Abstract
RNA deep sequencing technologies are revealing unexpected levels of complexity in bacterial transcriptomes with the discovery of abundant noncoding RNAs, antisense RNAs, long 5′ and 3′ untranslated regions, and alternative operon structures. Here, by applying deep RNA sequencing to both the long and short RNA fractions (<50 nucleotides) obtained from the major human pathogen Staphylococcus aureus, we have detected a collection of short RNAs that is generated genome-wide through the digestion of overlapping sense/antisense transcripts by RNase III endoribonuclease. At least 75% of sense RNAs from annotated genes are subject to this mechanism of antisense processing. Removal of RNase III activity reduces the amount of short RNAs and is accompanied by the accumulation of discrete antisense transcripts. These results suggest the production of pervasive but hidden antisense transcription used to process sense transcripts by means of creating double-stranded substrates. This process of RNase III-mediated digestion of overlapping transcripts can be observed in several evolutionarily diverse Gram-positive bacteria and is capable of providing a unique genome-wide posttranscriptional mechanism to adjust mRNA levels.
Keywords: antisense RNA, overlapping transcription, RNA processing, posttranscriptional regulation, microRNA
For many years, the catalog of transcripts (transcriptome) produced by bacterial cells was limited to the transcription products of known annotated genes (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA). In the past 10 years, the development of new approaches based on high-resolution tiling arrays and RNA deep sequencing (RNA-seq) has uncovered that a significant proportion (depending on the study, varying between 3% and >50%) of protein coding genes are also transcribed from the reverse complementary strand (1–17). In most cases, overlapping transcription generates a noncoding antisense transcript whose size can vary between various tens of nucleotides (cis-encoded small RNAs) to thousands of nucleotides (antisense RNAs). The antisense transcript can cover the 5′ end, 3′ end, middle, entire gene, or even various contiguous genes. Alternatively, overlapping transcription can also be due to the overlap between long 5′ or 3′ UTRs of mRNAs transcribed in the opposite direction. Independent of the mechanism by which it is generated, overlapping transcription has been proposed to affect the expression of the target gene at different levels [for review, see Thomason and Storz (18)]. These mechanisms include: (i) the overlapped transcript affects the stability of the target RNA by either promoting (RNA degradation) or blocking (RNA stabilization) cleavage by endoribonucleases or exoribonucleases; (ii) the overlapped transcript induces a change in the structure of the mRNA that affects transcription termination (transcription attenuation); (iii) the overlapped transcript prevents RNA polymerase from binding or extending the transcript encoded in the opposite strand (transcription interference); and (iv) the overlapping transcript affects protein synthesis either blocking or promoting ribosome binding (translational regulation). Although all these regulatory mechanisms have been proposed based on studies with specific sense–antisense partners, the presence of massive amounts of overlapping transcription strongly suggest that it might serve for a general purpose on bacterial gene expression (5, 18–24).
In this work, we used RNA sequencing to analyze both the long and short RNA fractions of the major human pathogen Staphylococcus aureus. S. aureus is a common asymptomatic colonizer of the skin, nasopharynx, and other mucosal surfaces of approximately one-fourth of the healthy human population. However, when S. aureus traverses the epithelial barrier, it becomes a leading cause of many diverse pathological syndromes, such as abscesses, bacteremia, endocarditis, osteomyelitis, and pneumonia (25). S. aureus has emerged as a model organism for the study of bacterial regulatory RNAs because key discoveries in bacterial regulatory RNAs have been achieved in this bacterium. In 1993, Novick and coworkers (26) identified the first example of a regulatory RNA (RNAIII) that controls the expression of virulence factors by pairing with the target mRNAs followed by degradation of the RNAIII–mRNA complex by the double-stranded specific RNase III (27). More recently, several studies using computational analysis of intergenic regions, microarray technology, and deep sequencing have allowed the identification of >140 small RNAs, including both trans- and cis-encoded antisense RNAs (10, 28–32). In this current study, we uncover the existence of an overlapping transcription process covering, in a genome-wide extent, the expressed protein coding genes. Base pairing between overlapping RNAs can create double-stranded substrates for RNase III endoribonuclease activity. Such duplex regions promote the cleavage of the double-stranded RNA and the generation of short RNAs (average size of 20 nt). Thus, a collection of stable small RNA molecules that symmetrically map both strands of every region with overlapping transcription is generated. The presence of an identical collection of short RNA molecules that symmetrically mapped both strands of annotated ORFs in Enterococcus faecalis, Listeria monocytogenes, and Bacillus subtilis indicated that this process is evolutionary conserved in Gram-positive bacteria.
