Abstract
Soft-tissue sarcomas are heterogeneous cancers that can present with tissue-specific differentiation markers. To examine the cellular basis for this histopathological variation and to identify sarcoma-relevant molecular pathways, we generated a chimeric mouse model in which sarcoma-associated genetic lesions can be introduced into discrete, muscle-resident myogenic and mesenchymal cell lineages. Expression of Kirsten rat sarcoma viral oncogene [Kras(G12V)] and disruption of cyclin-dependent kinase inhibitor 2A (CDKN2A; p16p19) in prospectively isolated satellite cells gave rise to pleomorphic rhabdomyosarcomas (MyoD-, Myogenin- and Desmin-positive), whereas introduction of the same oncogenetic hits in nonmyogenic progenitors induced pleomorphic sarcomas lacking myogenic features. Transcriptional profiling demonstrated that myogenic and nonmyogenic Kras; p16p19null sarcomas recapitulate gene-expression signatures of human rhabdomyosarcomas and identified a cluster of genes that is concordantly up-regulated in both mouse and human sarcomas. This cluster includes genes associated with Ras and mechanistic target of rapamycin (mTOR) signaling, a finding consistent with activation of the Ras and mTOR pathways both in Kras; p16p19null sarcomas and in 26–50% of human rhabdomyosarcomas surveyed. Moreover, chemical inhibition of Ras or mTOR signaling arrested the growth of mouse Kras; p16p19null sarcomas and of human rhabdomyosarcoma cells in vitro and in vivo. Taken together, these data demonstrate the critical importance of lineage commitment within the tumor cell-of-origin in determining sarcoma histotype and introduce an experimental platform for rapid dissection of sarcoma-relevant cellular and molecular events.
Keywords: cancer stem cell, sarcoma oncogenes, preclinical screening platform
Soft-tissue sarcomas (STS) are malignant tumors of mesodermal lineages that arise in nonepithelial, nonhematopoietic tissues, such as skeletal muscle. STS vary clinically and histopathologically (1, 2), and this heterogeneity may result from differences in the lineage commitment and differentiation state of the sarcoma cell-of-origin, from genetic or epigenetic changes that occur during transformation, or from a combination of these factors (3). Sarcomas with myogenic features constitute the large and varied category of rhabdomyosarcomas (RMS) (4, 5), for which satellite cells (6, 7), more differentiated muscle-lineage cells (3, 8), and undifferentiated mesenchymal cells (9) have been discussed as putative cells-of-origin. Still, the network of cellular and molecular events driving sarcomas in muscle is largely unknown (10). A number of oncogenetic lesions have been linked to these tumors. Activating Ras mutations can be found in up to 35% of human embryonal RMS (11, 12), and in up to 44% of human STS (13). Ectopic expression of oncogenic Kras induces RMS in zebrafish (6) and cooperates with loss of tumor protein p53 (Tp53) to induce RMS when activated broadly in whole mouse muscle (14). Like Ras, perturbations of the CDKN2A locus, encoding p16/p14Ink4A and p19ARF, also have been linked to sarcoma pathogenesis (15). Alterations in CDKN2A and its downstream effectors Retinoblastoma 1 (Rb1) and Tp53 have been noted in human STS (3, 16, 17), and one study reported reduced or absent expression of p16INK4A and/or p14ARF in human RMS (18). Together, these data implicate both oncogenic Kras and dysregulated p16p19 as clinically relevant, sarcoma-associated lesions.
In the studies described here, we developed a strategy to induce sarcomas in skeletal muscle to assess the effects of intrinsic and extrinsic cell states on the outcome of sarcoma-associated genetic lesions in muscle. By introducing relevant oncogenetic lesions [activation of Kras(G12V) and deletion of p16INK4Ap19ARF] into discrete mesodermal cell lineages purified directly from mouse muscle, we demonstrate that lineally distinct cell populations serve as tumor cells-of-origin for distinct sarcoma subtypes. However, despite notable differences in histological presentation, transcriptional profiling of these induced sarcomas revealed substantial overlap among up-regulated genes, including a common set of transcripts induced in Kras; p16p19null sarcomas and human RMS. This gene set is enriched in Ras-associated genes and genes linked to cell growth and proliferation, including the mTOR pathway, suggesting a potentially broad significance of these genes for sarcoma growth. Immunohistochemical (IHC) staining of both mouse Kras; p16p19null sarcomas and human RMS confirmed activation of Ras and mTOR effector molecules, and pharmacological inhibitors of Ras or mTOR (19–21) significantly impeded sarcoma cell growth. These data validate the utility of the Kras; p16p19null sarcoma system as a platform to identify critical cellular and molecular underpinnings of sarcomas.
Results
Mouse Skeletal Muscle Contains Myogenic and Nonmyogenic Precursors with Distinct Lineage Potential.
Skeletal muscle is composed of multinucleated myofibers as well as a variety of muscle-forming (myogenic) and nonmyogenic stem, progenitor, and mature cells. These distinct myofiber-associated (MFA) cell subsets can be isolated from dissociated mouse muscle by combinatorial staining for cell-surface markers and fluorescence-activated cell sorting (FACS) (Fig. 1A and Table S1) (22–24). To examine the contributions of individual MFA cell lineages to the formation of sarcomas in skeletal muscle, we sorted three distinct cell subsets directly from muscle (15): (i) CD45−MAC1−TER119−Sca1−β1-integrin+CXCR4+ cells, representing Pax7-expressing satellite cells highly enriched for myogenic stem cells (23, 24); (ii) CD45−MAC1−TER119−Sca1+ cells, hereafter designated “Sca1+ cells,” representing nonmyogenic progenitor cells (23, 24) with fibrogenic and adipogenic activity (Fig. 1B) (25–27); and (iii) CD45−MAC1−TER119−Sca1−CXCR4− cells, hereafter designated “CXCR4− cells,” a heterogeneous population containing both adipogenic and muscle-lineage cells (Fig. 1B). Previous studies show that the muscle-lineage cells within the CXCR4− population express muscle differentiation markers and lack in vitro clonogenic activity and in vivo myogenic engraftment capacity, consistent with a more differentiated phenotype (Table S1) (23, 24). Each of these three cell subsets could serve as a target for malignant transformation.
