Abstract
Prokaryotic DNA arrays arranged as clustered regularly interspaced short palindromic repeats (CRISPR), along with their associated proteins, provide prokaryotes with adaptive immunity by RNA-mediated targeting of alien DNA or RNA matching the sequences between the repeats. Here, we present a thorough screening system for the identification of bacterial proteins participating in immunity conferred by the Escherichia coli CRISPR system. We describe the identification of one such protein, high-temperature protein G (HtpG), a homolog of the eukaryotic chaperone heat-shock protein 90. We demonstrate that in the absence of htpG, the E. coli CRISPR system loses its suicidal activity against λ prophage and its ability to provide immunity from lysogenization. Transcomplementation of htpG restores CRISPR activity. We further show that inactivity of the CRISPR system attributable to htpG deficiency can be suppressed by expression of Cas3, a protein that is essential for its activity. Accordingly, we also find that the steady-state level of overexpressed Cas3 is significantly enhanced following HtpG expression. We conclude that HtpG is a newly identified positive modulator of the CRISPR system that is essential for maintaining functional levels of Cas3.
Keywords: defense mechanism, phage-host interactions, positive selection
The clustered regularly interspaced short palindromic repeats (CRISPR) system is a significant defense mechanism of prokaryotes against viruses and horizontally transferred DNA (1–3) and RNA (4). It is found in ∼90% of archaeal genomes and ∼40% of bacterial genomes, and consists of a CRISPR array, usually preceded by a leader DNA sequence, located near a cluster of CRISPR-associated (cas) genes (5–7). RNA transcribed from the CRISPR array (crRNA) is processed by Cas proteins and directs interfering proteins to target DNA/RNA matching the sequences between the repeats. These sequences, called spacers, often originate from plasmids and phages; thus, the system adaptively targets these invaders. High variability is found among bacterial species in the number, sequence, and length of the CRISPR arrays; the sequence and length of the leader DNA; and the number and sequence of the associated proteins. The CRISPR arrays are composed of 2 to ∼250 DNA repeats of ∼25 to ∼70 bp that flank similarly sized spacer sequences (8). The leader sequence is usually AT-rich and promotes transcription of the CRISPR array; it may also serve as a unique sequence that confers directional acquisition of new spacers into the arrays (1, 2, 9). The functions of most of the cas genes are not yet known, but many motifs, such as DNA-binding, RNA-binding, helicase, and nuclease domains, are present in their encoded proteins (10).
The cas genes have been mostly identified by bioinformatics. Systematic analysis of uncharacterized genes identified in various species in close proximity to the CRISPR arrays has led to the identification of many Cas protein families (11). One reported example of the experimental identification of a novel non-cas gene, which participates in the activity of the CRISPR system, is the rnc gene of Streptococcus pyogenes, encoding the enzyme RNase III (12). This enzyme is important for processing crRNA transcripts with the aid of an RNA molecule and the Cas protein, Cas9. H-NS and LeuO have been shown to regulate the CRISPR system in both Escherichia coli and Salmonella typhi, whereas LRP regulates the S. typhi CRISPR system (13–15). Other non-cas genes participating in the activity of the CRISPR system have not yet been identified in other bacteria. The identification of RNase III, as well as H-NS, LRP, and LeuO, suggests the existence of other non-cas gene products participating in the activity of the CRISPR system. Such gene products may have a direct role in CRISPR activity, such as DNA cleavage, crRNA processing, and mediating the encounter with incoming DNA, or they may have indirect supportive activities, such as proper folding of the involved proteins, energy supply for the system, regulation of the system, and bridging between protein machineries.
In this study, we established a system to search for genes whose products are required for activity of the E. coli CRISPR system. No such systematic search has ever been reported for any CRISPR system. The search is based on positive selection of E. coli colonies having disruptions in genes essential for activity of the CRISPR system. Our search coverage is thorough, consisting of 50,000 independent mutants covering over 10-fold the E. coli genome and identifying cas genes numerous times as essential for the CRISPR activity. We therefore believe that we have identified all the nonessential E. coli genes that are essential for the CRISPR activity. In particular, we identified high-temperature protein G (HtpG), a protein that is homologous to the eukaryotic chaperone heat-shock protein 90 (Hsp90) (16), as essential for maintaining functional levels of Cas3, a required protein for CRISPR interference with incoming DNA (3). A possible model for these interactions is discussed.
Results
Positive Selection System for E. coli Genes Required for a Functional CRISPR System.