Results
Pervasive Antisense Transcription in S. aureus.
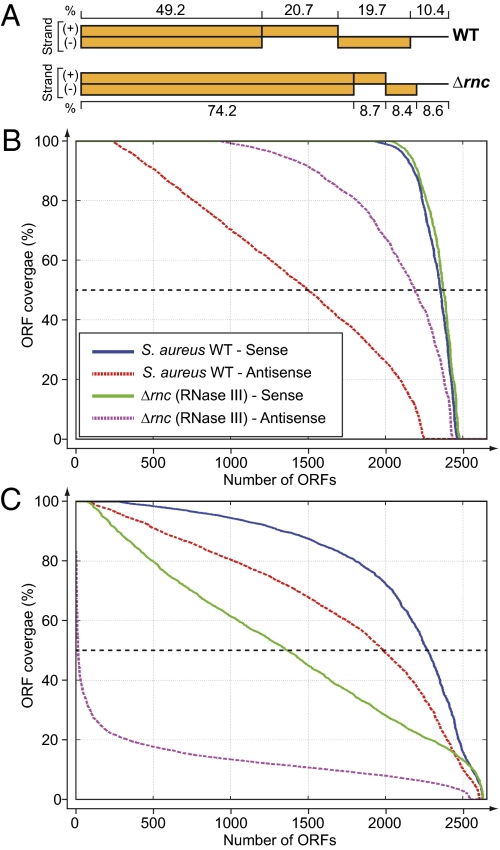
A systematic and hierarchical strategy (Fig. S1) to characterize both long and short RNA (<50 nt) fractions from log-phase growing S. aureus cells was developed. Long RNA sequencing was performed by using a cDNA synthesis procedure that preserves information about a transcript's direction based on the incorporation of deoxy-UTP during the second strand synthesis and subsequent destruction of the uridine-containing strand (33). The resulting 76-bp paired-end reads were mapped to the S. aureus NCTC 8325 reference genome. A total of 9.7 million uniquely mapped read pairs were identified (Fig. S1). 49.2% of the genome was covered by uniquely mapped reads on both strands, 40.4% by uniquely mapped reads on one of the strands, and 10.4% showed no coverage (Fig. 1A). Of the 2,653 annotated ORFs of the S. aureus genome, which covers ∼84% of the genome, we detected expression of 2,181 ORFs (coverage of >90%), of which 1,387 ORFs displayed 50% coverage on the antisense strand (Fig. 1B).
Fig. 1.
Genome-wide analysis of mapped reads from long and short RNA-seq libraries. (A) Percentage of the genome of S. aureus NCTC 8325 covered by uniquely mapped reads on both strands, reads on one of the strands, and showed no coverage, respectively. The long RNA-seq libraries were prepared from S. aureus 15981 wild-type strain (WT) and its corresponding RNase III mutant (Δrnc). (B and C) Comparison of the cumulative distribution of ORF coverage by long (B) and short (C) RNA reads. The plot represents the number of ORFs (x axis) found above the ORF coverage value (y axis). The coverage was computed from the collapsed reads uniquely mapped in the sense and antisense orientation to the ORFs. The dashed line represents 50% coverage.
Naturally occurring short RNAs were also sequenced in a strand-aware fashion by using a two-step adaptor ligation procedure to the 3′ and 5′ ends of the RNA molecules (34). The reads were aligned by algorithmically clipping off the 3′ adapter, and the remaining sequences of each read were mapped to the genome by using STAR (http://gingeraslab.cshl.edu/STAR/). For alignments of 10–19 bases long, up to one mismatch was allowed; for alignments >20 bases, up to two mismatches were allowed. Alignments of <10 bases were discarded, and spliced alignments were prohibited. This process yielded a total of 7,778,726 million reads mapped to the genome (Fig. S1). The average length of short RNA molecules was 20 nt (Fig. S1). The uniquely mapped short RNA sequences covered, in at least 50% of their length, 2,268 and 1,981 ORF regions on the sense and antisense strands, respectively (Fig. 1C). Thus, the percentage of ORFs covered in at least 50% of their length by reads in the antisense strand was higher in the case of short RNA (75%) than in the case of long RNA (56%), suggesting that short RNA libraries may prove to be a more sensitive way to detect antisense transcripts. Overall, and given that long and short RNA libraries were generated independently—that is to say from two fractions coming from the same RNA sample—these results provide evidence of the existence of antisense transcription not seen in the long RNA sequence analysis.
