Abstract
The rapid emergence of Escherichia coli O157:H7 from an unknown strain in 1982 to the dominant hemorrhagic E. coli serotype in the United States and the cause of widespread outbreaks of human food-borne illness highlights a need to evaluate critically the extent to which genomic plasticity of this important enteric pathogen contributes to its pathogenic potential and its evolution as well as its adaptation in different ecological niches. Aimed at a better understanding of the evolution of the E. coli O157:H7 pathogenome, the present study presents the high-quality sequencing and comparative phylogenomic analysis of a comprehensive panel of 25 E. coli O157:H7 strains associated with three nearly simultaneous food-borne outbreaks of human disease in the United States. Here we present a population genetic analysis of more than 200 related strains recovered from patients, contaminated produce, and zoonotic sources. High-resolution phylogenomic approaches allow the dynamics of pathogenome evolution to be followed at a high level of phylogenetic accuracy and resolution. SNP discovery and study of genome architecture and prophage content identified numerous biomarkers to assess the extent of genetic diversity within a set of clinical and environmental strains. A total of 1,225 SNPs were identified in the present study and are now available for typing of the E. coli O157:H7 lineage. These data should prove useful for the development of a refined phylogenomic framework for forensic, diagnostic, and epidemiological studies to define better risk in response to novel and emerging E. coli O157:H7 resistance and virulence phenotypes.
Keywords: foodborne pathogen, comparative phylogenomics, SNP typing, infectious disease
The human pathogen Shiga-toxin–producing, nonsorbitol fermenting, and β-glucuronidase–negative Escherichia coli O157:H7 is thought to have evolved from an O55:H7-like progenitor (1, 2). O157:H7 is the most common enterohemorrhagic E. coli serotype found in North America. Although causing disease in humans, E. coli O157:H7 does not appear to affect cattle, a major reservoir for this organism (3, 4). The E. coli O157:H7 lineage is distinguished from other E. coli serotypes by its highly homogenous population structure, comparable to clonal microbial species such as Yersinia pestis (5) or Bacillus anthracis (6). During the last 3 decades, E. coli O157:H7 has established itself as a major enteric pathogen, capable of causing large outbreaks of gastrointestinal disease. E. coli O157:H7 infections usually have a food-borne etiology (7). The virulence in E. coli O157:H7 has been attributed to the lateral acquisition of genetic determinants, such as the virulence plasmid pO157, the prevalence of a set of phylogenetically unrelated prophages (8) [in particular Shiga-like toxin (stx)–converting phages, which can carry different stx subtypes (9)], O-islands (10, 11), the locus of enterocyte effacement (LEE), the arginine translocation system (12), and adhesion factors (13, 14). However, little is known about the genomic diversity that exists among extant populations of E. coli O157:H7 or how various genotypes of this pathogen relate to development and severity of human disease, thus underscoring the need to understand the genome dynamics and plasticity of this important pathogen. In the context of this study, “population” is defined as a group of isolates temporally related and linked through strain-associated metadata, such as time (period), source, outbreak, or genotypic assignment. Infected patients present with a range of gastrointestinal morbidities such as severe abdominal cramping with little or no associated fever and a watery diarrhea, which can develop into severe bloody diarrhea (15). Although many infected with the pathogen remain asymptomatic, an estimated 15–20% of people infected with E. coli O157:H7 present with indications severe enough to require hospitalization. In such cases, symptoms may progress to hemolytic uremic syndrome (HUS), hemorrhagic colitis, and CNS failure with potentially lethal outcomes (16–18). Three widely publicized food-associated outbreaks of E. coli O157:H7 infections in 2006 captured the attention of the US Congress as well as the public health, forensic, and lay communities (19). E. coli O157:H7 continues to be a significant public health threat. Recent outbreaks have been associated with the emergence of apparently more virulent E. coli O157:H7 clones, with more than 50% of infected persons hospitalized and many patients presenting with more severe clinical and potentially life-threatening complications (19). These outbreaks were traced to ingestion of contaminated fresh produce and are referred to as the “spinach” (SP), “Taco Bell” (TB), and “Taco John” (TJ) outbreaks. The spinach incident was a multistate outbreak (26 states) that caused 199 illnesses and at least three deaths. Among the ill, 51% (102 patients) were hospitalized, and in 16% of the cases (31 patients), infection progressed to HUS and kidney failure (19). Much research has been done to understand the pathogenesis of E. coli O157:H7, but data addressing selection and adaptation of variants during the time course of a single outbreak of human disease are lacking. In this study the genetic analysis of 231 E. coli O157:H7 strains, the genome sequences of 26 isolates (18 of which were obtained for this study) allowed unprecedented insights into the genomic plasticity and genome dynamics of the genetically homogenous E. coli O157:H7 lineage. This extensive dataset enabled a detailed analysis of the types of host selection and adaptation occurring during the time course of a single outbreak on a genome- and population-wide scale. This study identified numerous biomarkers that aided in the development of a more refined and accurate phylogenomic framework and contributed to the investigation of the genetic relatedness of individual strains and their assignment into distinct virulence clades.
Results and Discussion
Phylogenomic Analysis of the E. coli O157:H7 Lineage.