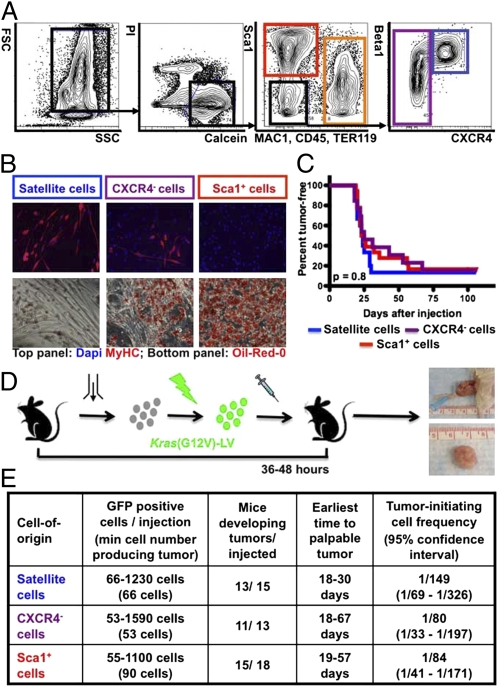
Fig. 1.
Tumor initiation by Kras(G12V)-expressing p16p19null MFA cells. (A) Mouse MFA cell subsets are discriminated by combinatorial staining for CD45, MAC1, Sca1, β1-integrin and CXCR4. (B) Differentiation of satellite cells (blue gate in A) and a subset of CXCR4− cells (purple gate in A) into myosin heavy chain-positive (MYHC+) myoblasts and myofibers (B, Upper). Differentiation of Sca1+ cells (red gate in A) and a subset of CXCR4− cells (purple gate in A) into Oil-Red-O+ adipocytes (B, Lower). (C and D) p16p19null MFA cells were infected with Kras(G12V) lentivirus and injected into the cardiotoxin-preinjured gastrocnemius muscles of NOD/SCID mice 36–48 h after cell isolation. Kaplan-Meier analysis (C) showed no differences in the percent of mice developing tumors (D) induced by individual cell types (P = 0.8). (E) Limiting dilution analysis showed equivalently high frequencies of tumor-initiating cells within each of the individual subsets of Kras(G12V)-transduced p16p19null MFA cells (P = 0.4). FSC, Forward Scatter; SSC, Side Scatter; Pi, Propidium Iodide; MAC1, Macrophage-1 antigen; TER119, antigen recognized by anti-Ly76 antibody; Sca1, stem cell antigen-1; CXCR4, C-X-C chemokine receptor type 4; Beta1, β1 integrin.
Tumors in Skeletal Muscle Can Originate from Different Lineages of Myogenic and Nonmyogenic Precursors.
To test the oncogenic effect of sarcoma-associated genetic lesions (6, 11, 12, 17, 18), we ectopically expressed oncogenic Kras in freshly isolated p16p19null satellite cells, CXCR4− cells, and Sca-1+ cells. Importantly, loss of p16p19 did not change muscle mass or alter the distribution or differentiation capacities of p16p19null MFA cells (Fig. S1 A and B and Table S1). Cells were transduced with oncogenic Kras(G12V) in a GFP-tagged pGIPZ vector or with GFP-tagged empty vector control virus. GFP expression, evaluated 5–7 d after infection, demonstrated effective transduction of each cell population (Fig. S1 C and D).
Intramuscular injection of Kras(G12V)-infected p16p19null satellite cells, Sca1+ cells, and CXCR4− cells into NOD/SCID recipients reproducibly generated palpable tumors 18–67 d after injection (Fig. 1 C and D). In contrast, p16p19null satellite cells, Sca1+ cells, and CXCR4− cells infected with control virus did not induce tumors in any of 18 transplanted recipients (followed for 78–127 d after transplantation) (Fig. S1E), arguing against the transformation of target cells caused by nonspecific integration of lentivirus. To assess the number of cells within each transduced population that can induce tumors, we performed limiting dilution transplantation analyses (28) with Kras(G12V)-transduced p16p19null satellite cells, Sca1+ cells, and CXCR4− cells (Fig. 1E). This analysis revealed equivalently high frequencies of tumor-initiating cells within each of the individual subsets of Kras(G12V)-transduced p16p19null MFA cells (P = 0.4; Fig. 1E). Likewise, Kaplan–Meier analysis showed no differences in the percent of mice developing tumors from Kras(G12V)-expressing p16p19null cell subsets (P = 0.8; Fig. 1C). Thus, despite clear differences in their differentiation status, the tumor-forming potential of these three MFA cell subsets appears indistinguishable.
Distinct Myogenic Marker Expression in Kras; p16p19null Sarcomas Arising from Different MFA Cell Subsets.
Kras; p16p19null tumors arising from each of the three cells-of-origin studied here presented as flesh-colored masses within the injected muscle (Fig. 1D) and were intensely positive for GFP [on average 62–67 ±7–16% (mean ± SD) of cells] (Fig. 2 and Fig. S2A). GFP+ cells sorted from tumors showed similar levels of induced expression of Kras by quantitative RT-PCR (qRT-PCR) (Fig. S2B), confirming tumor development from the injected, Kras-expressing, GFP-marked cells. All tumors were highly cellular and exhibited strong Ki67 positivity (Fig. 2). Histopathologically, tumors resembled pleomorphic, high-grade STS and consisted of intertwining bundles of tumor cells with large atypical nuclei and frequent mitotic figures (Fig. 2). Many tumors contained multinucleated strap cells, occasional large multinucleated giant cells, small areas of necrosis, and normal-appearing myofibers. In a blinded review of H&E-stained sections, morphologically distinct categories of Kras; p16p19null mouse tumors could not be distinguished reliably.