Symmetric Distribution of Short RNA Reads in both Strands of ORF Regions.
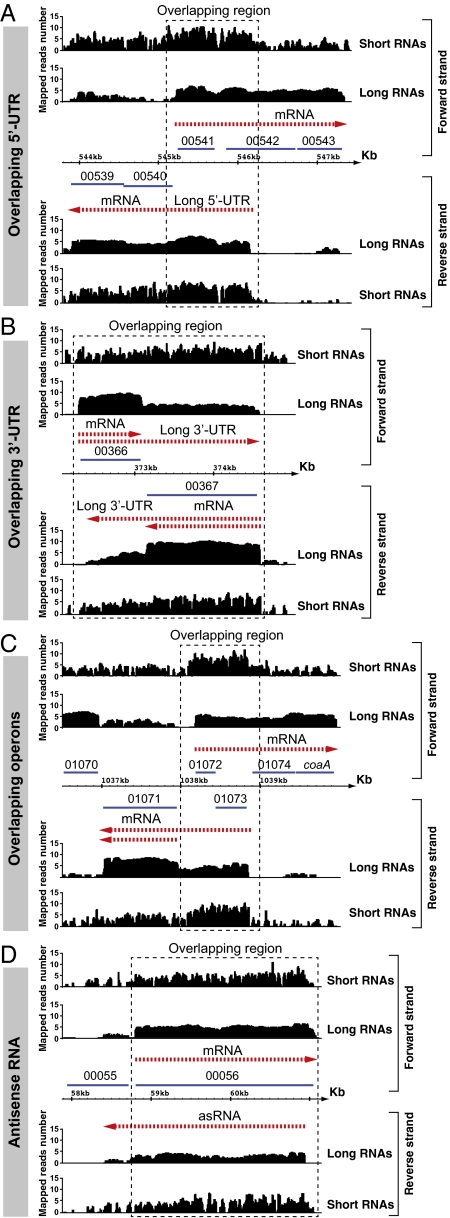
We next sought to determine whether the distribution of short and long reads for a given ORF were linked. For that, we visualized normalized log2 values representing the number of mapped reads per nucleotide using the Integrated Genome Browser (IGB) (35). Fig. 2 illustrates a randomly selected 30-kb region of the genome of S. aureus that represents 1% of the genome and depicts the uniquely mapped long and short RNAs. The results revealed that short RNA sequences were symmetrically distributed in both strands of the ORFs, whereas long RNA transcripts follow the expected biased distribution toward the sense strand. Intriguingly, the regions with detectable overlapped transcription between long RNA transcripts, such as those regions corresponding to antisense transcripts to ORFs (00056, 00061, sirABC operon), were covered with higher numbers of short RNA reads in both strands. Similar symmetrical accumulation of high levels of short RNAs was detected in every region of the genome where noticeable overlapping transcription occurs, such as 5′ and 3′ overlapping UTRs, overlapping operons (ORFs that, being located in the middle of an operon, are transcribed in opposite direction to the other genes of the operon), and antisense transcripts (see Fig. 3 and Figs. S2–S4 for additional examples). To most accurately demonstrate that the distribution of short RNA reads was symmetric genome-wide, we quantified the number of long and short RNAs mapping to the sense and antisense strands of each ORF. In accordance with the images observed with the IGB browser, the results revealed very similar numbers of short RNA reads genome-wide in both strands of ORF regions and the expected biased distribution of long RNA reads in the sense strand (Fig. 4 A and B). In summary, these results show that the S. aureus transcriptome contains both long and very short RNA molecules. The amount of long RNAs is, as expected, higher in the sense strand of each ORF. In contrast, short RNAs are equally distributed in both strands of each ORF and specially enriched in those regions with detectable overlapped transcription between long RNAs.
Fig. 2.
Long and short mapped reads distribution in S. aureus genome. The drawing is an IGB software image showing the uniquely mapped long and short RNAs in a 30-kb region (1%) of the genome of S. aureus NCTC 8325. Transcripts are represented as dashed red arrows. Genomic coordinates denote the position in kilobases of the S. aureus NCTC 8325 genome. Annotated ORFs are shown as blue lines. The number on the ORF indicates the gene identification. Long and short RNAs show the distribution of uniquely mapped reads of long and short RNA libraries. S. aureus 15981 (black) and S. aureus 15981 Δrnc (RNase III mutant) (green). The scale (log2) indicates the number of mapped reads per nucleotide position.