The strain history and associated metadata of all 231 strains analyzed in this study are summarized in SI Materials and Methods and Dataset S1. To resolve the genetically homogenous genome structure and study the genetic relatedness of E. coli O157:H7 strains, we deployed several strategies. Verotoxin profiling of 231 strains of E. coli O157:H7, including the largest number of E. coli O157:H7 obtained from a single human outbreak (Dataset S1), revealed that all 191 strains associated with the SP outbreak, three clinically derived strains that the Centers for Disease Control (CDC) considered by to be outliers and not associated with the SP outbreak, and all eight strains obtained from the TB outbreak share a characteristic stx1-, stx2+, stx2c+ Shiga toxin content. Notably among the collection examined, seven strains from unrelated human outbreaks (EC1574, EC1582, EC1585, EC1588, EC1592, EC1610, EC4002, and EC508) assigned to clade 8 according to Manning et al. (20) share this distinct verotoxin pattern, but other genotypic and phenotypic data (Datasets S1 and S2) suggest that these clade 8 strains are not directly phylogenetically related to the SP or TB outbreak strains.
SNP-Derived Phylogeny of E. coli O157:H7.
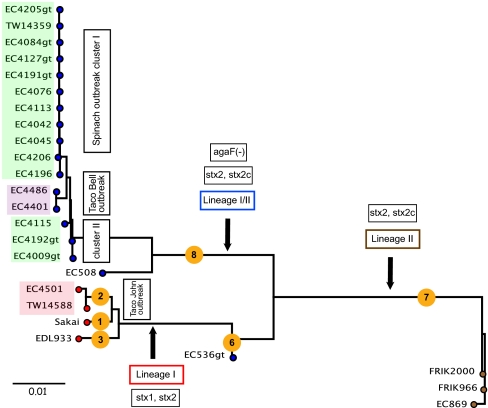
To establish a robust, high-resolution phylogenomic framework for E. coli O157:H7, 16 genomes were subjected to SNP discovery and SNP validation by comparing their genome sequences with the fully closed genome of a strain collected at the time of the 2006 SP outbreak, strain EC4115 (Dataset S1). This analysis yielded a panel of 1,225 high-quality SNPs (Dataset S2) distributed stochastically within the genome without any indication of mutational hotspots (Fig. 1A). Nine additional previously sequenced E. coli O157:H7 genomes were genotyped and placed in the phylogenetic tree by querying their sequences against the SNP panel. This bioinformatic pipeline takes into account the sequence read coverage and quality, genome coverage, and paralogous genomic regions. The SNP panel comprised 356 synonymous SNPs, 647 nonsynonymous SNPs, and 222 intergenic SNPs, far more than the SNP panel used previously for clade assignments of E. coli O157:H7 strains (20, 21). We note that the SNPs showed only two different alleles for every strain at each assayed position (Dataset S2), indicative of low homoplasy. The high resolution of the SNP-derived phylogenetic scheme allowed the placement of strains from the 2006 SP, TB, and TJ outbreaks onto distinct branches. Further, each E. coli O157:H7 strain was subjected to a lineage-specific polymorphism assay (LPSA) in six repetitive genomic loci (22), multilocus variable-number tandem repeat analysis (MLVA) (23), and in silico scoring of 96 SNPs previously used for clade typing (20) to assess genetic heterogeneity among these strains. We note that the tree architecture and branching is supported by the strain-specific metadata, such as the LSPA-derived lineage and SNP-derived clade assignment or the toxin content and MLVA types (Fig. 2 and Datasets S1 and S2) (24). TJ strains EC4501 and TW14588 form a cluster with the lineage I, clade 2 outbreak strains EDL933 and Sakai (Fig. 2), from which they are separated by 57 branch-specific SNPs carrying 13 and 2 strain-specific SNPs, respectively (Dataset S2). Lineage I strains are separated by 138 SNPs from the transitional lineage I/II strain EC536 (Dataset S2). The lineage II strains [i.e., strains EC869, FRIK2000, and FRIK966 (25)] are of bovine origin and cluster on a separate branch from lineage I and lineage I/II strains that harbor 298 unique SNPs (Dataset S2) (26). The phylogeny clearly reveals the homogenous genetic structure of the strains obtained during the SP outbreak and a close genetic relationship of the SP and TB outbreak strains. The lineage I/II TB strains EC4486 and EC4401 are separated by 24 TB-specific SNPs (Dataset S2) from the remainder of lineage I/II strains and carry three and a single strain-specific SNPs, respectively. Interestingly, although obtained from the same outbreak, the lineage I/II SP outbreak strains are split into two distinct genetically heterogeneous groups by SNP-based phylogenetic placement of the TB branch. Group A is separated by 16 lineage I/IIb-specific SNPs and comprises 8 of 11 typical SP outbreak strains featuring the dominant genotype, such as EC4045, and 3 of 11 bovine strains from California farms, all of which were obtained during the 2-mo period of the SP outbreak (Dataset S2). The SNP pattern among group A lineage I/IIb strains is identical except for a single strain-specific SNP in each California bovine strain (EC4205 and EC4206) at positions 532 and 182, respectively, revealing the close genetic relationship of these strains (Dataset S2). Besides the two bovine and two TB outbreak strains, the genotypes of three clinical outbreak strains (EC4196, EC4205, and EC4206) showed variations from a dominant SP outbreak group A strain, such as EC4205 (Dataset S2). The SP outbreak strains EC4009 and EC4192 form a distinct lineage I/group B cluster together with the Maine (ME) strain EC4115, which carries 15 strain-specific SNPs (Dataset S2). According to the CDC epidemiological investigation, strain EC4115 was considered an outlier and was not included as part of the SP outbreak. Our findings may support the hypothesis that an unrelated outbreak was caused by such EC4115-type strains or, alternatively, may indicate microevolution from the original outbreak isolates within the 2-mo timeframe of the SP outbreak.