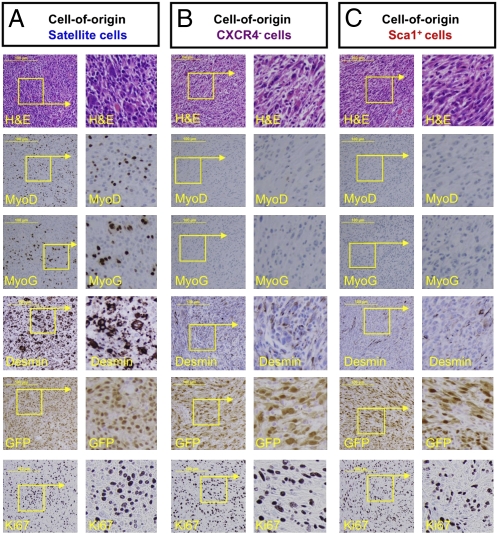
Fig. 2.
Morphological and IHC evaluation of tumors arising from Kras(G12V)-expressing p16p19null cells. H&E (Top row) and IHC staining of Kras(G12V);p16p19null tumors for MyoD (Second row), myogenin (MyoG) (Third row), desmin (Fourth row), GFP (Fifth row), and Ki67 (Bottom row). Representative images are shown for a MyoD-, myogenin-, and desmin-positive sarcoma arising from satellite cells (A), a MyoD- and myogenin-negative sarcoma arising from CXCR4− cells (B), and a MyoD- and myogenin-negative sarcoma arising from Sca1+ cells (C). Images were obtained at 20× magnification. (Scale bars, 100 μm.) Boxed areas are shown at threefold greater magnification at right.
To phenotype the Kras; p16p19null sarcomas further, we evaluated tumor cell expression of myogenic regulatory factors by IHC staining of 11 sarcomas of each category (Fig. 2). All but one of 11 sarcomas of satellite-cell origin exhibited clear nuclear staining for MyoD and myogenin in a patchy distribution throughout the tumors (Fig. 2A), similar to that described in a large Children's Oncology Group series of human nonalveolar RMS (5). The same 10 out of 11 sarcomas of satellite-cell origin also showed intense cytoplasmic staining for the muscle intermediate filament protein desmin (Fig. 2A). This pattern of myogenic marker staining in tumors arising from Kras(G12V)-expressing p16p19null satellite cells is consistent with RMS tumors of nonalveolar, pleomorphic histotype. In contrast, eight of 11 tumors of Sca1+-cell origin lacked staining for MyoD, myogenin, and desmin, with the exception of rare, isolated immunopositive nuclei, most of which appeared to localize within normal myofibers surrounding or within the tumors (Fig. 2C). Because of the predominant lack of myogenic features in the majority of sarcomas of Sca1+-cell origin, these tumors were classified as nonmyogenic sarcomas. Finally, tumors of CXCR4− origin exhibited variable levels of myogenic commitment (7 of 11 tumors were MyoD, myogenin, and desmin negative) (Fig. 2B), consistent with the heterogeneous lineage potential of this mixed cell population (Fig. 1B). Differences in expression of MyoD and myogenin in sorted GFP+ tumor cells also were evaluated by qRT-PCR (Fig. S2 C and D), which confirmed the presence of MyoD and myogenin in sarcomas of satellite-cell origin, the absence of MyoD and myogenin in sarcomas of Sca1+ origin (six tumors evaluated), and variable MyoD and myogenin levels in sarcomas of CXCR4− origin. Thus, the tumors arising from Kras(G12V)-expressing p16p19null MFA cells are high-grade STS tumors with marked differences in myogenic differentiation that correlate with their origination in distinct cell types within skeletal muscle.
Sarcomas Originating from Discrete Lineages of MFA Cells Share a Common Gene-Expression Profile with Human STS.
The molecular underpinnings of Kras; p16p19null sarcomas were evaluated by whole-genome microarray. We purified total RNA from the sorted GFP+ fraction of four RMS tumors of satellite-cell origin, four sarcomas of Sca1+-cell origin, and four sarcomas of CXCR4−-cell origin, generated in three biologically independent experiments. At three different filtering levels tested, we observed no clustering of tumors based on which cell type was transduced initially, and hierarchical clustering failed to identify any genes (including any myogenic regulatory factors) that could distinguish tumor subtypes based on cell-of-origin. These data indicate that the core transcriptional profile of Kras; p16p19null mouse sarcomas is remarkably similar. For this reason, our subsequent bioinformatic analyses (described below) considered all 12 sarcoma samples together in comparison with normal mouse skeletal muscle (the anatomic site from which the tumor cells-of-origin were isolated and where tumors emerged (Dataset S1) (29–32). This comparison revealed a common set of 1,116 differentially expressed probes in all three Kras; p16p19null sarcoma types compared with normal muscle. The gene IDs for those probes up-regulated >25-fold in Kras; p16p19null mouse sarcomas constitute the heatmaps shown in Fig. S3A and Fig. S4 A and F (sequences of probe identifiers are listed in Datasets S2, S3, and S4).
To determine whether Kras; p16p19null mouse sarcomas and human sarcomas exhibit concordantly up-regulated genes, we obtained published datasets (Dataset S1) from 78 embryonal RMS tumors, 70 alveolar RMS tumors, and 12 undifferentiated/indeterminate pediatric STS (33) and compared these datasets with human skeletal muscle (34). The genes up-regulated in Kras; p16p19null sarcomas (identified above) were up-regulated in human sarcomas but not in normal human muscle (Fig. S3A), and gene set enrichment analysis (GSEA) showed that these genes were enriched at up to four different fold-change cut-offs for human embryonal RMS [P = 0.002, false-discovery rate (FDR) = 0.004], human fusion-positive alveolar RMS (P < 0.001, FDR = 0), and human pediatric non-RMS STS (P = 0.02, FDR = 0.029) (Fig. 3 A–C and Table S2). Overlap of the top quartiles of probe sets up-regulated in Kras; p16p19null sarcomas and enriched in human sarcomas (Fig. 3 A–C, top quartiles marked in gray) identified 194 of 500 probe sets (corresponding to 144 genes) that were up-regulated in both the mouse Kras; p16p19null sarcomas and human sarcomas evaluated here (Fig. S3B; probe identifiers and gene IDs are listed in Table S3). Also, enrichment of Kras; p16p19null sarcoma–up-regulated genes in a broader panel of mouse sarcomas (including alveolar RMS) and human malignant fibrous histiocytomas confirmed similarities in their transcriptional profiles (Fig. S4 and Table S2). Thus, Kras; p16p19null mouse sarcomas recapitulate genetic events that are conserved in sarcomas across species. The cluster of genes conserved in these tumors may include targets of fundamental importance in sarcomas.
