF activates neuronal NOS with the nitrosylated GAPDH/seven in absentia (Siah) homolog complex translocating to the nucleus. Degradation of SUV39H1 by Siah facilitates histone H3 on lysine 9 acetylation, CREB binding to DNA, enhanced expression of CREB-regulated genes and neurite outgrowth.
Keywords: epigenetics, GAPDH
Neurotrophic factors, such as nerve-growth factor (NGF) and brain-derived neurotrophic factor (BDNF), act by turning on genetic programs, especially those associated with cAMP response element-binding (CREB) (1, 2). Phosphorylation of CREB at serine-133 has been thought to be important in the actions of numerous growth factors including the neurotrophins (3–5). Recent evidence has established that growth factors, including neurotrophic factors, act by enhancing histone acetylation, which in turn permits activation of CREB (2, 6, 7). Riccio and associates (7) described a specific pathway that mediates stimulation of histone acetylation. In this model, S-nitrosylation of histone deacetylase-2 induces chromatin remodeling in neurons, indicating an important role for NO in this process. Histone acetylation leads to increased transcriptional activity, whereas histone methylation down-regulates transcription (8).
During apoptotic cell death, GAPDH translocates to the nucleus in a number of cell systems (9). NO mediates a wide range of physiologic and pathophysiologic cellular functions. Nuclear signaling by NO in response to apoptotic cell stressors leads to cell death. In this cascade, nitrosylation of GAPDH enables it to bind to the ubiquitin E3 ligase, seven in absentia homolog 1 (hereafter designated Siah), translocating nitrosylated GAPDH (SNO-GAPDH) to the nucleus (10). In the nucleus SNO-GAPDH generated in response to apoptotic cell stressors binds to the acetylating enzyme CREB-binding protein (CBP)/p300, activating it and leading to augmentation of apoptotic proteins (11, 12).
In the present study, we describe a signaling pathway wherein down-regulation of histone methylation mediates physiologic actions of neurotrophins such as BDNF and NGF. The neurotrophins activate neuronal NOS (nNOS) with the generated NO-nitrosylating GAPDH, enabling it to bind to Siah and translocate to the nucleus. In the nucleus, the neurotrophin pathway diverges from the previously reported apoptotic cascade. Instead of interacting with CBP/p300 as in the apoptotic scheme, neurotrophin treatment leads to the GAPDH–Siah complex associating with the histone-methylating enzyme suppressor of variegation 3–9 homolog 1 (SUV39H1) in a ternary complex. In this complex, Siah, a known ubiquitin E3 ligase, ubiquitinates SUV39H1, which then is degraded. Loss of SUV39H1's methylating activity leads to less methylation of histone 3 on lysine 9 (H3K9) and increases its acetylation, facilitating activation of CREB target genes and augmenting dendrite outgrowth.
Results
Histone-Methylating Enzyme SUV39H1 Regulates Dendritic Outgrowth.
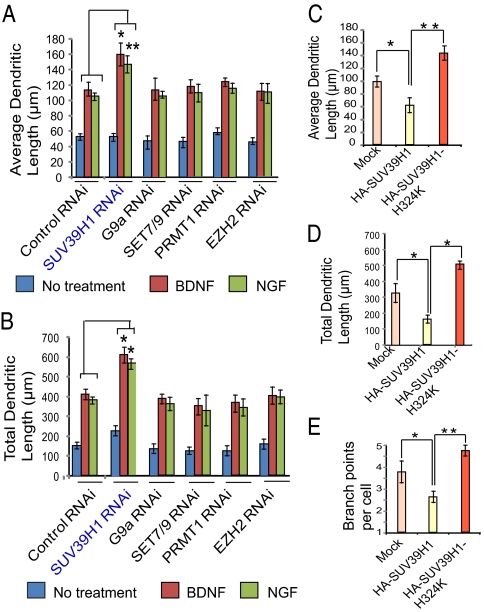
We wondered whether histone methylation, like histone acetylation, might play a role in neuronal process extension in response to neurotrophins. We investigated potential roles of the major histone-methylating enzymes SUV39H1 (13), G9a (14), SET domain-containing lysine methyltransferase 7/9 (SET7/9) (15), histone-lysine N-methyltransferase (EZH2) (16), and protein arginine N-methyltransferase 1 (PRMT1) (17). We depleted these five enzymes by RNAi using viral infection of cerebral cortical neuronal cultures with siRNA constructs (Fig. 1). Depletion of SUV39H1 elicits an approximately 50% increase in average (Fig. 1A) and total (Fig. 1B) dendritic length in preparations treated with either BDNF or NGF. By contrast, no alteration in dendritic length is associated with RNAi depletion of G9A, SET 7/9, EZH2, or PRMT1.