Fig. 3.
Examples of mapped reads distribution in regions with overlapping transcription of S. aureus. Drawings are images from IGB software showing different regions of the genome of S. aureus NCTC 8325. Examples of overlapping 5′ UTRs (A), overlapping 3′ UTRs (B), overlapping operons (C), and antisense RNA (D) are shown. Transcripts are represented as dashed red arrows. Genomic coordinates denote the position in kilobases of the S. aureus NCTC 8325 genome. Annotated ORFs are shown as blue lines. The number on the ORF indicates the gene identification. Long and short RNAs show the distribution of uniquely mapped reads of long and short RNA libraries in S. aureus 15981. The scale (log2) indicates the number of mapped reads per nucleotide position. Dashed rectangles highlight increased accumulation of short mapped reads in regions with overlapping transcription, according to long RNA reads.
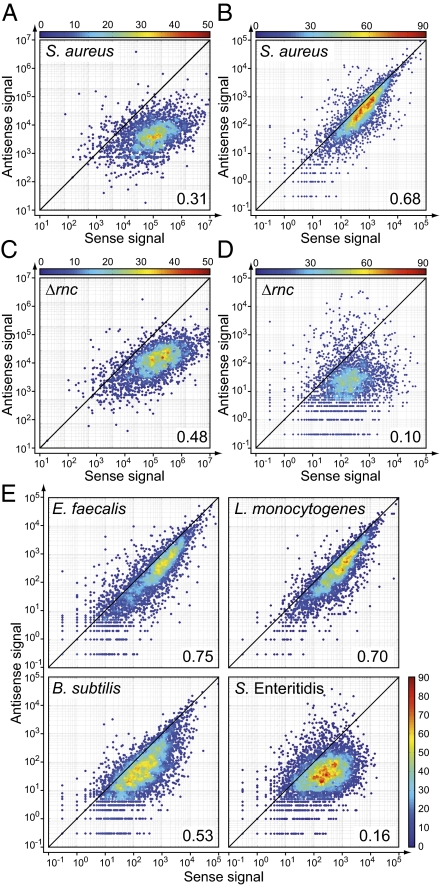
Fig. 4.
Expression levels of sense/antisense transcripts. (A and C) The plots show the dependence of the antisense vs. sense ORF-averaged signal in long RNA reads. Each dot corresponds to one ORF annotated in the S. aureus NCTC 8325 genome. S. aureus 15981 wild-type (A) and S. aureus 15981 Δrnc (RNase III mutant) (C). (B and D) The plots show the dependence of the number of uniquely mapped reads per ORF for the antisense strand vs. sense strand in the short RNA reads. S. aureus 15981 wild-type (B) and S. aureus 15981 Δrnc (RNase III mutant) (D). (E) Genome-wide analysis distribution of mapped reads from short RNA-seq libraries in different bacterial species. The plot shows the dependence of the number of uniquely mapped reads per ORF for the antisense strand vs. sense strand in the short RNA-seq libraries of E. faecalis, L. monocytogenes, B. subtilis, and Salmonella Enteritidis. The color scale represents the number of points within a ±20% window of each point. The number in the bottom right corner is the Spearman correlation coefficient R2.
RNase III Is Responsible for the Production of Symmetrically Distributed Short RNA Populations.
The fact that short RNAs display a symmetrical distribution in sense/antisense strands and accumulate in higher numbers in regions with noticeable overlapping transcription raised the possibility that short RNA molecules were derived from the cleavage of overlapping long sense/antisense primary transcripts. S. aureus genome has been reported to encode at least eight putative endoribonucleases and three exoribonucleases (32). Among them, the RNase III endoribonuclease is the only enzyme known to be able to degrade double-stranded RNA. Thus, we tested the possibility that RNase III might be responsible for processing the overlapping transcripts into symmetrically distributed sense and antisense short RNA populations. An RNase III mutant in the S. aureus 15981 background (S. aureus 15981 Δrnc) was constructed by using a described approach (36).