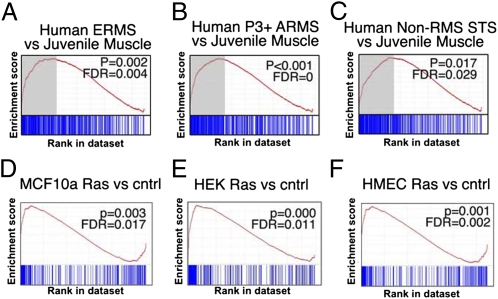
Fig. 3.
Transcriptional profiling of Kras; p16p19null mouse sarcomas. (A–C) GSEA analysis (details are given in Table S2) to test for enrichment of the set of Kras; p16p19null sarcoma-associated genes by comparing (A) embryonal RMS (ERMS), (B) Paired box 3-Forkhead Transcription Factor–positive (P3+) alveolar RMS (ARMS), and (C) non-RMS STS with normal muscle. The upper quartile of genes (A–C, marked in gray) identified by GSEA analyses shows 144 overlapping genes (194 overlapping probes), which are coordinately regulated in human RMS and non-RMS STS. (Fig. S3B and Table S3). (D–F) GSEA analysis demonstrates enrichment of this cluster of overlapping genes in (D) MCF10a cells transduced with Ras, (E) HEK cells transduced with Ras, and (F) HMECs transduced with Ras compared with nontransduced control cells but not in HMECs transduced with other oncogenes (Table S4).
Ras Pathway Activation Is Critical for the Growth of STS Tumors.
Consistent with tumor induction by activated Ras, Kras; p16p19null mouse sarcomas were positive for phosphorylated ERK (p-ERK) (Fig. S5A), a downstream effector of Ras/Rapidly Accelerated Fibrosarcoma (Raf)/Mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK) signaling. To evaluate the conserved cluster of sarcoma-associated genes described above (Fig. S3B and Table S3) in the context of Ras pathway activation, we obtained published datasets (Dataset S1) from oncogene-expressing human mammary epithelial cells (HMECs) (35), H-Ras–transduced HMECs (MCF10a), and H-Ras-transduced HEK cells (36, 37). GSEA demonstrated enrichment of the conserved cluster of sarcoma–up-regulated genes in H-Ras–transduced HMECs (P < 0.0001, FDR = 0), MCF10a (P = 0.003, FDR = 0.018), and HEK cells (P < 0.001, FDR = 0.010) (Fig. 3 D–F) but not in HMECs transduced with Myc (P = 0.011, FDR = 0.022), β-catenin (BCAT) (P = 0.037, FDR = 0.073), or E2F transcription factor 3 (P = 0.058, FDR = 0.131) (Table S4). Moreover, Kras; p16p19null sarcoma–up-regulated genes were enriched in nonsarcomatous malignancies associated with Ras-pathway activation (Table S2). Of the 194 probes (representing 144 genes) up-regulated in Kras; p16p19null sarcomas and human soft-tissue sarcomas, 132 are associated with Ras pathway activation, whereas 62 appear to be Ras independent (Table S3).
From these analyses, Ras signaling emerges as a major component of the conserved transcriptional signature induced in human and mouse sarcomas [including some malignancies, e.g., mouse synovial sarcoma (32) and human alveolar RMS (6, 33) in which Ras activating mutations have not been noted previously]. Indeed, immunostaining for p-ERK confirmed Ras pathway activation in 44% of human leiomyosarcomas (12 of 27) and 50% of human RMS tumors (12 of 24) evaluated (Fig. S5 B and C). Moreover, inhibition of MEK1/2 signaling by the highly selective inhibitor U0126 (38) impaired in vitro growth of Kras; p16p19null sarcoma cell lines established from GFP+ cell explants of Kras; p16p19null RMS tumors or non-myogenic sarcomas (two independently established cell lines evaluated for each tumor type; Fig. 4 A and B). U0126 also blocked expansion of the human RMS cell line RD (Fig. 4C), consistent with previous studies in xenografted mice (20). Thus, activated Ras/Raf/MEK/ERK signaling contributes to sarcoma growth, consistent with its prominence in our bioinformatic analyses (Fig. 3 and Table S3) (6, 11–13, 20).
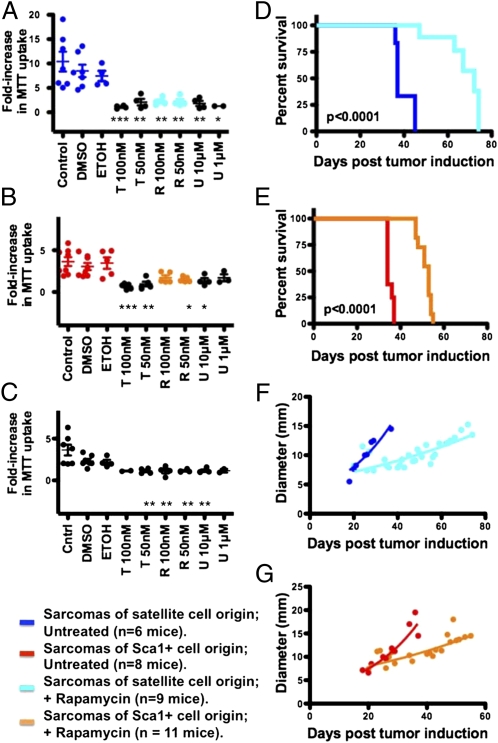
Fig. 4.