Fig. 1.
Influence of histone methyltransferases on dendritic outgrowth. (A) Average dendritic length of cortical neurons transfected with lentiviral siRNA constructs of SUV39H1, G9a, SET7/9, EZH2, or PRMT1. (B) Total dendritic length of cortical neurons transfected with lentiviral siRNA constructs of SUV39H1, G9a, SET7/9, EZH2, or PRMT1. (C–E) Average (C) and total (D) dendritic length and number of branch points per cell (E) were measured in cells transfected with wild-type SUV39H1 or with SUV39H1-H324K constructs. At least 30 neurons were analyzed for each condition for each experiment. *P < 0.01, **P < 0.001; n = 3; one-way ANOVA; mean ± SEM.
We then explored the influence of overexpressing SUV39H1 in cortical cultures, comparing it with catalytically inactive SUV39H1 with histidine-324 mutated to lysine (SUV39H1-H324K) (18). We monitored average (Fig. 1C) and total (Fig. 1D) dendritic length as well as the number of dendritic processes (Fig. 1E). Overexpressing SUV39H1 elicits a 40–50% reduction in average and total dendritic length and in the number of processes. By contrast, SUV39H1-H324K augments total and dendritic length by about 40% and process number by about 30%. Presumably, this catalytically inactive SUV39H1 acts as a dominant negative to prevent actions of endogenous SUV39H1, which, under basal conditions depress neuronal process extension. Thus, these findings indicate that the basal, steady-state disposition of neuronal processes is determined by SUV39H1.
SUV39H1 Mediates the Influence of Neurotrophins on CREB Genetic Programs.
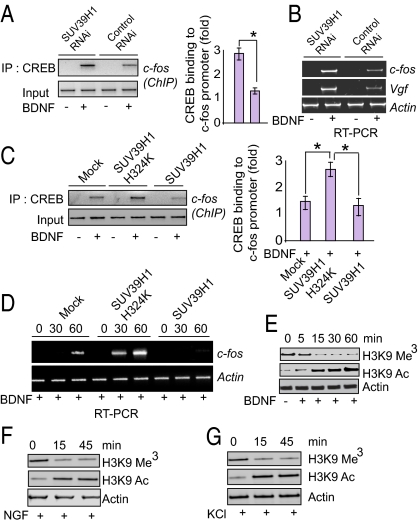
The neurotrophic actions of BDNF and NGF are known to be mediated by the CREB genetic program. We wondered whether the regulation of neuronal processes by SUV39H1 reflects an action of this methylating enzyme impacting CREB transcriptional events. We depleted SUV39H1 by viral-mediated RNAi in primary cortical neurons and examined the binding of CREB to the c-fos promoter by ChIP assay (Fig. 2A). BDNF stimulates CREB binding to the c-fos promoter. Depletion of SUV39H1 elicits a 2.7-fold increase in this binding, consistent with endogenous SUV39H1 physiologically suppressing this element of the CREB genetic program. Similarly, BDNF-induced augmentation of mRNA levels for the CREB target genes c-fos and Vgf is increased two- to threefold by depletion of SUV39H1 in cortical cultures (Fig. 2B).
Fig. 2.
Neurotrophin-induced CREB transcription is regulated by SUV39H1 and histone methylation. (A) PCR quantification of ChIP analysis of primary cortical neuronal cells depleted of SUV39H1 and stimulated with BDNF. CREB immunoprecipitation was followed by PCR analysis of the c-fos promoter. Densitometric analysis of CREB binding to c-fos promoter: *P < 0.01; n = 3; one-way ANOVA; mean ± SEM. (B) mRNA levels of c-fos, vaccinia nerve growth factor (vgf), and actin in primary cortical neuronal cells after depletion of SUV39H1. (C) PCR quantification of ChIP analysis of primary cortical neuronal cells transfected with wild-type SUV39H1 or SUV39H1-H324K, and stimulated with BDNF. CREB immunoprecipitation was followed by PCR analysis of the c-fos promoter. Densitometric analysis of CREB binding to c-fos promoter: *P < 0.01, n = 3, one-way ANOVA, mean ± SEM. (D) mRNA levels of c-fos and actin in primary cortical neuronal cells after transfection of SUV39H1 and SUV39H1-H324K constructs in primary cortical neurons. (E–F) Immunoblot analysis of methylated and acetylated H3K9 and actin in primary cortical neuronal cells after treatment with BDNF (E) and NGF (F). (G) Immunoblot analysis of methylated and acetylated H3K9 and actin in PC-12 cells after treatment with KCl.