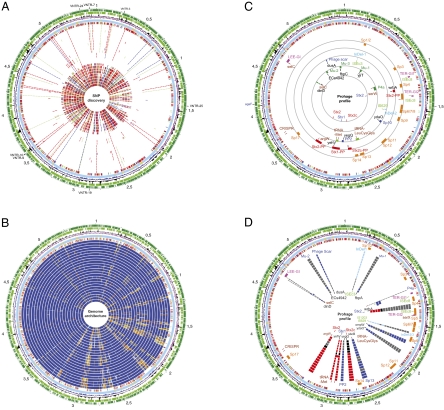
Fig. 1.
Genome organization of E. coli O157:H7. (A) SNP discovery. Circles (numbered from outer to inner circle): predicted ORFs of strain EC4115 encoded on the plus strands (circle 1) (with the replication initiation gene dnaA marked in red) and minus strands (circle 2). GC-skew (circle 3). χ2 indicating deviations in the GC content (circle 4). SNP-backbone of the E. coli O157:H7 lineage comprising 1,225 SNPs; 356 sSNPs (green), 647 nonsynonymous SNPS (nsSNPs) (red), and 222 intergenic SNPs (blue) (circle 5), SNP distribution, genome-wide distribution of identified SNPs using a window size of 5 kbp: 0–3 SNPs (light blue), 4–5 (gold), 6–7 (orange), 8–10 (red) (circle 6). Circles 7–30 show Individual SNP patterns referenced to EC4115 positions in strains EC4045 (7), EC4042 (8), EC4113 (9), EC4076 (10), EC4084 (11), EC4127 (12), EC4191 (13), EC4205 (14), TW14359 (15), EC4206 (16), EC4196 (17), EC4401 (18), EC4486 (19), EC4192 (20), EC4009 (21), EC508 (22), EC869 (23), FRIK2000 (24), FRIK966 (25), EC536 (26), EDL933 (27), Sakai (28), TW14588 (29), and EC4501 (30). VNTR (Variable Number Tandem Repeat) loci are marked in the outermost circle (1). (B) Genome architecture. Circles from outer to inner circle: predicted ORFs of strain EC4115 encoded on the plus (circle 1) and minus (circle 2) strands; GC-skew (circle 3); χ2 indicating deviations in the GC content (circle 4); SNP-backbone of the E. coli O157:H7 lineage comprising 1,225 SNPs; 356 sSNPs (green), 647 nsSNPs (red), and 222 intergenic SNPs (blue) (circle 5); SNP distribution, genome-wide distribution of identified SNPs using a window size of 5 bp, 0–3 (light blue), 4–5 (gold), 6–7 (orange), 8–10 (red) (circle 6). Circles 7–30 show comparative nucleotide analysis of EC4115 (7–30) with nucleotide identities as follows: 100–98% (dark blue), 98–95% (blue), 95–90% (orange), 90–85% (gold), 85–70% (yellow) to strains EC4045 (7), EC4042 (8), EC4113 (9), EC4076 (10), EC4084 (11), EC4127 (12), EC4191 (13), EC4205 (14), TW14359 (15), EC4206 (16), EC4196 (17), EC4401 (18), EC4486 (19), EC4192 (20), EC4009 (21), EC508 (22), EC869 (23), FRIK2000 (24), FRIK966 (25), EC536 (26), EDL933 (27), Sakai (28), TW14588 (29), and EC4501 (30). VNTR loci are marked in the outermost circle (1). (C) Phage content. Chromosomal key markers in the E. coli O157:H7 lineage are shown in circles (numbered outer to inner) 7–11. Genomic islands for enterocyte effacement (LEE) and tellurite (Ter) resistance (magenta), and CRISPR loci (brown). TER-GI1 is introduced by prophage P4a and thus is unique to EDL933 (circle 7). Common prophages in the E. coli O157:H7 lineage (orange) labeled according to ref. 40 (8). Potentially Stx-converting phages (red) (9). Signatures of the SP strains: prophages are shown in dark blue, genomic islands in light blue, and polymorphisms among the SP strains in light green (10). Identified isolate specific prophages in the studied population are shown in circle 11. Stx prevalence is shown in circle 12: Stx-positive loci in the SP strains are marked in red; inactive loci are marked in blue. Phage-attachment sites comprising tRNA loci are shown in brown, neighboring IS elements in green, and genes in white. (D) Phage prevalence. Chromosomal key markers in the E. coli O157:H7 lineage are shown in circle 7. Circles (numbered outer to inner) 8–23: show the common prophage pattern (orange) and islands (magenta) distinguishing phages EC4045 (8), EC4042 (9), EC4113 (10), EC4076 (11), TW14359 (12), EC4206 (13), EC4196 (14), EC4401 (15), EC4486 (16), EC4115 (17), EC508 (18), EC869 (19), EDL933) (20), Sakai (21), TW14588 (22), and EC4501 (23). Potentially Stx-converting phages are shown in red, other prophages and phage remnants in blue, and genomic islands in light blue; absent phages are highlighted in gray. Of note, strain EC869 lacks both Stx2-converting phages (black), whereas the TJ strains carry an stx prophage at both the argW and wrbA locus. Also, the EC869 P4-type phage is integrated within the borders of the LEE genomic islands at the selC locus. Phage-attachment sites comprising tRNA loci are shown in brown, neighboring IS elements in green, and genes in black. The unique additional fragment in strain EC4115 integrated within the borders of the Stx1-converting prophage at the yehV locus is a composite of the Sp9/Sp9’ prophages.