Ras/Raf/MEK/ERK and mTOR signaling regulate the growth of Kras; p16p19null sarcomas and the human RMS cell line RD. (A–C) Proliferation of two stable cell lines established from mouse Kras; p16p19null RMS tumors of satellite-cell origin (A), of two stable cell lines established from mouse Kras; p16p19null nonmyogenic sarcomas of Sca1+ origin (B), and of the human RMS cell line RD (C) was evaluated by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay in control cultures (dark blue in A and red in B) and after inhibiting mTOR by exposure to 50–100 nMol rapamycin (abbreviated R, light blue in A and orange in B) or to 50–100 nMol torin (abbreviated T) or by inhibiting Ras/Raf/MEK/ERK by exposure to 1–10 μMol U0216 (abbreviated U). (D–G) Mice with Kras; p16p19null RMS tumors of satellite-cell origin (D and F) or Kras; p16p19null nonmyogenic sarcomas of Sca1+ origin (E and G) were treated daily with rapamycin; treatment prolonged time until killing (P < 0.0001) and impeded growth of both myogenic and nonmyogenic sarcomas.
mTOR Signaling Regulates Sarcoma-Cell Growth.
The 144 genes up-regulated in both human and mouse sarcomas represent a unique list that likely includes targets of broad relevance in sarcomas. Of these 144 genes, 74 have been associated previously with human cancers, and 28 have been linked to sarcoma biology (Table S3). Gene ontology analyses predicted that 61 of the 144 sarcoma-associated genes contribute to cell growth and proliferation, two key cellular activities often regulated by mTOR. In fact, several mTOR targets [e.g., DAP1 (39), SLC2A1 (40), SKP2 (41), TIMP1 (42), and BCAT1 (43)] are included in this gene set, and all Kras; p16p19null sarcomas showed broad reactivity for phosphorylated S6 (p-S6), a downstream target of mTOR Complex 1 (mTORC1) signaling (Fig. S5D). Analogous p-S6 immunostaining of human tissue arrays likewise indicated activation of mTOR signaling in human STS tumors, including 33% of leiomyosarcomas (9 of 27) and 26% of RMS tumors (6 of 23) evaluated (Fig. S5 E and F). Blockade of mTOR signaling in Kras; p16p19null sarcoma cell lines and RD cells by exposure to rapamycin (an allosteric partial mTORC1 inhibitor) or torin 1 (an ATP-competitive inhibitor of mTOR) (44) substantially slowed the growth of two Kras; p16p19null RMS cell lines, two Sca1+ tumor cell-derived, nonmyogenic Kras; p16p19null sarcoma cell lines, and RD cells (Fig. 4 A–C), consistent with prior reports on the growth-inhibitory effects of temsirolimus (a rapamycin analog) on RD cells (21). Thus, the cluster of genes up-regulated in Kras; p16p19null sarcomas and shared by STS across species identifies critical cellular events that support sarcoma growth. We also treated mice with established Kras; p1619null sarcomas with rapamycin by daily i.p. injections (initiated at a dose of 10 mg/kg shortly after tumor detection). Rapamycin slowed tumor growth (Fig. 4 F and G) and prolonged survival of sarcoma-bearing mice (P < 0.0001; Fig. 4 D and E) but did not induce tumor regression. These data support the potential efficacy of mTOR targeting compounds in antisarcoma treatment strategies (19) and recapitulate limitations in efficacy seen in clinical trials designed to treat human sarcomas with mTOR inhibitors (45). Taken together, these findings validate that Kras; p16p19null sarcomas can serve as an effective preclinical test platform for functional evaluation of the effects of modulating sarcoma-relevant pathways.
Discussion
The sarcoma-induction strategy described here introduces a systematic approach to dissect the developmental origins of STS by ex vivo genetic manipulation of lineally distinct populations of cells freshly isolated from target tissue. Using a viral delivery system, we were able to compare sarcoma formation directly in three different target cell populations. This comparision could not be accomplished using direct Cre-mediated induction because appropriate lineage-specific Cre drivers are unavailable for two of the three cell populations studied here (i.e., Sca1+ and CXCR4− cells). Also, by prospectively evaluating the tumor-initiating effects of identical, sarcoma-associated oncogenetic lesions in freshly isolated cell populations, we avoided complications that can arise from uncontrolled preselection during in vitro passaging. Importantly, the transcriptional profile of Kras; p16p19null mouse sarcomas recapitulates genetic events that occur naturally in human sarcomas, reducing concerns about possible caveats in this system, such as oncogene overexpression or insertional mutagenesis caused by viral integration. Finally, this approach is attractive because it can be translated directly to produce human sarcoma models when methods (not yet available) for the prospective isolation of analogous human muscle cell subsets finally are developed.
Using this tumor-induction system, we found that satellite cells, as well as more differentiated myogenic cells (sorted as CXCR4− cells), give rise to myogenic sarcomas similar to human nonalveolar RMS with pleomorphic features. In contrast, Sca1+ cells, a population of bipotent fibrogenic/adipogenic precursors that lack myogenic activity but reside in skeletal muscle, induce sarcomas that lack myogenic differentiation features in most tumors. These findings support the origination of RMS in cells of the myogenic lineage (3, 7, 8, 46) and demonstrate that the preexisting lineage commitment of the target cell into which oncogenetic hits are introduced is critical in determining the phenotypic properties of sarcomas in muscle (3, 7, 8, 46). Similar studies performed in the hematopoietic system (47) suggest that the influence of cellular context on the outcome of oncogenesis is likely to be a conserved feature of both liquid and solid malignancies of mesodermal lineage, perhaps reflecting inherent differences in established epigenetic marks within distinct cancer cells-of-origin. In this regard, recent experience with induced pluripotent stem cells (iPSCs) may be relevant, because iPSCs produced from different cellular origins vary in their differentiation potential and propensity to form tumors in mice (48), likely as a result of persistent histone modifications (49). It is conceivable that such epigenetic “memory” in cancer cells-of-origin also translates into persistent lineage-specific differences in gene expression in malignancies produced from different cell types.