We explored whether overexpression of SUV39H1 might act in a fashion opposite to its depletion. Overexpression of SUV39H1 in cortical cultures decreases CREB binding to the c-fos promoter (Fig. 2C) and lowers levels of c-fos mRNA (Fig. 2D). By contrast, catalytically inactive SUV39H1–H324K markedly increases CREB binding to the c-fos promoter (Fig. 2C) and elicits a five- to 10-fold increase in c-fos mRNA (Fig. 2D). These findings establish that catalytic activity of SUV39H1 is important for BDNF-induced CREB transcriptional activity.
We investigated whether BDNF directly alters SUV39H1 actions. H3K9 is the sole physiologic substrate of SUV39H1, which trimethylates H3K9 (13). We monitored its trimethylation in response to BDNF (Fig. 2E), NGF (Fig. 2F), and potassium depolarization (KCl treatment) (Fig. 2G), all well known to stimulate nerve outgrowth (19). Each of the three treatments markedly reduced levels of trimethylated histone H3K9 in a time-dependent fashion. Histone methylation and acetylation are reciprocal processes (20). We observe that diminished H3K9 methylation is associated with increased acetylation (Fig. 2 E–G).
Neurotrophins Elicit Degradation of SUV39H1 by Increasing Its Ubiquitination by Siah.
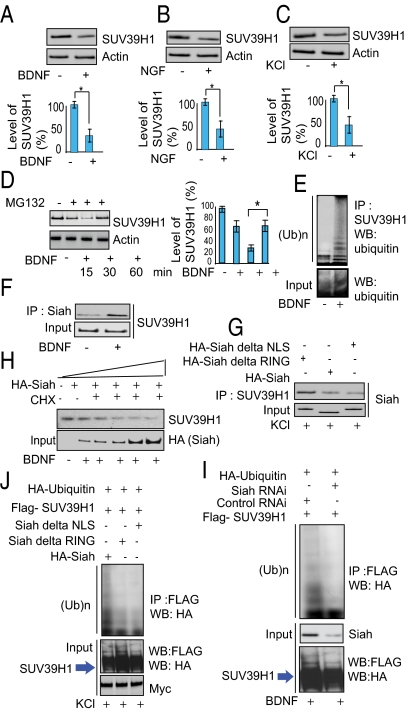
How might neurotrophins decrease levels of the methylated histone targets of SUV39H1? One possibility would be that neurotrophins somehow elicit degradation of SUV39H1 itself. In primary cortical cultures treatment with BDNF (Fig. 3A), NGF (Fig. 3B), or KCl (Fig. 3C) markedly reduces levels of SUV39H1 protein.
Fig. 3.
Siah is required for ubiquitination of SUV39H1. (A–C) Immunoblot analysis of SUV39H1 in BDNF- (A) and NGF- (B) treated primary cortical neuronal cells and in KCl-treated (C) PC-12 cells. Densitometric analysis of SUV39H1 in BDNF, NGF, and KCl treated cells: *P < 0.01, n = 3, one-way ANOVA, mean ± SEM. (D) Proteasome inhibition leads to SUV39H1 up-regulation. Primary neurons were treated with MG132 as indicated. Endogenous SUV39H1 and actin were detected by Western blots. *P < 0.01, n = 4, one-way ANOVA, mean ± SEM. (E) SUV39H1 is polyubiquitinated endogenously in primary neurons upon treatment with BDNF. Ten percent of the input lysate was analyzed by Western blot (WB) (Lower). IP, immunoprecipitation; (Ub)n, polyubiquitin chains. (F) Siah–SUV39H1 binding in primary neurons treated with BDNF. Cell lysates were immunoprecipitated (IP) with an anti-Siah lysine antibody, and the immunoprecipitates were analyzed by Western blotting with an anti-SUV39H1 antibody. (G) Siah lacking the NLS or RING domain does not bind SUV39H1. (H) SUV39H1 is degraded by Siah in PC-12 cells. SUV39H1 and actin proteins were detected by Western blot in cycloheximide (CHX)-treated cells with increasing concentrations of HA-Siah. (I) Depletion of Siah by RNAi leads to diminished ubiquitination of SUV39H1. (J) Siah lacking the NLS or RING domain does not elicit SUV39H1 ubiquitination. PC-12 cells were transfected as indicated, and their lysates were analyzed by immunoprecipitation and Western blotting. Ten percent of the input lysate was analyzed by Western blotting with an anti-HA and anti-FLAG antibody.