Fig. 2.
SNP-based phylogenetic tree of E. coli O157:H7. The phylogenetic tree is based on 1,225 intra- and intergenic SNPs identified in the studied E. coli O157:H7 population and reveals a clustering of the analyzed strains into distinct outbreak-associated clusters (SP, green; TB, purple; TJ, light red). Strains are colored according to lineages: lineage I (red circles); lineage II (brown circles), and lineage I/II (green circles). Specific features of each lineage are indicated in boxed text. Numbers in yellow circles indicate clade assignment.
Genotypic SNP-Profiling.
To validate our findings, we extended the genotypic profiling and tested a total of 229 E. coli O157:H7 strains (Dataset S3). Strains were subjected to pyrosequencing-based screening (27–29) of 19 canonical SNPs, which were chosen to differentiate the newly defined individual phylogenetic branches described above. The identified SNP panel allowed the genotypic grouping of closely related strains with high phylogenetic accuracy and resolution (SI Results and Discussion). However, SNP-based genotypic profiles are essential but not sufficient to establish genetic similarity. In this study we have fully characterized genetic states within group A and group B strains of lineage I/II by cataloguing genome architecture as well as prophage prevalence and location, revealing genetic heterogeneity among these strains.
Genome Organization in the E. coli O157:H7 Lineages.
The genome organization and plasticity of the E. coli genomes analyzed is shown in Fig. 1. We found a well-defined chromosomal GC bias defining the origin of replication at the dnaA gene. The fully closed reference genome sequence (EC4115) consists of a circular chromosome of 5,572,075 bp with a G+C content of 50% and coding density of 83% (Fig. 1A). Strain EC4115 harbors two plasmids, virulence plasmid pO157 (94,644 bp), a characteristic feature of the E. coli O157:H7 lineage, and pEC4115 (37,452 bp), a conjugal transfer plasmid (Fig. S1). PCR screening using pEC4115-specific primers showed that pEC4115 is unique to ME strains (EC4114, EC4115, and EC4116) and is absent from closely related strains also derived from the SP outbreak (Table S1). All the strains analyzed share a genome-wide synteny of the chromosomal backbone (Fig. 1B) indicative of the lack of major chromosomal rearrangements. Common features that define the E. coli O157:H7 lineage are two tellurite genomic islands (TER-GIs) and the LEE island. As evidenced in Fig. 1C, island TER-GI1 is part of the P4a-PP prophage and is unique to strain EDL933, whereas TER-GI2 is present in the remainder of E. coli O157:H7 strains analyzed. The high degree of homology among these multiple lambdoid phages is evident in the comparative nucleotide analysis shown in Fig. 1B. The overall synteny is disrupted by multiple insertion of prophages and genomic islands (30), a characteristic feature of the E. coli O157:H7 genome architecture (Fig. 1 C and D). Because of the highly homologous nature of the chromosomal backbone and the integrated but differently positioned lambdoid phages in the E. coli O157:H7 lineage, the genomic comparison makes sense only in the broader context of the overall genomic architecture and in the study of prophage and pathogenicity island prevalence and dynamics integrated at varying chromosomal locations (Fig. 1C). As evidenced in Fig. 1D, sequence-based prophage profiling led to the identification of numerous branch-specific prophage- and island-borne genome signatures that should serve as valuable biomarkers for future study of the E. coli O157:H7 lineage. We could identify four potential integration sites carrying Stx-converting phages (Fig. 1). The Stx2-converting phages target two chromosomal loci (wrbA and argW) and could differ in their regulatory circuits and verotoxin expression levels (31), Fig. 1C). On the other hand, unlike the Stx2 prophage, the Stx2c and Stx1 prophages seem to target preferentially the sbcB and yehV loci, respectively. These alterations in phage inventory and dynamics are mediated by unknown prophages and phage combinations as well as by varying integration loci (32), particularly in the Stx-converting phages (Fig. 1D).
Prophage Prevalence and Dynamics in the E. coli O157:H7 Lineage.
The high-quality sequencing and optical mapping methodologies used in this study were key for positioning the phage sequences and other laterally acquired genomic islands (Fig. 1 B and C), a task otherwise difficult because of the highly homologous nature of lambdoid prophages (33, 34). Comparative genomics helped tracking alterations of the stx gene state (Fig. 2), which, in part, could determine the virulence potential and ultimately disease severity of E. coli O157:H7 (Dataset S1).
Stx2c Locus.
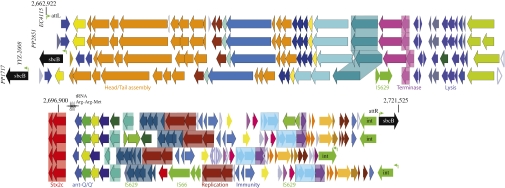
An second set of stx genes was found in lineage I/II clade 8 strains within the Stx2c-converting prophage integrated at the sbcB locus (ECH74115_2944) (Dataset S1and Fig. 1D). This Stx2c prophage is a unique genomic feature of the SP and TB outbreak strains and the HUS strain EC508 (19). As evidenced in Fig. 3, this 57,224-bp phage bears a striking resemblance in gene content and architecture to Enterobacteria phages derived from other E. coli O157:H7 strains, such as the 57,248-bp human feces phage 2851 isolated from strain CB2851 (35), the 62,147-bp phage PP1717 (NC_011357), and the 54,896-bp phage YYZ2008 (NC_011356) (Fig. 3 and Table S2).