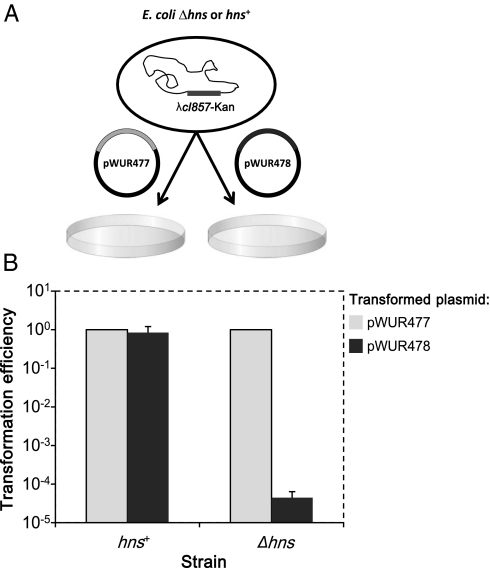
To identify genes that are essential to the activity of the CRISPR system, we established a genetic screen that positively selects E. coli mutants with an inactive CRISPR system. The selection principle is based on the suicidal activity of the CRISPR system when E. coli λ lysogens are transformed with a plasmid encoding spacers against the λ phage (17). In such a system, transposon mutagenesis can be applied to identify viable bacterial clones with transposon disruptions of genes essential for CRISPR activity. We selected E. coli strain BW25113Δhns, in which the CRISPR system is constitutively active (9, 14, 17, 18), for the genetic screening. This strain was lysogenized with the λ phage, making the bacteria incompatible, in a CRISPR-dependent manner, with the plasmid pWUR478 encoding spacers against the λ phage (3, 17). In addition, the strain was genetically engineered to carry an arabinose-inducible T7-RNA polymerase (RNAp) to enhance expression of the CRISPR spacers, encoded downstream of a T7-promoter in the pWUR478 plasmid. When this strain was transformed with plasmid pWUR478 encoding anti-λ spacers, the transformation efficiency, compared with that of plasmid pWUR477 encoding control spacers, was reduced over 10,000-fold (Fig. 1), indicating that the spacers against the λ phage kill the cells in a CRISPR-dependent manner. To demonstrate further that the CRISPR system is responsible for the reduced transformation efficiency, we used an isogenic control strain, with an intact hns gene encoding the H-NS protein that represses CRISPR-system activity in vivo (9, 14). This strain showed similar transformation efficiency with pWUR478 and the control plasmid (Fig. 1). We concluded that the apparent decrease in transformation efficiency of strain BW25113Δhns by pWUR478 is attributable to cell death as a result of CRISPR-system activity. This significant difference in transformation efficiency enabled us to establish a robust system for the positive selection procedure by transforming pWUR478 into transposon-mutagenized bacteria and examining gene disruptions in the viable colonies.
Fig. 1.
The E. coli CRISPR system specifically prevents establishment of plasmids encoding spacers against an integrated λ prophage. (A) Schematic representation of the transformation assay used throughout the study. The indicated bacteria, harboring a λ prophage, were transformed with the indicated plasmids encoding anti-λ (pWUR478) or control (pWUR477) spacers. Transformed bacteria were plated and counted after overnight incubation. (B) BW25113 λcI857-kan araB::T7-RNAp with (hns+) or without (Δhns) the hns gene were made competent and transformed with pWUR478 or pWUR477 as described in Materials and Methods. The relative efficiency of transformation was calculated by dividing the number of obtained pWUR478 or pWUR477 transformants by the number of pWUR477 transformants. Results are presented as the average ± SD of three independent experiments.
Identification of E. coli Genes Essential for CRISPR-System Activity.
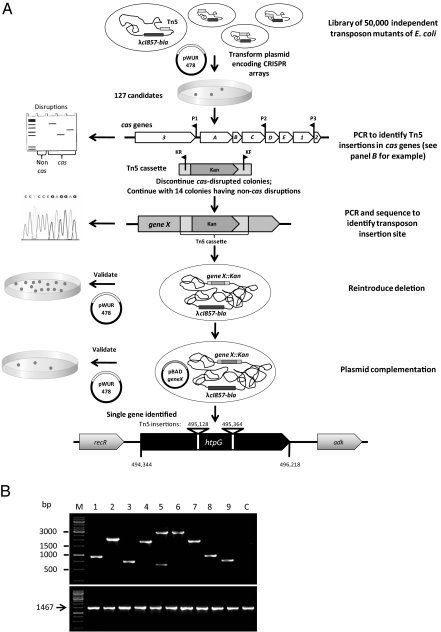
We next genetically engineered BW25113Δhns araB::T7-RNAp λcI857-bla, a strain similar to the above but with a λ prophage having a different resistance marker, to harbor transposon insertions at various genetic locations, and we followed the selection scheme depicted in Fig. 2A. The generated transposon library represented ∼50,000 independent events, providing over 10-fold coverage of the E. coli genome. This bacterial library was made competent, transformed with pWUR478 encoding anti-λ spacers, and selected on LB-agar plates supplemented with chloramphenicol, resistance to which is conferred by the plasmid. Bacteria able to harbor the plasmid were suspected of having an inactive CRISPR system because, as explained above, the lysogenic bacteria can only coexist with this plasmid in the absence of CRISPR activity. This procedure yielded 127 bacterial candidates that were further tested. We first eliminated transposition events in known CRISPR essential genes, the cas genes, from further analyses using multiplex PCR on these colonies. We used a mix of five oligonucleotides, three binding to the cas genes at 2- to 3.5-kb intervals, and two primers amplifying outward of the transposon (Fig. 2A). This procedure generates a PCR product when a transposon is inserted in one of the cas genes. Of the 127 candidates, 113 indeed harbored a transposon insertion in one of these genes, confirming the validity of the selection assay. A typical agarose gel showing PCR-amplified products from nine colonies selected for the ability to harbor pWUR478 is shown in Fig. 2B. Because cas1 and cas2 have been shown not to be required for CRISPR activity (3), and transposon insertion in cas1 cannot be excluded by the indicated PCR, we wanted to verify that, indeed, these genes do not contain any transposon insertions. As expected, PCR amplification of cas1-cas2 did