We explored whether proteasomal degradation by the ubiquitin system is responsible for the loss of SUV39H1 in response to neurotrophins. The time-dependent depletion of SUV39H1 in cortical cultures in response to BDNF is prevented by treatment with Z-Leu-Leu-Leu-CHO (MG132), an inhibitor of proteasomal activity (Fig. 3D). We directly assessed ubiquitination of SUV39H1 and observed substantial increases in cortical cultures treated with either BDNF (Fig. 3E) or NGF (Fig. S1A).
We wished to determine the identity of the enzyme(s) responsible for degradation of SUV39H1. In immunoprecipitation experiments, Siah coprecipitates with SUV39H1 (Fig. S1B), which possesses a consensus substrate sequence for Siah (Fig. S1C). Moreover, we directly demonstrate binding of Siah to SUV39H1, which is stimulated 3.2-fold by BDNF treatment of primary cortical cultures (Fig. 3F).
Catalytic activity of Siah requires the presence of a really interesting new gene (RING) domain and a nuclear-localization signal (NLS) domain. Binding of Siah to SUV39H1 in KCl-treated preparations is diminished markedly in experiments using Siah lacking the NLS or RING domain (Fig. 3G). We directly demonstrate Siah-elicited degradation of SUV39H1 in cortical cultures treated with increasing concentrations of Siah in the presence of cycloheximide, with virtual abolition of SUV39H1 in the presence of the highest levels of Siah (Fig. 3H).
Ubiquitination of SUV39H1 requires Siah (Fig. 3I). Thus, ubiquitination of SUV39H1 is reduced markedly in cortical cultures with viral-mediated depletion of Siah by RNAi. The NLS and RING domains of Siah are required for the ubiquitination process, because Siah lacking the NLS or RING domain fails to elicit ubiquitination of SUV39H1 in KCl-treated cells (Fig. 3J).
Neurotrophins Stabilize Siah by the Generation of NO, Which Nitrosylates GAPDH, Which in Turn Binds to and Stabilizes Siah.
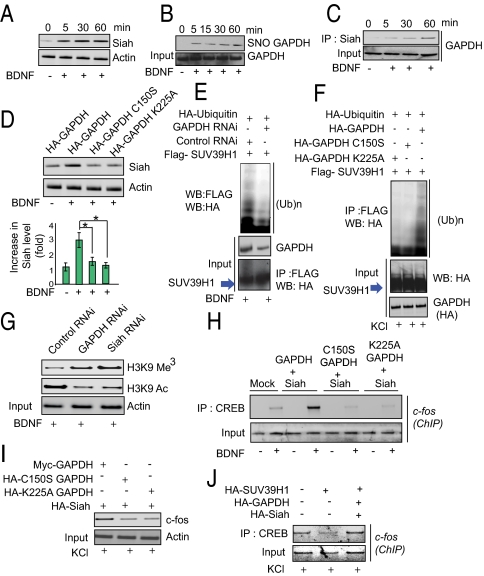
Up to this point our experiments established that neurotrophins stimulate neurite outgrowth by a signaling cascade wherein Siah binds SUV39H1, leading to its degradation and thereby diminishing histone H3K9 methylation and enhancing the CREB genetic program. We wondered how neurotrophins might stimulate Siah. In primary cultures BDNF (Fig. 4A), NGF (Fig. S2A), and KCl (Fig. S2B) increase Siah levels about threefold. Siah is a protein that turns over very rapidly, with increases in its levels often reflecting its stabilization via inhibition of its proteasomal degradation (21). Previously we reported NO is generated that in response to cell stressors, leading to nitrosylation of GAPDH with SNO-GAPDH binding to Siah and stabilizing it (10). We investigated whether a similar process takes place in response to neurotrophins. Both BDNF and NGF increase nitrosylation of GAPDH in cortical cultures (Fig. 4B and Fig. S2 C and D), an effect lost in nNOS-deleted mice.
Fig. 4.