Fig. 3.
Architecture and plasticity of the Stx2c-converting phage. The four compared Stx2c-converting prophages reveal high homology and synteny in structural and enzymatic key components and occupy different genomic locations. The Stx2c prophage ranges in size from 54,896–62,147 bp and preferentially targets the sbcB locus (black) (Table S2). Comprehensive analyses revealed several polymorphic regions and identified IS element insertions as microevolutionary drivers. Corresponding genes are colored and shaded: hypothetical genes in light gray, conserved hypothetical genes in dark gray, gene length variation in the phages caused by differences in annotation in white, and mobility genes, such as transposases and phage integrases in light green. Other genes without specific function are colored to highlight the syntenic nature of these phages.
Stx2 Locus.
Unlike the Stx1 and Stx2c phages, the Stx2-converting phage is found integrated at two distinct chromosomal loci in the E. coli O157:H7 lineages (Fig. 1 C and D). In lineage I, the stx2 genes of strains EDL933 and Sakai are found in the 62-kb prophage CP-933-W at the wrbA locus (Z1423/Z1504) harboring phage integrase intW (Z1424). The wrbA gene locus is unoccupied in lineage I/II strains carrying this toxin type, but stx2 is found at the argW gene locus (ECH74115_3581) (Fig. S2A). Verotoxin 2 is introduced by a unique prophage variant, Sp15-PPV (62,282 bp), that uses a phylogenetically unrelated integrase (ECH74115_3579). This phage is homologous in architecture and gene content to the 60,238-bp phage 86 found in the human pathogenic E. coli O86 strain DIJI from Japan (AB255436). Its borders are defined by 25-bp perfect direct-repeat attL/R sequences (Table S2), and the integrases share 99% nucleotide identity. The target region of this prophage is marked by a change in G+C content and features genes coding for a phage integrase and two phage-related DNA injection proteins (ECH74115_3582–ECH74115_3584) adjacent to the argW gene locus. The genetic composition of this phage integration site makes it prone to lateral acquisition of foreign DNA. Analysis of the wrbA and argW Stx2 subtypes revealed four major polymorphic sites that appear to drive microevolution of this stx2 prophage subtype (Fig. S2A): the insertion of two genes in phage 86 (Stx2-86_gp06/05), the altered gene content, the organization between antitermination and replication loci (ECH74115_3543–ECH74115_3552), and location in a region neighboring the replication exonuclease (ECH74115_3566–ECH74115_3573). Of note, the stx2 phage in the lineage I/II strains contains a structural pseudogene disrupted by an insertion sequence (IS) element, IS629, absent in phage 86, which may alter the surface-exposed phage structures and thus bacteria-host interactions. In silico analysis showed yet another polymorphic verotoxin state in the TJ strains EC4486 and EC4401 that carry two copies of the stx2 prophage insertions at both the wrbA and argW loci (Fig. 1D and Dataset S2).
Stx1 Locus.
In stx1-positive lineage I strains of E. coli O157:H7, such as EDL933, stx1 genes are found within the cryptic prophage Sp15-PPV (48,738 bp), which is characterized by its intV integrase (ECSP_2997, ECH74115_3251) and is located near the yehV gene and triplet tRNA loci. In lineage I/II strains, we identified two distinct prophage variants at the yehV gene locus that readily explain the characteristic stx1-negative genotype of lineage I/II strains (Fig. S2B). The first, slightly smaller Sp15-PPV variant (48,144 bp), was characterized by the deletion of stx1 genes from the prophage. A second, larger stx1-negative phage variant (93,567 bp) was distinguished by an additional 45,632-bp phage-related fragment that encodes integrase intO (ECH74115_3178); this fragment is inserted at the tRNA Met locus within phage SP15-PPV. This prophage variant is characteristic of the three ME strains (EC4113, EC4115, and EC4116), whereas the smaller variant of 48,144 bp is carried in the remainder of lineage I/II strains examined in the present study. It is most likely that this phage organization arose by duplication and transposition of phage Sp9 (cpO) targeting phage Sp15. Comparative genomics shows that both variants are organized syntenically; i.e., compared with the stx1-positive variant, the stx1-negative variant is a complex composite of two joined fragments of Sp9 and a unique Sp9’ prophage. We speculate that the adjacent triplet tRNA loci may have facilitated homologous recombination events and rearrangements of these joined prophage fragments. Interestingly, we also identified several polymorphic regions (Fig. S2B, shaded in red) that most likely arose through recombinatorial events facilitated by the homologous nature of lambdoid phages. Of note are the fragmented repP phage replication pseudogenes resulting from disruption by IS629 in the lineage I/II-associated stx1-negative phage subtypes; this fragmentation may result in an impaired replication and immobilization of these phages in the SP and TB outbreak strains as well as in strain EC508. Strain EC4115 carries a unique lambdoid prophage termed “cpO’ ” (45,449 bp) inserted at the right border of the cpO prophage (58,298 bp) at the ompW locus (Fig. S3A). The cpOO’ tandem phage architecture explains the characteristic stx1-negative genotype of lineage I/II ME strains. We note here that optical mapping was key to confirm this unique phage architecture in the two other collected ME strains, EC4114 and EC4116. This intrachromosomal duplication and translocation led to a complex cpOO’ tandem phage architecture flanked by 117-bp direct repeats (Table S2).
Non-Shiga Toxin-Carrying Phage Markers and Genomic Islands in E. coli O157:H7.