not reveal any transposon insertion in these genes (Fig. 2B). These mutants were not examined further. The transposon insertion site was identified in the other 14 candidates by semirandom PCR amplification followed by sequencing, as described previously (19). Redundant candidates were eliminated from further analyses, and P1 transductions generating “clean deletions” were carried out for representatives of each disrupted gene. These reconstructed deletion mutants were then tested for transformation efficiency of pWUR478 compared with pWUR477, and only one showed significant restoration of transformation efficiency, suggesting that the CRISPR system is nonfunctional in the absence of this gene. Thus, the other originally isolated colonies probably carried secondary mutations (perhaps in the cas genes), or the transposon somehow modified the gene's activity such that it could not be reconstituted by deletions.
Fig. 2.
(A) Screening system for identifying E. coli genes required for CRISPR activity. A random transposon-insertion library, consisting of ∼50,000 cfu, was prepared on the genetic background of BW25113Δhns λcI857-bla araB::T7-RNAp as described in (31) and in SI Materials and Methods. Bacterial cells were transformed with pWUR478, encoding spacers against the λ prophage; 127 of the surviving colonies were subjected to colony-PCR assay to rule out transposon insertions in the cas genes, as follows: Three oligonucleotides (P1, P2, and P3) at 2- to 3.5-kb intervals in the cas genes were used for one direction, and two oligonucleotides, KF and KR, amplifying both ends of the transposon, were used for the other direction. Colonies showing DNA product of this PCR amplification presumably have transposon insertions in one of the cas genes and were not studied further. The Tn5-insertion site was identified in 14 colonies having disruptions in non-cas genes. The disrupted gene was reintroduced by transduction from the Keio collection, and the newly developed strain was retested for transformation efficiency of pWUR478. Positive clones were complemented by a plasmid encoding the disrupted gene. A single gene, htpG, was validated using the above procedure. This gene had two independent Tn5 insertions in 4 of the identified colonies. (B) PCR amplification of DNA template from 9 random colonies identified by the screening procedure. (Upper) Products from the multiplex PCR assay illustrated in A. The presence of amplified product(s) indicates that the transposon inserted at a distance equivalent to the product length from any of the oligonucleotides used. Note similar sizes of several of the products, suggesting either hot spots for transposon insertion, that sibling colonies harboring the same insertion were selected, or both. (Lower) PCR amplification of the region encompassing cas1-cas2. The expected size when no transposon is inserted is 1,467 bp, and this was the size observed in all the tested colonies, indicating no transposon insertion in these genes. Colonies are numbered 1–9. M, marker; C, control (a random transposon mutant selected for in a control reaction transformed with pWUR477).
Validating the Essentiality of the Identified Genes to CRISPR-System Functionality.
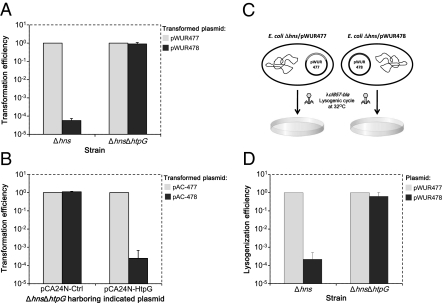
The deleted gene showing consistent restoration of transformation efficiency represented four selected colonies having disruptions in htpG, two at position 495,128 and two at position 495,364 (Fig. 2). As described above, to verify that disruption of htpG is responsible for the lack of CRISPR activity, and that there are no secondary mutations causing this effect, we reconstructed a deletion of htpG in BW25113Δhns araB::T7-RNAp by P1 transduction of the htpG::kan cassette (20), and after removing the kan cassette, we lysogenized these cells with λ-cI857-kan, yielding BW25113Δhns ΔhtpG araB::T7-RNAp λ-cI857-kan. The newly constructed strain was tested for transformation efficiency of pWUR478 compared with pWUR477. As shown in Fig. 3A, the transformation efficiency of pWUR478, encoding anti-λ spacers, into the htpG deletion mutant was comparable to that of pWUR477, encoding control spacers, indicating that the CRISPR system does not function in the absence of htpG. To verify further the essentiality of htpG to CRISPR interference and to exclude, for example, polar effects of htpG deletion on other genes, we complemented the ΔhtpG mutant with a plasmid encoding htpG under an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter. As shown in Fig. 3B, transformation efficiency of pAC-478, encoding anti-λ spacers, into the ΔhtpG mutant harboring the HtpG-encoding plasmid was over 1,000-fold lower than that with the control pAC-477, or in comparison to pAC-478 transformation into an ΔhtpG mutant harboring a control vector. These results indicated that expression of HtpG from a plasmid can restore CRISPR activity to ΔhtpG cells, unequivocally confirming that htpG is essential for CRISPR-system activity.