However, despite the differences in myogenic differentiation observed among sarcomas generated from distinct cells-of-origin, expression profiling of Kras; p16p19null mouse sarcoma subtypes revealed a highly overlapping set of transcripts that were up-regulated in comparison with normal mouse muscle. This observation parallels genomic analyses of human STS histotypes, suggesting that distinct human sarcoma entities also share profound similarities on a global genomic level (50). Moreover, our studies identified a Ras-predominated gene-expression signature shared by Kras; p16p19null mouse sarcomas and human embryonal and alveolar RMS [previous studies linked Ras pathway activation only to embryonal RMS (6)] and demonstrated activation of the Ras/Raf/MEK/ERK pathway in Kras; p16p19null mouse sarcomas and in 50% of human RMS. Consistent with a central role of Ras signaling in these tumors, inhibition of Ras/Raf/MEK/ERK signaling reduced the proliferation of mouse and human sarcoma cells (20). Thus, although the present study does not specifically interrogate the relative importance of Ras pathway activation compared with other possible sarcoma-relevant oncogenic events (2), our findings highlight the important contribution of aberrant Ras signaling to the growth and malignancy of STS (6, 11, 12, 20).
Several of the 144 sarcoma-associated genes identified in this study have been associated with human malignancies, including sarcomas (Table S3). For example, the membrane-cytoskeleton linker Ezrin has been implicated in the metastatic behavior of RMS (51). Several other candidate genes have been linked to mTOR signaling (51), and positive p-S6 immunostaining confirmed enhanced mTOR signaling in Kras; p16p19null sarcomas and in 26–33% of human RMS and leiomyosarcomas. Similarly, phosphorylation of AKT (another mTOR pathway component) was reported in 43–55% of human embryonal and alveolar RMS (52). Like Ras, pharmacological inhibition of mTOR signaling in mouse and human sarcoma cells impaired tumor growth, consistent with prior studies using RMS cell lines and xenografts (19, 21) and with beneficial effects seen in some patients with advanced sarcomas (45). Together, these findings validate the utility of the Kras; p16p19null mouse sarcomas as a platform to study the functional role of candidate sarcoma genes and pathways. Future studies of the additional genes identified here likely will provide further insights into the events that promote and sustain STS tumors and identify new targets for therapeutic intervention.
Materials and Methods
See SI Materials and Methods for additional details regarding experimental methods.
Mice.
C57BL6/J wild-type and NOD/CB17-Prkdcscid/J (NOD/SCID) mice were purchased from the Jackson Laboratory. p16p19null mice (B6.129 background) were obtained from the National Institutes of Health/Mouse Models of Human Cancer Consortium. Mice were bred and maintained at the Joslin Diabetes Center Animal Facility. All animal experiments were approved by the Joslin Diabetes Center Institutional Animal Care and Use Committee.
FACS Sorting of MFA Cells and Differentiation Assays.
Isolation of MFA cell subsets from 3- to 8-wk-old C57BL6/J wild-type and p16p19null mice was performed as described (23, 24). All cell populations were double-sorted to maximize purity, typically resulting in recovery of cell populations in which >98% of cells exhibit the desired marker profile (23, 24). In vitro myogenic and adipogenic differentiation assays were performed as previously described (23, 53).
Sarcoma-Induction Assays.
The Kras(G12V)-IRES-GFP pGIPZ plasmid was a gift from Junhao Mao (University of Massachusetts, Worcester, MA). Control pGIPZ plasmid was from Open Biosystems. Freshly isolated MFA cell subsets were transduced within 1–2 h after sorting and were harvested from culture within 36–48 h of sorting. Cells were washed and injected at defined numbers into the gastrocnemius muscles of 1- to 3-mo-old, anesthesized NOD/SCID mice. Recipient muscles were preinjured 24 h before cell implantation by injection of 25 μL of a 0.03 mg/mL solution of cardiotoxin (from Naja naja mossambica; Sigma) to enhance cell engraftment. Mice were monitored for up to 4 mo following cell transplantation for the emergence of tumors. For mice that did not develop palpable tumors, extremity muscles were dissected 4 mo posttransplantion to confirm tumor absence.
Histopathological Evaluation of Tumors.
Tumors harvested from mice were fixed in 4% (vol/vol) paraformaldehyde for 2 h and were embedded in paraffin. Human tissue arrays were purchased from US Biomax (S02081). Sections were stained with H&E and for myogenin (1:100; M3559; Dako ), MyoD1 (1:50; M3512; Dako ), desmin (1:50; M0760; Dako), GFP (1:1,500; 632381; Clontech), Ki67 (1:250; VP-K451; Vector Labs), p-ERK (1:200; 4370; Cell Signaling Technology) and p-S6 (1:200; 4060; Cell Signaling Technology).
Microarrays.
Tumors were FACS-sorted to isolate GFP+ cells. Total RNA was isolated by TRIzol extraction, labeled, and hybridized to Affymetrix microarrays (Mu430v2) as previously described (6). Dataset S1 lists previously published datasets used in our analyses. Microarray data obtained from Kras; p16p19null sarcomas were deposited in the National Center for Biotechnology Information database (accession no. GSE22841).
Proliferation Assays.
Stable mouse sarcoma cell lines were established from two Kras; p16p19null RMS tumors of satellite-cell origin (MyoD/myogenin/desmin positive) and two Kras; p16p19null sarcomas of Sca1+-cell origin (MyoD/myogenin/desmin negative). Human RD cells were purchased from ATCC. All cell lines were maintained in DMEM with 10% FBS and 1% penicillin/streptomycin. Cells were exposed to 50–100 nMol rapamycin (R-5000; LC Laboratories), 50–100 nMol torin 1 (kindly provided by Nathaniel Gray, Dana-Farber Cancer Institute, Boston, MA), 1–10 μMol U0126 (9903; Signaling Technology), or vehicle (ethanol, DMSO). Cell growth was determined by MTT assay (Cayman Chemical) 4 d after first exposure to compounds and was quantified as fold-increase in MTT uptake compared with baseline. All assays were performed in triplicate and replicated in two to four independent experiments.
In Vivo Sarcoma-Growth Assays.