Analysis of the uniquely mapped reads from long RNA-seq of the RNase III mutant revealed that the percentage of the genome covered by reads on both strands increased up to 74.2% compared with wild-type strain (49.2%; Fig. 1A). This increase was mainly due to a significantly higher coverage of the antisense strand (82% ORFs displayed 50% coverage on the antisense strand; Fig. 1B). In contrast, the number of the short RNA reads was drastically reduced, especially in the antisense strand (Fig. 1C), reducing the percentage of the genome that was covered on both strands by short RNAs to only 6%. Of note, the median length of the short RNA molecules in RNase III mutant was 15 nt, suggesting the possibility that short RNAs detected in the RNase III mutant were produced by another RNA processing pathway (Fig. S1). Visualization of the distribution of mapped reads by using IGB confirmed that short RNA sequences had lost their symmetric distribution, whereas long RNA transcripts followed the expected biased distribution toward the sense strand (Fig. 2 and Figs. S2–S4). Accordingly, the correlation first observed in the wild-type strain between the numbers of short RNA reads in sense and antisense strands per annotated ORF region disappeared in the analysis of the RNase III mutant (Fig. 4 C and D). Together, these results indicate that a large majority of short RNA molecules present in the transcriptome of S. aureus are produced by the cleavage activity of double-stranded RNase III enzyme.
Because the pattern and cleavage frequency by RNase III is unknown, the short RNA molecules might be direct products of RNase III activity or processed products of larger RNA fragments generated by RNase III. Pnp is the most important 3′–5′ exoribonuclease activity in bacteria. S. aureus contains a gene (SAOUHSC_01251) encoding a protein that shares 66% identity with Pnp of B. subtilis. We produced libraries from short RNA fraction of S. aureus 15981 Δpnp. Analysis of the mapped reads from S. aureus 15981 Δpnp mutant revealed that the distribution and size of the short RNAs followed a pattern indistinguishable from that of the wild-type strain (Fig. S1), suggesting that Pnp activity is not required for subsequent processing of the short RNA molecules generated by RNase III activity.
Abundance of Short RNAs Correlates with the Levels of Double-Stranded Sense/Antisense Transcripts.
One prediction of the model that short RNAs are produced from the processing of genome-wide overlapping regions of transcription is that the abundance of short RNAs detected should be proportional to the abundance of either the sense/antisense transcripts, depending upon which transcribed strand is less abundant and available for processing. To explore this prediction, we analyzed the short and long RNA complements from a sigma B (sigB) mutant (S. aureus ΔsigB). The transcription factor Sigma B drives the transcription activity of genes under specific environmental conditions. We analyzed ORF regions for which the long antisense transcripts contained a consensus SigB promoter box (Fig. S5), and their expression was significantly suppressed in the sigB mutant (>50% reduction in the ΔsigB/WT antisense transcript ratio based on the long RNA libraries). Consistent with the hypothesis that the abundance of short RNAs depends on the levels of double-stranded RNA, knockdown of the antisense transcripts levels in sigB mutant correlates with a decrease in the amount of short RNAs produced from both strands (Fig. S5). These results indicated that the short RNA abundances at ORF regions are strongly correlated with the less abundant levels of overlapping long RNA capable of forming double-stranded RNA.
Detection of Occurrence and Abundance of Antisense Transcripts in RNase III Mutant.
Detection of antisense transcripts has been difficult in bacteria, and only the presence of a few antisense transcripts has been confirmed by Northern blot techniques. RNAse III cleavage of overlapping RNA transcripts into short RNA molecules could explain at least in part the paucity of antisense transcripts detected so far. Only minimal amounts of antisense transcripts would be maintained in the cell by RNAse III activity. To explore this hypothesis, we performed Northern hybridizations with strand-specific probes to interrogate sense and antisense transcripts of several individual genes in wild-type and RNase III mutant strains. The candidate genes were selected based on their relevance to different aspects of S. aureus virulence (sarA, agrBCDA, saePQRS, clpP) or biology (lexA, recF, yhcSR) (Fig. S6). The results of the Northern analyses indicated a specific absence and presumed degradation of most full-length antisense transcripts in the steady-state condition of the wild-type strain (Fig. 5), whereas the presence of discrete-size antisense transcripts was clearly detectable in the RNase III mutant for all genes tested. It is worth noting that these results confirm the existence of antisense transcripts for genes that have been thoroughly studied because of their impact on S. aureus virulence and antibiotic resistance. For some genes, these hybridizations showed that the RNA levels of the sense strand (lexA, clpP, saePQRS) increased in the RNase III mutant, suggesting that the absence of RNase III cleavage can slightly modulate the expression levels of sense transcripts (Fig. 5). To confirm that the presence of antisense RNA was restricted to those regions where short RNAs were detected, we selected two genes (SAOUHSC_00086 and SAOUHSC_00410) for which very few short RNAs were detectable in the wild-type strain (Fig. S7). In both cases, no transcript antisense to these genes was detectable in the RNase III mutant. Overall, these results uncover the existence of long antisense RNAs transcripts for most ORFs of the S. aureus genome. These long antisense transcripts are underrepresented in the wild-type strain because of the double-stranded RNase activity of RNase III.