GAPDH regulates Siah stabilization, histone methylation, and CREB transcription. (A) Immunoblot analysis of Siah in BDNF-treated primary cortical neurons. (B) BDNF stimulates GAPDH nitrosylation. Cortical neurons were stimulated with BDNF for the indicated times. Protein extracts were subjected to the biotin-switch assay followed by streptavidin precipitation and Western blotting for GAPDH. (C) BDNF increases the binding of GAPDH to Siah in a time-dependent manner. Cell lysates were immunoprecipitated with anti-Siah1 antibody. (D) Siah is stabilized by overexpression of wild-type GAPDH but not GAPDH-C150S or GAPDH-K225A. (E) Depletion of GAPDH and Siah prevents SUV39H1 ubiquitination in primary cortical neurons. (F) Overexpression of GAPDH-C150S or GAPDH-K225A prevents ubiquitination of SUV39H1 in PC-12 cells upon KCl treatment. (G) Depletion of GAPDH or Siah by RNAi decreases histone methylation and increases histone acetylation. Immunoblot analysis of histone methylation and acetylation in GAPDH- or Siah-depleted cells upon treatment with BDNF. (H) CREB binding to c-fos promoter is enhanced by GAPDH (in BDNF-treated cells) but not by GAPDH-C150S or GAPDH-K225A. PCR quantification of ChIP analysis of primary neuronal cells transfected with various GAPDH and Siah vectors as indicated and stimulated with BDNF. CREB immunoprecipitation was followed by PCR analysis of the c-fos promoter. (I) C-fos levels are depleted by GAPDH-C150S and GAPDH-K225A. Immunoblot analysis of c-fos in BDNF-treated primary neuronal cells after overexpression of GAPDH, C150S-GAPDH or K225A-GAPDH. (J) Reduced CREB binding to c-fos promoter elicited by SUV39H1 is reversed by overexpressed GAPDH and Siah. PCR quantification of ChIP analysis of primary neuronal cells transfected with GAPDH and Siah vectors in SUV39H1-overexpressing cells stimulated with BDNF. CREB immunoprecipitation was followed by PCR analysis of the c-fos promoter.
Nitrosylation of GAPDH by cell stressors occurs at cysteine150 (C150), which is critical for catalytic activity (10). Nitrosylation of GAPDH in response to BDNF also is abolished by C150S mutation (Fig. S2E) in primary cortical neurons. Cell stressors elicit binding of GAPDH to Siah (10, 11). BDNF stimulation also enhances GAPDH–Siah binding, with stimulation evident as early as 5 min after BDNF treatment (Fig. 4C). The GAPDH–Siah binding is dependent on NO, being lost in nNOS-knockout primary cortical neurons (Fig. S2F). This binding is critically dependent on GAPDH lysine 225 (K225), whose mutation abolishes binding (Fig. S2). We examined the influence of mutating C150 and K225 of GAPDH upon Siah levels (Fig. 4D). Both mutations prevent BDNF and NGF enhancement of Siah levels.
These data are consistent with a model in which neurotrophins stimulate NO formation, as previously reported (6), leading to nitrosylation of GAPDH, which binds to Siah and stabilizes it with the SNO-GAPDH–Siah complex translocating to the nucleus and binding to SUV39H1 in a ternary complex. In this complex Siah ubiquitinates SUV39H1, eliciting its degradation so that histone H3K9 trimethylation is markedly diminished, leading to augmentation of the transcriptional activity of CREB and neuronal process extension.
If this model is valid, then perturbations of GAPDH that influence its binding to Siah should alter the overall signaling cascade. Depletion of GAPDH by RNAi prevents the ubiquitination of SUV39H1 (Fig. 4E). This process is dependent on the binding of GAPDH to Siah, because SUV39H1 ubiquitination is abolished by the GAPDH-K225A mutation, which prevents GAPDH binding to Siah (Fig. 4F). SUV39H1 ubiquitination also is prevented by the GAPDH-C150S mutation (Fig. 4F). The importance of GAPDH and Siah for downregulating histone H3K9 trimethylation is evident in the substantial increase of this methylation in cortical cultures in which GAPDH and Siah are depleted by RNAi (Fig. 4G and Fig. S3A). The increased methylation is associated with decreased H3K9 acetylation (Fig. 4G). Moreover, histone H3K9 trimethylation is augmented by both GAPDH-C150S and GAPDH-K225A mutation (Fig. S3B). The influence of BDNF on histone H3K9 acetylation and methylation is diminished markedly when Siah lacks its NLS or RING domain (Fig. S3C).
The GAPDH–Siah interaction is required for the transcriptional activation of CREB, because CREB interactions with the c-fos promoter are enhanced by GAPDH but not by GAPDH-C150S or GAPDH-K225A (Fig. 4H). Moreover, c-fos protein levels are reduced substantially with GAPDH-C150S or GAPDH-K225A as compared with wild-type GAPDH (Fig. 4I). Additionally, in PC-12 cells, overexpression of GAPDH and Siah reverses the decreased binding of CREB to the c-fos promoter elicited by SUV39H1 (Fig. 4J).