We cataloged numerous non–verotoxin-related mobile genomic regions and variations thereof, which in many instances are accompanied by insertion-specific repeats (Table S2). These regions comprise newly identified prophages (Fig. S3 A and B) and phage scars (Fig. S3 D and E) as well as genomic islands ranging in size from 2 to 15 kbp (Fig. S3 F and G and SI Results and Discussion). Interestingly, their distribution among the strains analyzed does not necessarily follow the phylogenetic relationships. This discrepancy is caused by the mobile nature of these elements, and although these lateral regions present valuable biomarkers, the findings highlight the need to evaluate the mobilome of a species to reconstruct a species’ evolutionary history accurately. Typing of these lateral regions allowed us not only to differentiate the outbreak strains (e.g., SP, TB, and TJ) from each other and from the remainder of the analyzed strains but also to distinguish among closely related strains derived from the same outbreak. For example, some of these phage polymorphic loci were key in establishing genetic heterogeneity among the closely related strains in the SP outbreak as evidenced by the ME-specific cpOO’ tandem prophage (Fig. S3A) or the EC4076-specific deletion of the Sp13 prophage (Fig. S3C).
Prophage Microevolution.
Comparison of the phylogenetically related Shiga toxin-converting prophages in the E. coli O157:H7 lineage revealed insights into microevolutionary processes that drive phage evolution and account for the observed length of polymorphisms (Fig. 3 and Fig. S2A). We identified numerous hypervariable regions among Enterobacteria phages, often associated with the insertion of the IS elements IS629 and IS66 that are present in multiple copies in the E. coli O157:H7 genomes. The transposable IS elements IS629 and IS66 are major drivers of Stx prophage microevolution, because these mobile elements disrupt structural and enzymatic components and mediate the influx of new genetic information (Fig. 3, Fig. S3B, and SI Results and Discussion). We speculate that IS insertion and propagation might alter viable prophage function, because we observed the targeting of structural and enzymatic phage key components, thus controlling phage dynamics in the diverging E. coli O157:H7 subtypes. Moreover, these IS elements introduce new genetic information and create fragmented pseudogenes, which together might alter the phage-borne gene content and expression.
Genetic Alterations in Metabolic Properties.
Phenotypic Biolog characterization and genotypic analyses of 231 strains, which originate from different clinical and environmental sources (bovine host reservoir and contaminated spinach bags), from the E. coli O157:H7 culture collection maintained by the Food and Drug Administration (Dataset S2) revealed that all 194 strains associated with the SP outbreak, all 10 bovine strains from the California farms, and all eight strains of the TB outbreak displayed a characteristic N-acetyl-d-galactosamine–negative/d-galactosamine–negative phenotype because of a single SNP (Dataset S3 and SI Results and Discussion). Of note, only a single unrelated outbreak lineage I/II strain, EC508, shared this point mutation in the agaF gene with SP and TB strains (Dataset S2). The observed prevalence of this unique phosphotransferase transport system-deficient phenotype and the Shiga toxin state support the phylogenetic placement of EC508 within lineage I/II and its close relatedness to the strains associated with the SP and TB outbreaks (Fig. 2). SNP discovery further revealed a unique point mutation in the sucrose transporter gene of strain EC4115 (ECH74115_3591) that was verified in all analyzed ME strains and results in a pseudogene (Dataset S3). The truncated pseudogene product underlies the Biolog-observed ME-specific d-sucrose–negative phenotype (SI Results and Discussion). Their unique phylogenetic position is supported by the ME-specific genome architecture, which distinguishes these strains from the remainder of lineage I/II strains (Figs. S1 and S2B). This study clearly demonstrates the power of combining genomic and metabolic profiling in analyzing closely related strains.
Conclusion
Understanding the genetic diversity and genome dynamics in E. coli O157:H7 provides critical insights into this pathogen's evolutionary patterns and ecological niches. The identification of numerous biomarkers (of the core and in the mobilome) for the E. coli O157:H7 lineage allowed high-resolution typing with a unprecedented discriminatory power, thus revealing genomic heterogeneity, although the overall genome composition of this serotype is genetically homogenous, comparable with clonal microbial species such as Yersinia pestis or Bacillus anthracis. As evidenced in this study, the methodologies applied clearly differ in their discriminative power; however we found the complementary use of different typing approaches, including SNP genotyping, most effective in achieving high phylogenetic accuracy and resolution. The phylogenomic analyses used accounted for fine polymorphisms in the E. coli O157:H7 backbone and overall genome organization and dynamics, as well as recent evolution of the Shiga-like toxin gene state and metabolic capabilities. Triaging these methodologies enabled us to elucidate the phylogenetic relatedness of individual strains and identified multiple microevolutionary traits that lead to lineage-specific genome characteristics. Although the current molecular assays used in public health microbiology laboratories may be adequate for routine surveillance and identification of E. coli O157:H7, they lack the discriminatory power needed to resolve its genetically homogenous population structure and ultimately for linking human E. coli O157:H7 infection to its source with high accuracy. This study provides a high-resolution phylogenomic framework for E. coli O157:H7 with great accuracy and high resolution. This study established a panel of 1,225 SNPs that now is available for the typing of E. coli O157:H7 and can be optimized further by careful review of covariant loci that possess redundant phylogenetic information. These covariant loci include the presence of plasmids, prophage combinations and positions that by themselves potentially define a distinct phylogenetic branch, as discussed in greater detail for the SP strains associated with the Maine outbreak. Identified polymorphisms in prophage profiles and dynamics were shown to arise through recombination, duplication, and translocation events of cryptic phages and microevolutionary gene loss and acquisition. IS targeting of phage-related regions seems to be a common phenomenon in E. coli O157:H7. This study clearly demonstrates that the collecting and sequencing multiple strains from each source or patient from a disease outbreak would provide a better basis for assessing the dynamics of genomic plasticity within outbreak-derived strains. In accordance with the present but limited plasticity in the E. coli O157:H7 strains associated with the SP outbreak, this conclusion also is supported by the genetic heterogeneity reported in several serotype O104 strains derived from the German outbreak (36–38). Even in an outbreak from an apparent single source, nucleotide variations were detected among recovered strains; moreover, under laboratory cultivation we found three instances of microevolutionary alterations comprising two SNPs and a 1.9 kbp inversion (Fig. S4, SI Results and Discussion). It is not clear, if observed genotypic differences are the result of changes occurring in strains during the course of the outbreak, resulting in new clone(s) from the typical isolate, or are caused by the mixture of strains in the source of the outbreak. The latter possibility could have major implications in the study of the epidemiology of outbreaks and suggests that the microbial populations in an outbreak should be studied, rather than relying on single archetypal reference outbreak strain. Some of the identified genomic signatures, such as the Shiga toxin virulence states (Stx1, Stx2+, Stx2c+) of the lineage I/II-derived strains responsible for the SP and TB outbreaks, appear to be associated intimately with an increased pathogenic potential (19). These data provide an accurate and refined depiction of the processes driving the evolution of the E. coli O157:H7 pathogenome.