Fig. 5.
Expression levels of sense/antisense transcripts. Northern blot analysis of RNA harvested from S. aureus 15981 wild-type and its corresponding S. aureus 15981 Δrnc. The blot was probed with a riboprobe specific for sense and antisense transcripts. The positions of RNA standards in kilobases are indicated. The time of exposure of the autoradiographies are indicated in hours (h) or days (d).
Analysis of Short RNA Complement Present in Diverse Bacterial Species.
To investigate whether this genome-wide sense/antisense overlapping transcript processing mechanism is specific to S. aureus or is active in other bacterial species, we characterized the short RNA complement present in three representative Gram-positive (E. faecalis, L. monocytogenes, and B. subtilis) and one Gram-negative (Salmonella enterica serovar Enteritidis) bacteria (Fig. 4E). Short RNA libraries were produced, sequenced, and mapped by using the described protocol (Fig. S1). Analysis of the distribution of short RNAs in sense and antisense strands of ORF regions revealed a highly significant correlation between quantities of short RNAs in sense/antisense strands for the three low-GC content Gram-positive bacteria, mirroring the observations in S. aureus. In contrast, the results obtained from the analysis of Salmonella demonstrated the absence of such a correlation, indicating the existence of a different processing pattern of overlapping RNA pairs in Gram-negative bacteria. Previous transcriptome analysis has allowed the identification of antisense transcripts in L. monocytogenes (5), B. subtilis (17), and E. faecalis (37). Analysis of the distribution of short RNAs in those regions with recognized antisense transcription confirmed the accumulation of high amounts of short RNA in every region with overlapping transcription, indicating that genome-wide digestion of overlapping sense/antisense transcripts is conserved at least in Gram-positive bacteria (Fig. S8).
Discussion
Development of RNA-seq technology is allowing the characterization of the multiple types of RNA molecules present in a living cell. The application of this technology in bacteria has primarily been restricted to the analysis of long RNA molecules because of the difficulty in removing highly abundant small-sized rRNA and tRNA molecules. Here, we have used two methods developed for microRNA analysis in eukaryotic cells to analyze the RNA fraction of <50 nt of the human pathogen S. aureus. The short RNA fraction was purified by size fractionation electrophoresis, and libraries for RNA-seq were generated by following a protocol that preserves the information about a transcript's direction developed for the direct cloning of microRNA in Drosophila (34).
The analysis of the distribution of short RNA molecules revealed several unexpected results. First, the sense strand of 2,268 ORFs and the antisense strand of 1,981 ORFs were covered with unique short RNA reads in at least 50% of their length, indicating the existence of antisense transcription from both strands of most ORFs in S. aureus genome. Second, similar numbers of short RNAs were mapping to sense and antisense strands of each ORF, irrespective of the transcription levels of the sense strand. Third, short RNA reads accumulated in higher numbers in regions with noticeable overlapping transcription between long RNA transcripts. The simplest interpretation for these observations was that short RNAs were products of the processing activity of a RNase on the double-stranded overlapping RNA transcripts. In support of this explanation, knockout of the rnc gene, which encodes for the only known double-stranded RNase (RNase III) contained in the S. aureus genome, caused a significant decrease in the number of short RNAs, the loss of the symmetric distribution of short RNAs in sense/antisense strands of each ORF, and the accumulation of long RNA molecules (Fig. 1) that, in the case of antisense transcripts, emerge as defined visible bands in Northern hybridizations. Other evidence supporting the hypothesis that short RNA molecules are generated by cleavage of overlapping RNA transcripts was obtained by the analysis of the short and long RNA fractions of the S. aureus ΔsigB mutant. Because the expression of some antisense transcripts requires the presence of the SigB transcription factor, the expression of these antisense transcripts decreases in the sigB mutant strain. The analysis of the distribution of short RNAs for those ORFs in which the expression of the antisense transcript is SigB-dependent revealed a decrease in the number of short RNAs that specifically mapped with the sense and antisense strands of these ORFs indicating that the levels of shorts RNAs is limited by the amount of double-stranded RNA. Notably, in most of these ORF regions, the decrease expression of the antisense RNA correlates with an increase in the expression level of the sense transcript, suggesting that RNase III-dependent sense/antisense cleavage process might serve to modulate the levels of the sense RNA.