Fig. 3.
htpG is essential for activity of the CRISPR system. (A) “Clean” htpG deletion was introduced into BW25113Δhns T7-RNAp, and cells were then lysogenized with λcI857-kan. BW25113Δhns ΔhtpG λcI857-kan T7-RNAp was made competent and transformed with plasmids encoding spacers against the λ prophage (pWUR478) or control spacers (pWUR477) as described in Materials and Methods. The relative efficiency of transformation was calculated by dividing the number of obtained pWUR478 or pWUR477 transformants by the number of pWUR477 transformants. Results are presented as the average ± SD of three independent experiments. (B) Plasmid encoding htpG (pCA24N-HtpG) or control vector (pCA24N-Ctrl) was transformed into BW25113Δhns ΔhtpG λcI857-kan T7-RNAp. These cells were then transformed with plasmids encoding spacers against the λ prophage (pAC-478) or control spacers (pAC-477) as described in Materials and Methods. The relative efficiency of transformation was calculated by dividing the number of obtained pAC-478 or pAC-477 transformants by the number of pAC-477 transformants. Results are presented as the average ± SD of three independent experiments. (C) Schematic diagram of the assay used to test CRISPR protection from lysogenization. (D) BW25113Δhns or BW25113Δhns ΔhtpG transformed with either pWUR477 or pWUR478 was lysogenized at 32 °C with λcI857-bla, conferring ampicillin resistance. Bacteria were then serially diluted and plated on LB-agar plates supplemented with ampicillin. Lysogenization efficiency was calculated by dividing the number of lysogenized bacteria in a given plate by the number of lysogenized bacteria obtained in its respective parental bacteria harboring the pWUR477 plasmid. Results are presented as the average ± SD of three independent experiments.
We have previously shown that the CRISPR system is active against the establishment of lysogens (17). We wished to demonstrate that deficiency of htpG prevents this activity against lysogenization. We therefore transformed pWUR478 or pWUR477 into E. coli BW25113Δhns ΔhtpG araB::T7-RNAp or into BW25113Δhns araB::T7-RNAp as a control. These strains were tested for lysogenization efficiency by infecting them with λcI857-bla phage at 32 °C, conditions favoring the lysogenic cycle, and then determining the number of colonies acquiring antibiotic resistance encoded by the phage (Fig. 3C). In this assay as well, lack of htpG significantly decreased the CRISPR-dependent protection of the bacteria from lysogenization by over 1,000-fold compared with the isogenic strain carrying htpG (Fig. 3D). These results indicate that htpG is also essential for the E. coli CRISPR system's prevention of phage lysogenization.
Expression of Cas3 Restores CRISPR Activity in the ΔhtpG Mutant.
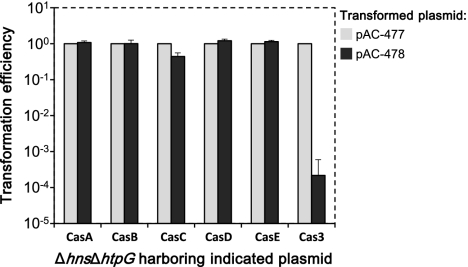
It is important to note that in both the lysogenization and transformation-efficiency assays described above for E. coli ΔhtpG, the size of the colonies harboring pWUR478 was smaller than that of those harboring pWUR477. These results led us to assume that CRISPR activity is not completely silent in the absence of htpG and that HtpG somehow modulates the CRISPR system's functionality. HtpG is the bacterial homolog of the eukaryotic Hsp90 protein, which is a well-characterized chaperone (16). Because we established that HtpG is essential for CRISPR activity, we speculated that one or more of the Cas proteins are client proteins of this chaperone. Consequently, if HtpG is indeed essential for the accumulation of an active Cas protein, htpG deficiency should be suppressed by expression of this client protein in higher amounts than its endogenous expression. This speculation assumes that htpG deficiency results in a limiting amount of an active Cas protein and that this might be overcome by overexpressing this Cas protein from a plasmid. To test this possibility and to identify the putative client protein, we transformed plasmids from the ASKA library (21) encoding each of the six proteins known to be essential for CRISPR interference into E. coli BW25113Δhns ΔhtpG araB::T7-RNAp. We then measured transformation efficiencies with a plasmid encoding anti-λ spacers, pAC-478, compared with a control plasmid, pAC-477. As shown in Fig. 4, expression of Cas3 was able to relieve the dependency of CRISPR activity on HtpG. In the absence of HtpG, increased expression of Cas3, but not of any other Cas protein, fully restored the CRISPR system's ability to decrease the transformation efficiency of pAC-478 by over 1,000-fold. These results indicate that sufficient expression of Cas3 can overcome the absence of HtpG, suggesting that under htpG deficiency, Cas3 protein is the only Cas component whose level is limiting for CRISPR activity. The results suggest that HtpG either helps in recruiting Cas3 protein to the CRISPR interference stage or is involved in this protein's folding and/or in preventing its aggregation, thereby maintaining functional levels of Cas3.