Kras; p16p19null sarcomas were induced as described above. Shortly after tumor detection, mice were exposed to rapamycin (10 mg/kg in sterile PBS) by daily i.p. injections. Tumor size was determined at least once per week by measuring the maximum diameter of tumor-bearing extremities using a caliper. Mice were killed once tumor size reached 1 cm3, if the vascular supply of the extremity was impaired, or if animals were ill.
Statistics.
Limiting dilution analyses were performed based on Bonnefoix et al. (28) using the limdil function of the StatMod package (author G. K. Smyth, http://bioinf.wehi.edu.au/software/limdil/).
Supplementary Material
Acknowledgments
We thank G. Buruzula, J. LaVecchio, and S. Harris at The Joslin Diabetes and Endocrinology Research Center (DERC) Flow Cytometry Core for flow cytometry support; A. Pinkhasov at The Joslin DERC Histology Core for help with tumor sectioning; J. Schroeder at The Joslin DERC Genomics Core for microarray processing (funded by National Institutes of Health Grant P30DK036836); A. Perez-Atayde for reviewing histopathological findings, C. L. Unitt, T. Caron, and A. Kreshock at The Dana-Farber/Harvard Cancer Center Histopathology Core for help with immunohistochemistry; N. Gray for providing torin 1, and L. Wang, D. Kirsch, and C. Keller for helpful comments on the manuscript. This work was funded in part by grants from the Burroughs–Wellcome Fund, Harvard Stem Cell Institute, and The Beckman Foundation (to A.J.W.), and from Hope Street Kids, Patients Assistance League and Service Bermuda/St. Baldrick's, Alex Lemonade Stand Foundation, and Bear Necessities (to S.H.). D.M.L. is supported by the Sarcoma Foundation of America, ALSF, and National Institutes of Health Grants K01AR055619, R01CA154923, and R21CA156056.
Footnotes
The authors declare no conflict of interest.
Data deposition: Microarray datasets have been deposited in the National Center for Biotechnology Information database (accession no. GSE22841).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111733108/-/DCSupplemental.
References
- 1.Clark MA, Fisher C, Judson I, Thomas JM. Soft-tissue sarcomas in adults. N Engl J Med. 2005;353(21):701–711. doi: 10.1056/NEJMra041866. [DOI] [PubMed] [Google Scholar]
- 2.Parham DM, Ellison DA. Rhabdomyosarcomas in adults and children: An update. Arch Pathol Lab Med. 2006;130:1454–1465. doi: 10.5858/2006-130-1454-RIAACA. [DOI] [PubMed] [Google Scholar]
- 3.Rubin BP, et al. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell. 2011;19(2):177–191. doi: 10.1016/j.ccr.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fletcher CD, Gustafson P, Rydholm A, Willén H, Akerman M. Clinicopathologic re-evaluation of 100 malignant fibrous histiocytomas: Prognostic relevance of subclassification. J Clin Oncol. 2001;19:3045–3050. doi: 10.1200/JCO.2001.19.12.3045. [DOI] [PubMed] [Google Scholar]
- 5.Morotti RA, et al. Children's Oncology Group An immunohistochemical algorithm to facilitate diagnosis and subtyping of rhabdomyosarcoma: The Children's Oncology Group experience. Am J Surg Pathol. 2006;30:962–968. doi: 10.1097/00000478-200608000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Langenau DM, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21:1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiffin N, Williams RD, Shipley J, Pritchard-Jones K. PAX7 expression in embryonal rhabdomyosarcoma suggests an origin in muscle satellite cells. Br J Cancer. 2003;89:327–332. doi: 10.1038/sj.bjc.6601040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller C, et al. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: Cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 2004;18:2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren YX, et al. Mouse mesenchymal stem cells expressing PAX-FKHR form alveolar rhabdomyosarcomas by cooperating with secondary mutations. Cancer Res. 2008;68:6587–6597. doi: 10.1158/0008-5472.CAN-08-0859. [DOI] [PubMed] [Google Scholar]
- 10.Hettmer S, Wagers AJ. Muscling in: Uncovering the origins of rhabdomyosarcoma. Nat Med. 2010;16(2):171–173. doi: 10.1038/nm0210-171. [DOI] [PubMed] [Google Scholar]
- 11.Martinelli S, et al. RAS signaling dysregulation in human embryonal Rhabdomyosarcoma. Genes Chromosomes Cancer. 2009;48:975–982. doi: 10.1002/gcc.20702. [DOI] [PubMed] [Google Scholar]
- 12.Stratton MR, Fisher C, Gusterson BA, Cooper CS. Detection of point mutations in N-ras and K-ras genes of human embryonal rhabdomyosarcomas using oligonucleotide probes and the polymerase chain reaction. Cancer Res. 1989;49:6324–6327. [PubMed] [Google Scholar]
- 13.Yoo J, Robinson RA, Lee JY. H-ras and K-ras gene mutations in primary human soft tissue sarcoma: Concomitant mutations of the ras genes. Mod Pathol. 1999;12:775–780. [PubMed] [Google Scholar]
- 14.Tsumura H, Yoshida T, Saito H, Imanaka-Yoshida K, Suzuki N. Cooperation of oncogenic K-ras and p53 deficiency in pleomorphic rhabdomyosarcoma development in adult mice. Oncogene. 2006;25:7673–7679. doi: 10.1038/sj.onc.1209749. [DOI] [PubMed] [Google Scholar]
- 15.Serrano M, et al. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85(1):27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 16.Kohashi K, et al. Alterations of RB1 gene in embryonal and alveolar rhabdomyosarcoma: Special reference to utility of pRB immunoreactivity in differential diagnosis of rhabdomyosarcoma subtype. J Cancer Res Clin Oncol. 2008;134:1097–1103. doi: 10.1007/s00432-008-0385-3. [DOI] [PubMed] [Google Scholar]
- 17.Obana K, et al. Aberrations of p16INK4A, p14ARF and p15INK4B genes in pediatric solid tumors. Int J Oncol. 2003;23:1151–1157. [PubMed] [Google Scholar]
- 18.Chen Y, et al. Mutation and expression analyses of the MET and CDKN2A genes in rhabdomyosarcoma with emphasis on MET overexpression. Genes Chromosomes Cancer. 2007;46:348–358. doi: 10.1002/gcc.20416. [DOI] [PubMed] [Google Scholar]
- 19.Houghton PJ, et al. Initial testing (stage 1) of the mTOR inhibitor rapamycin by the pediatric preclinical testing program. Pediatr Blood Cancer. 2008;50:799–805. doi: 10.1002/pbc.21296. [DOI] [PubMed] [Google Scholar]