Current RNA sequencing techniques needs microgram amounts of total RNA for analysis, which corresponds to millions of bacterial cells. The discovery of the existence of overlapping transcripts in a bacterial population does not mean that sense/antisense transcripts are simultaneously present in the same bacteria. Indeed, the transcriptome map will be identical if a subgroup in the bacterial population will synthesize the sense transcript and another subgroup will synthesize the antisense transcript. Our results imply that overlapping transcription occurs extensively in the same cell because RNase III can only produce short RNAs when both transcripts are present and emphasize that overlapping transcription plays a role in posttranscriptional regulation of RNA.
One question that emerges from these results concerns the biological role of the pervasive overlapping transcription and RNase III-mediated processing. At least two important biological consequences for this process are suggested from these results. First, antisense transcription and RNase III activity could provide a means for the removal of transcriptional noise. In this circumstance, RNase III would digest low-level expression of sense RNA transcription whose expression, if left unchecked, could unnecessarily compete with the processing and translation of required transcripts. When the transcription of the gene is increased in a regulated fashion, the sense RNA levels would exceed that of the antisense expression, leaving the unpaired sense transcripts impervious to RNase III activity and possible productive translation. Such a model predicts that the levels of expression of sense and antisense transcripts would be coordinated to achieve this threshold effect. Second, this mechanism would also permit the fine tuning of the sense transcript levels by adjusting the levels of antisense transcription to levels that allow for more or less final sense transcripts. By controlling which regions within multiple gene operons are subject to overlapping transcription, selection of which genes will be ultimately expressed at the protein level could be regulated. It is worth noting that implementation of this mechanism could be used to avoid simultaneous expression of 5′ or 3′ overlapping transcripts in the same cell.
Another interesting aspect raised by these results is the fate and functional role(s) of the stable short RNAs derived from the processing of overlapping long RNA transcripts. Although the answer to this question clearly requires additional studies, it is important to recall the processing of long RNA precursors into short microRNAs and into short interfering RNA molecules as guided by the eukaryotic RNase III-related ortholog enzymes and consider whether there are similar or related genome-wide regulatory mechanisms ongoing in eukaryotic cells.
Materials and Methods
The strains and oligonucleotides used in this study are listed in Table S1. Methods for bacterial growth, chromosomal gene deletion, RNA extraction, riboprobe synthesis, Northern blot assays, read mapping, and statistics analysis are described in detail in SI Materials and Methods. Short RNA libraries were prepared from an RNA fraction containing RNAs of <50 nt by adapting a described method (34). This fraction was obtained from total RNA with the flashPAGE fractionator (Ambion). Long RNA libraries were constructed by adapting the described procedure based on the incorporation of deoxy-UTP during the second strand cDNA synthesis (33). Detailed protocols for short and long RNA libraries construction are presented in SI Materials and Methods. Short and long RNA libraries were sequenced by using Illumina Genome Analyzer II at the Cold Spring Harbor Laboratory facilities.
Supplementary Material
Acknowledgments
We thank Pascale Romby for providing plasmid pLUG519; Philippe Batut, Carries Davis, and Johann Schlesinger for many fruitful discussions; and Juan Valcarcel for critical reading of the manuscript. During his sabbatical stay in Cold Spring Harbor Laboratory, I.L. was supported by a “Salvador Madariaga” fellowship from the Spanish Ministry of Science and Innovation. A.T.-A. and J.V. were supported by Spanish Ministry of Science and Innovation “Ramon y Cajal” contracts. M.V. was supported by a Consejo Superior de Investigaciones Científicas JAE Predoctoral research contract. This work was supported by Spanish Ministry of Science and Innovation Grants BIO2008-05284-C02-01 and ERA-NET Pathogenomics PIM2010EPA-00606.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Short Read Archive (accession no. SRP003288.1).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113521108/-/DCSupplemental.