Fig. 4.
Expression of Cas3, but not of other Cas proteins, restores CRISPR activity in a ΔhtpG strain. BW25113Δhns ΔhtpG λcI857-kan araB::T7-RNAp was transformed with the indicated pCA24N-based plasmids, containing the indicated genes under the lac promoter (19). Cells were made competent and transformed with plasmids encoding spacers against the λ prophage (pAC-478) or control spacers (pAC-477) as described in Materials and Methods. The relative efficiency of transformation was calculated by dividing the number of obtained pAC-478 or pAC-477 transformants by the number of pAC-477 transformants. Results are presented as the average ± SD of three independent experiments.
HtpG Expression Significantly Increases Levels of Overexpressed Cas3.
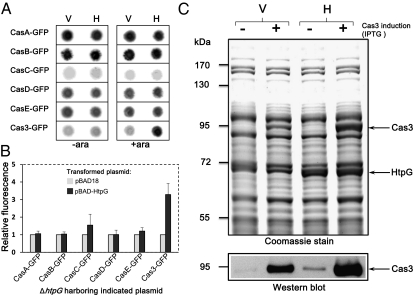
Finally, if HtpG is indeed essential for maintaining functional levels of Cas3, its expression should support higher accumulation of Cas3. We first tested this possibility in vivo by transforming cas3 fused to GFP into E. coli BW25113ΔhtpG with either a plasmid encoding htpG or a control plasmid. As negative controls, we transformed the other cas genes required for CRISPR interference, fused to GFP, with either a plasmid encoding HtpG or a control plasmid. These bacteria were grown overnight on LB-agar plates supplemented with appropriate antibiotics, with or without l-arabinose for HtpG induction. To detect the level of fluorescence proportional to the levels of the GFP-fused proteins, plates were imaged and quantified as described in Materials and Methods. The measured fluorescence intensities indicated that the level of Cas3 increases over threefold when HtpG is expressed (Fig. 5 A and B). The increase in Cas3 level was only observed when HtpG expression was induced by l-arabinose, further validating that the increase in Cas3 levels is HtpG-dependent. The levels of the other tested Cas-GFP proteins were unaffected by the expression of HtpG, indicating that, of all the known CRISPR-effector proteins, HtpG specifically increases the steady-state level of Cas3-GFP in the bacterial cells. To confirm this in vivo observation further, we used Cas3 without GFP. Cas3 was overexpressed from a plasmid in BW25113ΔhtpG with or without HtpG, and its protein level was analyzed. As shown by SDS-gel electrophoresis of total cell extracts, coexpression of HtpG and Cas3 resulted, in this case as well, in greater than a threefold increase in the level of Cas3 compared with its level without HtpG. The identity of Cas3 in the gel was confirmed by Western blot analysis against the His-tag preceding this protein (Fig. 5C). These findings confirmed that HtpG increases the steady-state level of Cas3. Taken together, our results show that HtpG's essentiality to CRISPR-system activity is mediated by its effect on Cas3.
Fig. 5.
Cas3 steady-state level increases with coexpression of HtpG. (A) BW25113ΔhtpG was cotransformed with either pBAD18 (V) or pBAD-HtpG (H) along with pCA24N-based plasmids encoding the indicated protein fused to GFP. Bacteria were replicated on agar plates with the appropriate antibiotics, with (+ara) or without (−ara) 0.2% l-arabinose, for induction of HtpG expression as described in Materials and Methods. The plates were scanned for GFP fluorescence using a Typhoon 9400. A representative image from three independent experiments with similar results is shown. (B) Results of the l-arabinose–induced samples were quantified by ImageJ-based densitometry. Results are the average ± SD of three independent experiments. (C) BW25113ΔhtpG was transformed with either pBAD18 (V) or pBAD-HtpG (H) along with pCA24N-Cas3 (no GFP, from ASKA library). Cell extracts were prepared as described in Materials and Methods, and 20 μg of total protein was loaded on a 10% wt/vol polyacrylamide gel. (Upper) Protein separation by gel electrophoresis was carried out, followed by Coomassie blue staining and scanning. The gel is representative of three independent experiments showing similar results. (Lower) Western blot analysis using an antibody against the His-tag preceding Cas3.