- 20.Marampon F, et al. MEK/ERK inhibitor U0126 affects in vitro and in vivo growth of embryonal rhabdomyosarcoma. Mol Cancer Ther. 2009;8:543–551. doi: 10.1158/1535-7163.MCT-08-0570. [DOI] [PubMed] [Google Scholar]
- 21.Petricoin EF, 3rd, et al. Phosphoprotein pathway mapping: Akt/mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Res. 2007;67:3431–3440. doi: 10.1158/0008-5472.CAN-06-1344. [DOI] [PubMed] [Google Scholar]
- 22.Wagers AJ, Conboy IM. Cellular and molecular signatures of muscle regeneration: Current concepts and controversies in adult myogenesis. Cell. 2005;122:659–667. doi: 10.1016/j.cell.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 23.Cerletti M, et al. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134(1):37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherwood RI, et al. Isolation of adult mouse myogenic progenitors: Functional heterogeneity of cells within and engrafting skeletal muscle. Cell. 2004;119(4):543–554. doi: 10.1016/j.cell.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 25.Joe AW, et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12(2):153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulz TJ, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci USA. 2011;108(1):143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol. 2010;12(2):143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 28.Bonnefoix T, Bonnefoix P, Verdiel P, Sotto JJ. Fitting limiting dilution experiments with generalized linear models results in a test of the single-hit Poisson assumption. J Immunol Methods. 1996;194(2):113–119. doi: 10.1016/0022-1759(96)00077-4. [DOI] [PubMed] [Google Scholar]
- 29.Thorrez L, et al. Using ribosomal protein genes as reference: A tale of caution. PLoS One. 2008;3(3):e1854. doi: 10.1371/journal.pone.0001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishijo K, et al. Credentialing a preclinical mouse model of alveolar rhabdomyosarcoma. Cancer Res. 2009;69:2902–2911. doi: 10.1158/0008-5472.CAN-08-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mito JK, et al. Cross species genomic analysis identifies a mouse model as undifferentiated pleomorphic sarcoma/malignant fibrous histiocytoma. PLoS ONE. 2009;4:e8075. doi: 10.1371/journal.pone.0008075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: Insights into a myogenic origin. Cancer Cell. 2007;11:375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Davicioni E, et al. Molecular classification of rhabdomyosarcoma—genotypic and phenotypic determinants of diagnosis: A report from the Children's Oncology Group. Am J Pathol. 2009;174:550–564. doi: 10.2353/ajpath.2009.080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang PB, et al. Variations in gene expression among different types of human skeletal muscle. Muscle Nerve. 2005;32:483–491. doi: 10.1002/mus.20356. [DOI] [PubMed] [Google Scholar]
- 35.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 36.Chang JT, et al. A genomic strategy to elucidate modules of oncogenic pathway signaling networks. Mol Cell. 2009;34(1):104–114. doi: 10.1016/j.molcel.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoeflich KP, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009;15:4649–4664. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 38.Favata MF, et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 39.Koren I, Reem E, Kimchi A. DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr Biol. 2010;20:1093–1098. doi: 10.1016/j.cub.2010.04.041. [DOI] [PubMed] [Google Scholar]
- 40.Buller CL, et al. A GSK-3/TSC2/mTOR pathway regulates glucose uptake and GLUT1 glucose transporter expression. Am J Physiol Cell Physiol. 2008;295:C836–C843. doi: 10.1152/ajpcell.00554.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shapira M, Kakiashvili E, Rosenberg T, Hershko DD. The mTOR inhibitor rapamycin down-regulates the expression of the ubiquitin ligase subunit Skp2 in breast cancer cells. Breast Cancer Res. 2006;8:R46. doi: 10.1186/bcr1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karam AK, et al. Cisplatin and PI3kinase inhibition decrease invasion and migration of human ovarian carcinoma cells and regulate matrix-metalloproteinase expression. Cytoskeleton (Hoboken) 2010;67:535–544. doi: 10.1002/cm.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.She P, et al. Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell Metab. 2007;6:181–194. doi: 10.1016/j.cmet.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agulnik M. New developments in mammalian target of rapamycin inhibitors for the treatment of sarcoma. Cancer. 2011 doi: 10.1002/cncr.26361. 10.1002/cncr.26361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linardic CM, Downie DL, Qualman S, Bentley RC, Counter CM. Genetic modeling of human rhabdomyosarcoma. Cancer Res. 2005;65:4490–4495. doi: 10.1158/0008-5472.CAN-04-3194. [DOI] [PubMed] [Google Scholar]
- 47.Stubbs MC, Armstrong SA. Therapeutic implications of leukemia stem cell development. Clin Cancer Res. 2007;13:3439–3442. doi: 10.1158/1078-0432.CCR-06-3090. [DOI] [PubMed] [Google Scholar]
- 48.Miura K, et al. Variation in the safety of induced pluripotent stem cell lines. Nat Biotechnol. 2009;27:743–745. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]
- 49.Polo JM, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carneiro A, et al. Indistinguishable genomic profiles and shared prognostic markers in undifferentiated pleomorphic sarcoma and leiomyosarcoma: Different sides of a single coin? Lab Invest. 2009;89:668–675. doi: 10.1038/labinvest.2009.18. [DOI] [PubMed] [Google Scholar]
- 51.Yu Y, et al. Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulators. Nat Med. 2004;10(2):175–181. doi: 10.1038/nm966. [DOI] [PubMed] [Google Scholar]
- 52.Cen L, et al. PDK-1/AKT pathway as a novel therapeutic target in rhabdomyosarcoma cells using OSU-03012 compound. Br J Cancer. 2007;97:785–791. doi: 10.1038/sj.bjc.6603952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6(2):105–114. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.