References
- 1.Selinger DW, et al. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat Biotechnol. 2000;18:1262–1268. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- 2.Cho BK, et al. The transcription unit architecture of the Escherichia coli genome. Nat Biotechnol. 2009;27:1043–1049. doi: 10.1038/nbt.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Güell M, et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 4.Liu JM, et al. Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acids Res. 2009;37:e46. doi: 10.1093/nar/gkp080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toledo-Arana A, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 6.Mendoza-Vargas A, et al. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS ONE. 2009;4:e7526. doi: 10.1371/journal.pone.0007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 8.Filiatrault MJ, et al. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J Bacteriol. 2010;192:2359–2372. doi: 10.1128/JB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wurtzel O, et al. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–141. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaume M, et al. Cartography of methicillin-resistant S. aureus transcripts: Detection, orientation and temporal expression during growth phase and stress conditions. PLoS ONE. 2010;5:e10725. doi: 10.1371/journal.pone.0010725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jäger D, et al. Deep sequencing analysis of the Methanosarcina mazei Gö1 transcriptome in response to nitrogen availability. Proc Natl Acad Sci USA. 2009;106:21878–21882. doi: 10.1073/pnas.0909051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res. 2010;38:868–877. doi: 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin J, Zhu W, Passalacqua KD, Bergman N, Borodovsky M. Bacillus anthracis genome organization in light of whole transcriptome sequencing. BMC Bioinformatics. 2010;11(Suppl 3):S10. doi: 10.1186/1471-2105-11-S3-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georg J, et al. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol. 2009;5:305. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitschke J, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA. 2011;108:2124–2129. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dornenburg JE, Devita AM, Palumbo MJ, Wade JT. Widespread antisense transcription in Escherichia coli. MBio. 2010;1:e00024-10. doi: 10.1128/mBio.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasmussen S, Nielsen HB, Jarmer H. The transcriptionally active regions in the genome of Bacillus subtilis. Mol Microbiol. 2009;73:1043–1057. doi: 10.1111/j.1365-2958.2009.06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomason MK, Storz G. Bacterial antisense RNAs: How many are there, and what are they doing? Annu Rev Genet. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner EG, Flärdh K. Antisense RNAs everywhere? Trends Genet. 2002;18:223–226. doi: 10.1016/s0168-9525(02)02658-6. [DOI] [PubMed] [Google Scholar]
- 20.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorek R, Cossart P. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat Rev Genet. 2010;11:9–16. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- 23.Gripenland J, et al. RNAs: Regulators of bacterial virulence. Nat Rev Microbiol. 2010;8:857–866. doi: 10.1038/nrmicro2457. [DOI] [PubMed] [Google Scholar]
- 24.Toledo-Arana A, Solano C. Deciphering the physiological blueprint of a bacterial cell: revelations of unanticipated complexity in transcriptome and proteome. Bioessays. 2010;32:461–467. doi: 10.1002/bies.201000020. [DOI] [PubMed] [Google Scholar]
- 25.Klevens RM, et al. Active Bacterial Core Surveillance of the Emerging Infections Program Network Community-associated methicillin-resistant Staphylococcus aureus and healthcare risk factors. Emerg Infect Dis. 2006;12:1991–1993. doi: 10.3201/eid1212.060505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novick RP, et al. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boisset S, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pichon C, Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc Natl Acad Sci USA. 2005;102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geissmann T, et al. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009;37:7239–7257. doi: 10.1093/nar/gkp668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abu-Qatouseh LF, et al. Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. J Mol Med (Berl) 2010;88:565–575. doi: 10.1007/s00109-010-0597-2. [DOI] [PubMed] [Google Scholar]
- 31.Bohn C, et al. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010;38:6620–6636. doi: 10.1093/nar/gkq462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson KL, Dunman PM. Messenger RNA Turnover Processes in Escherichia coli, Bacillus subtilis, and Emerging Studies in Staphylococcus aureus. Int J Microbiol. 2009;2009:525491. doi: 10.1155/2009/525491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkhomchuk D, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Czech B, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huntzinger E, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fouquier d'Hérouel A, et al. A simple and efficient method to search for selected primary transcripts: Non-coding and antisense RNAs in the human pathogen Enterococcus faecalis. Nucleic Acids Res. 2011;39:e46. doi: 10.1093/nar/gkr012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.