Discussion
Earlier, we defined an apoptotic signaling cascade in which cell stressors activate NO formation, which nitrosylates GAPDH, enabling it to bind to Siah, leading to nuclear translocation of the GAPDH–Siah complex and augmented acetylation of p300/CBP to activate apoptotic targets such as p53 (11). The present study demonstrates that nonapoptotic, physiologic stimuli, the growth factors BDNF and NGF, also signal via the GAPDH–Siah pathway. Although nitrosylation of GAPDH in response to BDNF is robust, it is less substantial than that elicited by concentrations of the NO donor nitrosoglutathione (GSNO) that lead to cell death (Fig. S4A). This result accords with abundant evidence that modest levels of NO are physiologic, whereas high concentrations are pathophysiologic (22). The apoptotic actions of GSNO involve its stimulating the binding of GAPDH and p300/CBP (Fig. S4B). This process is competitive with interactions of GAPDH and SUV39H1 whose overexpression decreases the binding of p300/CBP to GAPDH. Conversely, treatment of cells with GSNO at concentrations that elicit apoptosis leads to decreased SUV39H1 levels (Fig. S4C). In another instance of competitive interactions in the SNO-GAPDH signaling cascade, binding of SNO-GAPDH to p300/CBP abrogates p300/CBP–CREB interactions (Fig. S4D). Another physiologic nuclear action of GAPDH involves SNO-GAPDH transnitrosylating nuclear proteins such as histone deacetylase 2 (HDAC-2) and DNA-PK (23).
The signaling cascade that we have established operates in the following fashion (Fig. S4E). BDNF activates nNOS, and the generated NO nitrosylates GAPDH to enable it to bind to Siah and translocate to the nucleus. In the nucleus the GAPDH–Siah complex associates with the methylating enzyme SUV39H1 in a ternary complex. In the ternary complex Siah ubiquitinates SUV39H1, leading to its degradation. Loss of SUV39H1's methylating activity leads to decreased methylation of histone H3K9 and its enhanced acetylation, in turn facilitating the actions of CREB which, in association with acetylated histone H3K9, binds to promoters such as c-fos to stimulate neurite outgrowth. The GAPDH–Siah–SUV39H1 cascade appears to mediate a major portion of the neurotrophic actions of BDNF. Thus, the enhancement of dendritic outgrowth elicited by BDNF is reduced by half after GAPDH depletion. Overall, our findings show that the influences of neurotrophins upon heterochromatin and neural process disposition are mediated by regulation of SUV39H1 levels.
How do our findings fit with earlier studies reporting BDNF enhancement of NO formation and stimulation of nitrosylation of the protein deacetylase HDAC-2 (7)? Nitrosylation of HDAC-2 leads to its dissociation from chromatin so that acetylation of histones can be augmented, thereby facilitating the actions of CREB. In this way, nitrosylation of HDAC-2 and of GAPDH play complementary roles, both leading to increased acetylation of histone H3K9 and thereby augmented effects of CREB.
In terms of functional effects of BDNF, we have presented data on neurite outgrowth. BDNF exerts numerous other actions. For instance, it is required for the therapeutic effects of the major antidepressant drugs (24) as well as the rewarding actions of cocaine (25, 26). Numerous investigators have reported possible links between BDNF and central nervous system disturbances such as schizophrenia, obsessive-compulsive disorder, Alzheimer's disease, Huntington disease, and Rett's syndrome (27).
Might our findings have therapeutic implications? The BDNF signaling pathway we describe depends on the integrity of the ternary complex of GAPDH–Siah–SUV39H1. Agents that enhance the stability of this complex to facilitate the selective degradation of SUV39H1 within the complex would be expected to elicit some of the therapeutic benefits associated with BDNF.
Materials and Methods
Cell Cultures.
Embryonic cortical mouse neurons were cultured as previously described (28). Before BDNF or NGF treatments, cells were placed in low-serum medium [3% (vol/vol) FBS] containing 50 μM dl-2-amino-5-phosphonovaleric acid for 12–16 h. PC-12 cells were maintained in DMEM supplemented with 10% (vol/vol) FBS and 5% horse serum. Cells were transfected with Lipofectamine 2000 containing 24 μg of DNA 2000 (Invitrogen) and incubated for 8–10 h before the transfection medium was replaced with serum-containing medium. Before KCl treatment, PC-12 cells were starved overnight in medium containing 0.5% horse serum.