Materials and Methods
Genomic DNA of the E. coli O157:H7 strains was subject to random shotgun sequencing and closure strategies using a combination of Sanger and 454 sequencing as previously described (5). Draft genome sequences were assembled using Celera Assembler (39) and manually annotated using the MANATEE system (http://manatee.sourceforge.net/). Details of bacterial strains, genome sequencing, assembly, annotation, comparative genome analysis, and the different typing assays are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported with federal funds from the National Institute of Allergy and Infectious Diseases (NAID), National Institutes of Health, US Department of Health and Human Services, under NIAID Contract N01 AI- 30071.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition The sequences reported in this paper have been deposited in the GenBank database. Accession numbers are listed in the SI.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107176108/-/DCSupplemental.
References
- 1.Leopold SR, et al. A precise reconstruction of the emergence and constrained radiations of Escherichia coli O157 portrayed by backbone concatenomic analysis. Proc Natl Acad Sci USA. 2009;106:8713–8718. doi: 10.1073/pnas.0812949106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogura Y, et al. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci USA. 2009;106:17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowe RM, et al. Escherichia coli O157:H7 strain origin, lineage, and Shiga toxin 2 expression affect colonization of cattle. Appl Environ Microbiol. 2009;75:5074–5081. doi: 10.1128/AEM.00391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutin L. Emerging enterohaemorrhagic Escherichia coli, causes and effects of the rise of a human pathogen. J Vet Med B Infect Dis Vet Public Health. 2006;53:299–305. doi: 10.1111/j.1439-0450.2006.00968.x. [DOI] [PubMed] [Google Scholar]
- 5.Eppinger M, et al. Genome sequence of the deep-rooted Yersinia pestis strain Angola reveals new insights into the evolution and pangenome of the plague bacterium. J Bacteriol. 2010;192:1685–1699. doi: 10.1128/JB.01518-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Mead PS, Griffin PM. Escherichia coli O157:H7. Lancet. 1998;352:1207–1212. doi: 10.1016/S0140-6736(98)01267-7. [DOI] [PubMed] [Google Scholar]
- 8.Makino K, et al. Complete nucleotide sequence of the prophage VT2-Sakai carrying the verotoxin 2 genes of the enterohemorrhagic Escherichia coli O157:H7 derived from the Sakai outbreak. Genes Genet Syst. 1999;74:227–239. doi: 10.1266/ggs.74.227. [DOI] [PubMed] [Google Scholar]
- 9.Bertin Y, Boukhors K, Pradel N, Livrelli V, Martin C. Stx2 subtyping of Shiga toxin-producing Escherichia coli isolated from cattle in France: Detection of a new Stx2 subtype and correlation with additional virulence factors. J Clin Microbiol. 2001;39:3060–3065. doi: 10.1128/JCM.39.9.3060-3065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor DE, et al. Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J Bacteriol. 2002;184:4690–4698. doi: 10.1128/JB.184.17.4690-4698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen S, Mascarenhas M, Morgan R, Rahn K, Karmali MA. Identification of four fimbria-encoding genomic islands that are highly specific for verocytotoxin-producing Escherichia coli serotype O157 strains. J Clin Microbiol. 2005;43:3840–3850. doi: 10.1128/JCM.43.8.3840-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pradel N, et al. Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2003;71:4908–4916. doi: 10.1128/IAI.71.9.4908-4916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xicohtencatl-Cortes J, et al. Intestinal adherence associated with type IV pili of enterohemorrhagic Escherichia coli O157:H7. J Clin Invest. 2007;117:3519–3529. doi: 10.1172/JCI30727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wells TJ, et al. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ Microbiol. 2008;10:589–604. doi: 10.1111/j.1462-2920.2007.01479.x. [DOI] [PubMed] [Google Scholar]
- 15.Griffin PM, et al. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann Intern Med. 1988;109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- 16.Riley DG, Gray JT, Loneragan GH, Barling KS, Chase CC., Jr Escherichia coli O157:H7 prevalence in fecal samples of cattle from a southeastern beef cow-calf herd. J Food Prot. 2003;66:1778–1782. doi: 10.4315/0362-028x-66.10.1778. [DOI] [PubMed] [Google Scholar]
- 17.Cimolai N, Morrison BJ, Carter JE. Risk factors for the central nervous system manifestations of gastroenteritis-associated hemolytic-uremic syndrome. Pediatrics. 1992;90:616–621. [PubMed] [Google Scholar]
- 18.Besser RE, Griffin PM, Slutsker L. Escherichia coli O157:H7 gastroenteritis and the hemolytic uremic syndrome: An emerging infectious disease. Annu Rev Med. 1999;50:355–367. doi: 10.1146/annurev.med.50.1.355. [DOI] [PubMed] [Google Scholar]