Discussion
We carried out a thorough experimental search for non-cas genes essential for the activity of the E. coli CRISPR system. This search yielded the htpG gene, which was confirmed to be essential for the suicidal activity of plasmid-encoded spacers against λ prophage, as well as for protection against lysogenization by λ phage.
For the CRISPR system to interfere efficiently with invading DNA, it must go through most, if not all, of the following steps: (i) recognize alien DNA entry, (ii) induce transcription of cas genes, (iii) correctly fold or modify the translated Cas proteins, and (iv) process the crRNA and mediate binding of the processed crRNA-Cas proteins to the invading DNA/RNA, resulting in (v) cleavage and destruction of the target DNA/RNA. Some of these steps, such as crRNA maturation by the CasABCDE proteins in the E. coli system, are fairly well understood (3), whereas others, such as sensing DNA entry, are poorly understood (22). Our screen to identify genes participating in the above steps contributes an additional piece of information to one of the interference steps in E. coli by establishing an important role for HtpG in maintaining levels of Cas3, a crucial component in CRISPR interference.
HtpG, a homolog of the ubiquitous eukaryotic Hsp90, is a protein that has been conserved from bacteria to humans (23), suggesting it might play a role in prokaryotic CRISPR systems other than the E. coli system as well. The precise role of the E. coli HtpG has remained elusive, compared with our detailed knowledge of its eukaryotic homolog. It is ubiquitous in E. coli, and its level is elevated following heat stress (hence, its name). HtpG-deletion mutants suffer from slow growth at high temperatures, as well as slight protein aggregation (24). HtpG has been shown to be involved with two client proteins, the ribosomal protein L2 and a linker polypeptide in the phycobilisome of Synechococcus elongatus, in an activity that characterizes it as a molecular chaperone (25–27). Recently, HtpG has also been shown to promote reactivation of heat-denatured luciferase in a reaction that requires the activity of another E. coli chaperone, DnaK (26).
We demonstrate that overexpression of Cas3 can relieve the dependency of CRISPR-system activity on HtpG, indicating that Cas3 is a limiting factor for this activity in the absence of HtpG. Furthermore, we show that coexpression of HtpG with Cas3 increases steady-state levels of the latter. Our results are not the first to suggest that HtpG is involved in stabilizing large protein complexes, and they are in line with a recent study showing that HtpG is required to stabilize the S. elongatus phycobilisome, a large protein complex, by specifically interacting with some of its components (27). Thus, in addition to the unique role of HtpG in the activity of the CRISPR system, we shed further light on its role as a chaperone by identifying its stabilization of another putative client protein.
Analyses of bacterial databases to identify a direct link between co-occurrence of Cas3 and HtpG are complicated by the lack of a straightforward way of identifying functional vs. nonfunctional CRISPR systems, because such co-occurrence is predicted based on the functionality of both proteins together. Nevertheless, basic analysis of bacterial databases using EcoCyc (28) indicates that of 33 E. coli strains, only 2 lack an htpG ortholog, and these 2 also lack cas3. All 7 examined Salmonella strains carry htpG, and 5 of them also encode cas3. Furthermore, all examined Mycoplasma and Staphylococcus strains and most of the Streptococcus strains lack both htpG and cas3. On the other hand, all the examined Lactobacillus strains encode cas3 but lack htpG. In addition, ∼50% of the archaea encode cas3 (29), but almost all of them lack htpG (30). This basic analysis indicates that co-occurrence of HtpG and Cas3 is not easily predictable and that interactions between these two proteins must be experimentally tested in each strain. Lack of co-occurrence between these two proteins in all systems is not surprising because Cas3 has low sequence similarity among different strains, suggesting different modes of action in different systems. It is tempting to speculate that in organisms lacking HtpG, such as the archaea, another non-Cas protein interacts with Cas3. The complexity of interactions of non-Cas proteins with the CRISPR system components is also evidenced by LRP, which regulates the Salmonella but not the E. coli CRISPR system, whereas other proteins (H-NS and LeuO) have similar regulatory functions in these strains.
Although thorough, genetic searches based on transposon insertions, such as those performed here, have some limitations. For instance, they cannot be relied on to identify genes that are essential for both the function of the CRISPR system and the viability of the transposed E. coli strain, because these genes, by definition, cannot be fully disrupted but only partially mutated in nonessential residues. Nevertheless, the experimental details provided herein may serve as a basis for a similar search for essential E. coli non-cas genes that are required for CRISPR-system activity. Such searches might make use of other mutagenesis methods generating, for example, point mutations. It is reasonable to assume that some essential E. coli genes also participate in the activity of the CRISPR system, and such searches may therefore prove valuable.