For RNAi experiments, medium for PC-12 cells was supplemented with 10 mM pyruvate, and cells were transfected with 167 nM GAPDH or control siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. After overnight starvation with 0.5% horse serum (24 h after transfection), PC-12 cells were incubated in the presence of KCl for an additional 1 h. Immunofluorescent staining of cells for confocal microscopy was carried out as described previously (10). We used lentiviral particles (Santa Cruz) for knockdown of SUV39H1 (sc-38464-V), PRMT1 (sc-41070-V), and G9a (sc-43777-V). For overexpression in primary neurons, we used lentiviral particles and followed the manufacturer's protocol (Invitrogen).
Identification of Interactor Proteins with SUV39H1.
Flag-conjugated SUV39H1 was incubated with cell lysates of BDNF-treated primary cortical neurons. Proteins were visualized by Coomassie stain, and the identity of Siah was verified by mass spectrometric analysis.
Dendritic Length Assay.
The dendritic length assay was performed as described previously (29), with modifications. Briefly, primary neurons were transfected with various constructs of GAPDH, Siah, and SUV39H1 along with GFP, and images of neuronal morphologies were captured based on immunoreactivities against GFP, using the 510 META confocal laser-scanning microscope (LSM) system (Zeiss). Dendrites and axons were identified by standard morphological criteria. Because the majority of neurons in our cortical culture preparation possessed only one clearly classifiable axon and one or more dendrites, neurons with nonpyramidal morphological features (such as multiple axons or no classifiable processes) were excluded from analyses. The average and total length were determined manually using Neuron J1.0.0 (30) plug-in software for ImageJ (National Institutes of Health). Representative images were acquired using the 510 META confocal LSM with a 40× objective. All analyses were performed by an observer blinded to the identity of the transfected constructs.
Quantitative Real-Time PCR.
Quantitative real-time PCR was performed as described previously (7). PCR reactions (25 μL) contained 12.5 μL of PCR Sybr Green mix (New England BioLabs) with 0.3-μM primers. At the end of the 35 cycles of amplification, a dissociation curve was performed in which Sybr Green was measured at 1-°C intervals between 50 °C and 100 °C. Results were normalized using total input DNA and expressed as bound/Input (percentage).
S-Nitrosylation Biotin-Switch Assay.
The S-nitrosylation biotin-switch assay was performed as described (31). In brief, cells were lysed, and reduced cysteines were blocked with 4 mM methyl methanethionsulphonate. Subsequently, S-nitrosylated cysteines were reduced with 1 mM ascorbate and biotinylated with 1 mM Biotin-HPDP (Pierce). The biotinylated proteins were pulled down with streptavidin agarose and analyzed by Western blotting.
ChIP Assay.
ChIP assay was performed as described previously (7, 32). In brief, intact cells were treated with 2 mM disuccinimidyl glutarate (Pierce) to cross-link protein complexes and then were treated with formaldehyde to link protein to DNA covalently. Cells were lysed, the nucleoprotein complexes were sonicated, and the cross-linked DNA–protein complexes were enriched by immunoprecipitation with specific antibodies. The retrieved complexes then were analyzed by PCR amplification to detect and quantify specific DNA targets. For real-time PCR we used Brilliant SYBR green master mix (Stratagene) according to the manufacturer's protocol.
Extraction of Nuclear and Cytoplasmic Proteins.
Nuclear and cytoplasmic extracts were prepared using the Biovision nuclear/cytosol extraction kit according to the manufacturer's instructions.
Ubiquitination of SUV39H1.
FLAG-SUV39H1 and HA-ubiquitin were coexpressed with various constructs of GAPDH and/or Siah in PC-12 cells. Forty-eight hours after transfection, cells were treated with BDNF or were left untreated. Cells were harvested and lysed, and equal amounts of proteins were loaded on SDS-PAGE. FLAG-SUV39H1 was immunoprecipitated, and ubiquitination was detected by anti-HA antibody. PC-12 cells expressing FLAG-SUV39H1were treated with 20 μM MG132 for 5 h after BDNF treatment. SUV39H1 was immunoprecipitated using an anti-FLAG antibody and was run on SDS/PAGE. Ubiquitinated SUV39H1 was detected with an anti-FLAG antibody.
Cycloheximide Treatment.
Cycloheximide (20 μg/mL) was added for the indicated time periods to cells overexpressing HA-Siah in a concentration (1, 5, 10, 15, 24 μg)-dependent manner. Cells were lysed, and equal amounts of proteins were loaded on SDS/PAGE.
Coimmunoprecipitation.