- 19.Kulasekara BR, et al. Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect Immun. 2009;77:3713–3721. doi: 10.1128/IAI.00198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manning SD, et al. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc Natl Acad Sci USA. 2008;105:4868–4873. doi: 10.1073/pnas.0710834105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, et al. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res. 2006;16:757–767. doi: 10.1101/gr.4759706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z, et al. Identification of common subpopulations of non-sorbitol-fermenting, beta-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl Environ Microbiol. 2004;70:6846–6854. doi: 10.1128/AEM.70.11.6846-6854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noller AC, McEllistrem MC, Harrison LH. Genotyping primers for fully automated multilocus variable-number tandem repeat analysis of Escherichia coli O157:H7. J Clin Microbiol. 2004;42:3908. doi: 10.1128/JCM.42.8.3908.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee A, Mammel MK, LeClerc JE, Cebula TA. Altered utilization of N-acetyl-D-galactosamine by Escherichia coli O157:H7 from the 2006 spinach outbreak. J Bacteriol. 2008;190:1710–1717. doi: 10.1128/JB.01737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dowd SE, et al. Microarray analysis and draft genomes of two Escherichia coli O157:H7 lineage II cattle isolates FRIK966 and FRIK2000 investigating lack of Shiga toxin expression. Foodborne Pathog Dis. 2010;7:763–773. doi: 10.1089/fpd.2009.0482. [DOI] [PubMed] [Google Scholar]
- 26.Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. Genome signatures of Escherichia coli O157:H7 isolates from the bovine host reservoir. Appl Environ Microbiol. 2011;77:2916–2925. doi: 10.1128/AEM.02554-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson SA, et al. Interrogating genomic diversity of E. coli O157:H7 using DNA tiling arrays. Forensic Sci Int. 2007;168:183–199. doi: 10.1016/j.forsciint.2006.06.079. [DOI] [PubMed] [Google Scholar]
- 28.Kotewicz ML, Mammel MK, LeClerc JE, Cebula TA. Optical mapping and 454 sequencing of Escherichia coli O157 : H7 isolates linked to the US 2006 spinach-associated outbreak. Microbiology. 2008;154:3518–3528. doi: 10.1099/mic.0.2008/019026-0. [DOI] [PubMed] [Google Scholar]
- 29.Davis MA, Hancock DD, Besser TE, Call DR. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7. J Clin Microbiol. 2003;41:1843–1849. doi: 10.1128/JCM.41.5.1843-1849.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darling AE, Miklós I, Ragan MA. Dynamics of genome rearrangement in bacterial populations. PLoS Genet. 2008;4:e1000128. doi: 10.1371/journal.pgen.1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Creuzburg K, et al. Genetic structure and chromosomal integration site of the cryptic prophage CP-1639 encoding Shiga toxin 1. Microbiology. 2005;151:941–950. doi: 10.1099/mic.0.27632-0. [DOI] [PubMed] [Google Scholar]
- 32.Karch H, Schmidt H, Janetzki-Mittmann C, Scheef J, Kröger M. Shiga toxins even when different are encoded at identical positions in the genomes of related temperate bacteriophages. Mol Gen Genet. 1999;262:600–607. doi: 10.1007/s004380051122. [DOI] [PubMed] [Google Scholar]
- 33.Kotewicz ML, Jackson SA, LeClerc JE, Cebula TA. Optical maps distinguish individual strains of Escherichia coli O157 : H7. Microbiology. 2007;153:1720–1733. doi: 10.1099/mic.0.2006/004507-0. [DOI] [PubMed] [Google Scholar]
- 34.Johansen BK, Wasteson Y, Granum PE, Brynestad S. Mosaic structure of Shiga-toxin-2-encoding phages isolated from Escherichia coli O157:H7 indicates frequent gene exchange between lambdoid phage genomes. Microbiology. 2001;147:1929–1936. doi: 10.1099/00221287-147-7-1929. [DOI] [PubMed] [Google Scholar]
- 35.Strauch E, et al. Bacteriophage 2851 is a prototype phage for dissemination of the Shiga toxin variant gene 2c in Escherichia coli O157:H7. Infect Immun. 2008;76:5466–5477. doi: 10.1128/IAI.00875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasko DA, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mellmann A, et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS ONE. 2011;6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brzuszkiewicz E, et al. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC) Arch Microbiol. 2011;193:883–891. doi: 10.1007/s00203-011-0725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huson DH, et al. Design of a compartmentalized shotgun assembler for the human genome. Bioinformatics. 2001;17(Suppl 1):S132–S139. doi: 10.1093/bioinformatics/17.suppl_1.s132. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi T, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.