To summarize, we identified HtpG as a non-Cas protein involved in CRISPR activity using a unique screening method tailored to the CRISPR system. In future studies, we will refine and modify our screening method to identify additional regulatory proteins in this system. We believe that regulatory genes thus identified and characterized will shed more light on the regulation of this intriguing system.
Materials and Methods
Reagents, Strains, Plasmids, and Construction Procedures.
Reagents, strains, plasmids, and construction of strains and plasmids are detailed in SI Materials and Methods.
CRISPR-Dependent Restriction of Transformation.
E. coli BW25113Δhns araB::T7-RNAp λ-cI857-kan was diluted 1:50 from an overnight culture and aerated at 32 °C in LB medium containing 25 μg/mL kanamycin to an OD600 of 0.4–0.6. Bacteria were then centrifuged at 4,000 × g at 4 °C, the supernatant was disposed of, and the bacteria were resuspended in 1 mL of ice-cold double-distilled water (DDW) and transferred to a 1.5-mL Eppendorf tube. The cells were spun down for 1 min at 13,000 × g at 4 °C. After an additional washing step, the cells were suspended in 150 μL of ice-cold DDW.
Cells (50 μL) were then mixed in an ice-cold 0.2-mm electroporation cuvette (Bio-Rad) with 20 ng of pWUR478 or pWUR477. The mixture was pulsed in a Bio-Rad micropulser at 200 Ω, 25 μF, 1.8 kV. 2YT broth (1.6% wt/vol tryptone, 1.0% yeast extract, 0.5% NaCl) (0.2 mL) containing 0.2% l-arabinose and 0.1 mM IPTG was immediately added, and the cells were aerated for 1 h at 32 °C. Various dilutions were plated on LB-agar plates supplemented with 25 μg/mL kanamycin, 35 μg/mL chloramphenicol, 0.2% l-arabinose, and 0.1 mM IPTG. Plates were incubated overnight at 32 °C. Colonies emerging on selection plates were counted. Efficiency of transformation of pWUR478 plasmid was compared with that of the control pWUR477 plasmid. In several cases, the assay was carried out using specific deletion mutants (e.g., ΔhtpG) complemented with various plasmids (e.g., a plasmid encoding htpG or each of the cas genes), as indicated. When resistance was incompatible with the other plasmids, a different set of plasmids, pAC-477 and pAC-478 harboring similar spacers but with an ampicillin rather than chloramphenicol resistance marker, was used as indicated. In experiments in which cas genes were supplied on plasmids, no IPTG was added because expression also occurs without the inducer (21).
CRISPR-Dependent Restriction of Lysogenization.
Overnight cultures of E. coli Δhns araB::T7-RNAp harboring pWUR478 or pWUR477 were diluted 1:25 in LB medium containing 35 μg/mL chloramphenicol, 10 mM MgSO4, and 0.2% maltose. When the culture reached an OD600 of 0.6–0.8, 100 μL was mixed with 10 μL of phage λ carrying a kanamycin resistance gene in a 1.5-mL tube and incubated at room temperature for 20 min. Bacteria were then inoculated on LB-agar plates supplemented with 0.2% maltose, 0.1 mM IPTG, 10 μg/mL tetracycline, 35 μg/mL chloramphenicol, and 50 μg/mL kanamycin, and incubated overnight at 32 °C. Lysogenization assays of E. coli BW25113Δhns ΔhtpG were carried out at 10-fold larger cell and phage volumes because of low lysogenization efficiency of this strain compared with BW25113Δhns. Nevertheless, absence of htpG does not, by itself, reduce lysogenization efficiency by λ (relative efficiency of lysogenization of BW25113ΔhtpG compared with BW25113 at a multiplicity of infection of ∼1 is 1.25 ± 0.08), indicating that HtpG does not specifically affect λ lysogenization (because of effects on, e.g., the temperature-sensitive CI857).
Quantification of Cas Proteins Following Coexpression with HtpG.
To examine the steady-state level of Cas3 following HtpG overexpression, BW25113ΔhtpG and either pBAD18 (control) or pBAD-HtpG, along with pCA24N-Cas3-GFP [ASKA library (21)] or other specified control plasmids from the ASKA library, were cotransformed into E. coli. Cells were grown overnight in LB supplemented with the appropriate antibiotics at 37 °C. Aliquots of 0.1 mL were transferred into a 96-well microtiter plate, and the bacteria were then replicated on an agar plate with the appropriate antibiotics, with or without 0.2% l-arabinose for induction of HtpG expression. The pCA24N-based plasmids contain a leaky lac promoter, and there is a sufficient amount of GFP for monitoring purposes, even without IPTG induction (21). After overnight growth on plates, the replicated bacteria were scanned for GFP fluorescence using a Typhoon 9400 (GE Healthcare) (488-nm excitation wavelength, 520-nm emission wavelength). The presented image was digitally optimized for background omission and uniform expression levels. Results were quantified using an ImageJ-based (National Institutes of Health) densitometer. To analyze protein quantities by gel electrophoresis, E. coli BW25113ΔhtpG was transformed with either pBAD18 or pBAD-HtpG along with pCA24N-Cas3 (no GFP, from ASKA library). Cells were grown to the logarithmic phase and induced with 0.2% l-arabinose. After 20 min of induction, the cultures were divided and IPTG was added to half of the sample. After an additional 1.5 h of growth, the cells were transferred to ice. Cell pellets (∼5 × 109 cells) were suspended in 1 mL of buffer A [1 mM EDTA, 50 mM Tris⋅HCl (pH 8), 100 mM NaCl) containing protease inhibitor mixture (Roche). Protein extracts were prepared by three cycles of brief 10-s sonication at 20-s intervals on ice. Equal amounts of total protein (20 μg) were separated on a 10% wt/vol polyacrylamide gel and stained with Coomassie blue. Western blotting was carried out using anti–His-tag antibody (Sigma) according to the manufacturer's instructions.