Cells were lysed in lysis buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 0.1% CHAPS, 100 μM deferoxamine, and 1 mM EDTA) and homogenized by passage through a 26-gauge needle. Crude lysates were cleared of insoluble debris by centrifugation at 14,000 × g. Immunoprecipitation buffer {50 mM Tris (pH 7.4), 150 mM NaCl, 0.1% CHAPS, 100 μM deferoxamine, 1 mM EDTA, and 0.1 mg/mL BSA] was added to 100 μg of cell lysates to bring samples to a total volume of 1,000 μL. Anti-Siah and anti-SUV39H1 antibody and 30 μL of protein G agarose were added and incubated on a rotator at 4 °C. The protein G agarose was washed four times with lysis buffer and quenched with 30 μL of SDS sample buffer. Coimmunoprecipitates were resolved by SDS-PAGE and analyzed by Western blotting with anti-GAPDH and anti-SUV39H1 antibodies.
Statistical Analysis.
P values were calculated by one-way ANOVA using MINITAB 13 (Minitab).
Supplementary Material
Acknowledgments
This study was supported by US Public Health Service Grant MH18501 (to S.H.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117820108/-/DCSupplemental.
References
- 1.Walton MR, Dragunow I. Is CREB a key to neuronal survival? Trends Neurosci. 2000;23:48–53. doi: 10.1016/s0166-2236(99)01500-3. [DOI] [PubMed] [Google Scholar]
- 2.Riccio A. Dynamic epigenetic regulation in neurons: Enzymes, stimuli and signaling pathways. Nat Neurosci. 2010;13:1330–1337. doi: 10.1038/nn.2671. [DOI] [PubMed] [Google Scholar]
- 3.Kornhauser JM, et al. CREB transcriptional activity in neurons is regulated by multiple, calcium-specific phosphorylation events. Neuron. 2002;34:221–233. doi: 10.1016/s0896-6273(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 4.Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Shaywitz AJ, Greenberg ME. CREB: A stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 6.Riccio A, et al. A nitric oxide signaling pathway controls CREB-mediated gene expression in neurons. Mol Cell. 2006;21:283–294. doi: 10.1016/j.molcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 9.Chuang DM, Hough C, Senatorov VV. Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2005;45:269–290. doi: 10.1146/annurev.pharmtox.45.120403.095902. [DOI] [PubMed] [Google Scholar]
- 10.Hara MR, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 11.Sen N, et al. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sen N, Snyder SH. Protein modifications involved in neurotransmitter and gasotransmitter signaling. Trends Neurosci. 2010;33:493–502. doi: 10.1016/j.tins.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 14.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 16.Nolz JC, Gomez TS, Billadeau DD. The Ezh2 methyltransferase complex: Actin up in the cytosol. Trends Cell Biol. 2005;15:514–517. doi: 10.1016/j.tcb.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 18.Mal AK. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006;25:3323–3334. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redmond L, Kashani AH, Ghosh A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34:999–1010. doi: 10.1016/s0896-6273(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 20.Rice JC, Allis CD. Histone methylation versus histone acetylation: New insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 21.Hu G, Fearon ER. Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol Cell Biol. 1999;19:724–732. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: A current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornberg MD, et al. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;12:1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brunoni AR, Lopes M, Fregni F. A systematic review and meta-analysis of clinical studies on major depression and BDNF levels: Implications for the role of neuroplasticity in depression. Int J Neuropsychopharmacol. 2008;11:1169–1180. doi: 10.1017/S1461145708009309. [DOI] [PubMed] [Google Scholar]
- 25.Graham DL, et al. Dynamic BDNF activity in nucleus accumbens with cocaine use increases self-administration and relapse. Nat Neurosci. 2007;10:1029–1037. doi: 10.1038/nn1929. [DOI] [PubMed] [Google Scholar]
- 26.Maze I, Nestler EJ. The epigenetic landscape of addiction. Ann N Y Acad Sci. 2011;1216:99–113. doi: 10.1111/j.1749-6632.2010.05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattson MP. Glutamate and neurotrophic factors in neuronal plasticity and disease. Ann N Y Acad Sci. 2008;1144:97–112. doi: 10.1196/annals.1418.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sen N, et al. GOSPEL: A neuroprotective protein that binds to GAPDH upon S-nitrosylation. Neuron. 2009;63(1):81–91. doi: 10.1016/j.neuron.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takemoto-Kimura S, et al. Regulation of dendritogenesis via a lipid-raft-associated Ca2+/calmodulin-dependent protein kinase CLICK-III/CaMKIgamma. Neuron. 2007;54(5):755–770. doi: 10.1016/j.neuron.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 30.Meijering E, et al. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58(2):167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 31.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3(2):193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 32.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39(5):715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.