Supplementary Material
Acknowledgments
We thank John van der Oost and Stan Brouns for providing the plasmids encoding the CRISPR spacers, William Metcalf for providing the plasmid for generation of transposon insertions, David Ze'evi for technical help, and Lynn C. Thomason for providing the λcI857-kan phage. We also thank Nir Osherov for critical reading of the manuscript and Camille Vainstein for professional language editing. This research was supported by the Israel Science Foundation (Grant 611/10), the Binational Science Foundation (Grant 2009218), the German-Israel Foundation (Grant 2061/2009), and Marie Curie International Reintegration Grants (PIRG-GA-2009-256340 and GA-2010-266717).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113519108/-/DCSupplemental.
References
- 1.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 3.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 6.Sorek R, Kunin V, Hugenholtz P. CRISPR—A widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 7.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11(3):181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heidelberg JF, Nelson WC, Schoenfeld T, Bhaya D. Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes. PLoS ONE. 2009;4:e4169. doi: 10.1371/journal.pone.0004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pougach K, et al. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol Microbiol. 2010;77:1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLOS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medina-Aparicio L, et al. The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi. J Bacteriol. 2011;193:2396–2407. doi: 10.1128/JB.01480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pul U, et al. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol. 2010;75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 15.Westra ER, et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol Microbiol. 2010;77:1380–1393. doi: 10.1111/j.1365-2958.2010.07315.x. [DOI] [PubMed] [Google Scholar]
- 16.Bardwell JC, Craig EA. Eukaryotic Mr 83,000 heat shock protein has a homologue in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:5177–5181. doi: 10.1073/pnas.84.15.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar R, Qimron U. The Escherichia coli CRISPR system protects from λ lysogenization, lysogens, and prophage induction. J Bacteriol. 2010;192:6291–6294. doi: 10.1128/JB.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker CB, et al. The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J Bacteriol. 2009;191:5793–5801. doi: 10.1128/JB.00356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006–0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Rodriguez R, et al. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol Microbiol. 2011;79:584–599. doi: 10.1111/j.1365-2958.2010.07482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta RS. Phylogenetic analysis of the 90 kD heat shock family of protein sequences and an examination of the relationship among animals, plants, and fungi species. Mol Biol Evol. 1995;12:1063–1073. doi: 10.1093/oxfordjournals.molbev.a040281. [DOI] [PubMed] [Google Scholar]
- 24.Bardwell JC, Craig EA. Ancient heat shock gene is dispensable. J Bacteriol. 1988;170:2977–2983. doi: 10.1128/jb.170.7.2977-2983.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motojima-Miyazaki Y, Yoshida M, Motojima F. Ribosomal protein L2 associates with E. coli HtpG and activates its ATPase activity. Biochem Biophys Res Commun. 2010;400:241–245. doi: 10.1016/j.bbrc.2010.08.047. [DOI] [PubMed] [Google Scholar]
- 26.Genest O, Hoskins JR, Camberg JL, Doyle SM, Wickner S. Heat shock protein 90 from Escherichia coli collaborates with the DnaK chaperone system in client protein remodeling. Proc Natl Acad Sci USA. 2011;108:8206–8211. doi: 10.1073/pnas.1104703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato T, Minagawa S, Kojima E, Okamoto N, Nakamoto H. HtpG, the prokaryotic homologue of Hsp90, stabilizes a phycobilisome protein in the cyanobacterium Synechococcus elongatus PCC 7942. Mol Microbiol. 2010;76:576–589. doi: 10.1111/j.1365-2958.2010.07139.x. [DOI] [PubMed] [Google Scholar]
- 28.Keseler IM, et al. EcoCyc: A comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39(Database issue):D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen B, Zhong D, Monteiro A. Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. BMC Genomics. 2006;7:156. doi: 10.1186/1471-2164-7-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen RA, Wilson MM, Guss AM, Metcalf WW. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol. 2002;178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.