Abstract
Targeting chromatin and its basic components through epigenetic drug therapy has become an increased focus in the treatment of complex diseases. This boost calls for the implementation of high-throughput cell-based assays that exploit the increasing knowledge about epigenetic mechanisms and their interventions for genotoxicity testing of epigenetic drugs. 3D quantitative DNA methylation imaging is a novel approach for detecting drug-induced DNA demethylation and concurrent heterochromatin decondensation/reorganization in cells through the analysis of differential nuclear distribution patterns of methylcytosine and gDNA visualized by fluorescence and processed by machine-learning algorithms. Utilizing 3D DNA methylation patterns is a powerful precursor to a series of fully automatable assays that employ chromatin structure and higher organization as novel pharmacodynamic biomarkers for various epigenetic drug actions.
Keywords: cell-based assay, DNA methylation imaging, epigenetic drug, heterochromatin, nuclear topology
Therapeutic targeting of chromatin
Chromatin conformation is an important factor in the global orchestration as well the locus-specific regulation of gene expression in the human cell nucleus [1,2]. The primary structure of chromatin consists of the basic repeat units – ~147 bp of dsDNA wrapped around a set of core histones, known as the nucleo-some – connected to a stretch of histone-free linker DNA (~80 bp), which form the so-called beads-on-a-string fiber [3,4]. This structure is then folded through several iterations into higher order chromatin [5]. Disruption of epigenetic processes such as DNA methylation, histone modifications and the metabolism of ncRNAs that are involved in the dynamic structure and function of this large polymer and its highly packed structure in the nucleus, is associated with many complex human diseases, including several types of cancer [6–8]. Pharmacologic targeting of chromatin and its basic components has become one of the major strategies in the treatment of cancer and other multifactorial traits such as metabolic, cardiovascular, autoimmune, neurodegenerative and behavioral disorders, and will likely expand into other pathologies [9]. Chromatin modifications and related signaling are highly complex, making the identification of more specific compounds challenging. Nevertheless, a steady increase in the development of epigenetic therapies for several human diseases can be perceived. In this respect, oncology constitutes the application area, in which currently the highest volume of activity is experienced. Cancer therapy is envisioned to gain the highest benefit from chromatin targeting drugs, specifically agents that could restore a normal DNA methylation equilibrium in cancerous cells.
Aberrant methylation of cytosine in the human genome is a hallmark of many cancers. The often coexisting alterations fall into two categories: abnormally high levels of methylation of a few percent (on average) of gene-specific promoters mostly in gene-rich genomic regions termed CpG-islands that cause gene silencing; and genome-wide hypomethylation, a large portion of which occurs in repetitive elements [10–12]. Relevant concepts in oncological therapies involve the use of inhibitors of DNA methylation and histone modifications. DNA methyltransferase inhibitors (DNMTi) and their demethylating effect on aberrantly hypermethylated gene promoters in cancer cells were discovered more than two decades ago [13,14]. Several DNA methylation inhibitors of different categories have been designed. Among these two DNMTi, 5-azacytidine (5-AZA; vidaza) and its analogue 5-aza-2-deoxycytidine (decitabine), have been approved for treatment of patients with myelodysplatic syndrome and hematologic malignancies [15–17]. Unfortunately, most of these agents turned out to be cytotoxic and genotoxic by demethylating the genome in a global fashion that can cause unwanted hypomethylation of oncogene promoters and the heterochromatin with its residing repetitive elements such as satellite DNAs and retrotransposons. These three classes of DNA are suppressed by methylation in normal cells, and have been found to be hypomethylated in transformed cells. Therefore treatment with demethylating agents bear an even stronger threat of genome instability to malignant cells that already harbor methylation-compromised genomic regions [18]. Possible adverse effects may comprise of transcriptional activation of oncogenes, imbalances in the copy numbers of satellite DNAs, translocation of latent retrotransposons such as long interspersed nuclear elements (LINEs) and short interspersed elements that can induce mutations, and telomere elongation of chromosomes [19–23]. Newer second-generation DNMTi are in development that should be more sequence-specific, including small molecules that are envisioned to be significantly less toxic [24]. On the other hand two histone deacetylase inhibitors (HDACi), suberoylanilide hydroxamic acid (vorinostat) and romidepsin (FK228), have been approved for treatment of patients with rare cutaneous T cell lymphoma and also other hematological malignancies; in addition, numerous other HDACi are undergoing preclinical testing as single agents as well as in combination with DNMTi for cancer therapy [25–27]. Lines of results have shown that combinations of these two classes of drugs exert synergistic effects on gene expression and tumor growth [28–30]. Nevertheless, the bilateral relationship between DNA methylation and chromatin modifications has an important implication in toxicology [31]. Even though HDACi directly influence histone modification, it has been shown in vitro that they can ultimately lead to DNA demethylation [32–34]. The antidepressant and antiepileptic drug valproic acid exemplifies this phenomenon. These effects have raised concerns as these classes of drugs may exhibit immediate genetic toxicity but also may lead to long-term chronic effects on the genome through compounding, as the reversible nature of epigenetic corrections may require longer if not lifetime exposure (substance intake) to these types of chemicals for maintaining epigenetic homeostasis. Other classes of chromatin modifying enzymes such as histone methyltransferases, histone demethylases, and sirtuins together with DNA demethylases – a novel class of enzymes that has gained increased attention because of its property to actively demethylate CG-dinucleotides – are also under investigation for their impact on DNA methylation and chromatin remodeling [13,24,35]. The fact that these chemicals exert an enormous complexity of actions, it is becoming increasingly important to consider their impact on the druggable genome and to define new end points in exploratory and regulatory testing of the currently applied drugs as well as the new generation of compounds in development. Adverse drug reactions (ADRs) are a major problem in drug therapy and drug development. The potential influence of epigenetics on adverse drug response can be divided into three categories: environmental factors that influence pharmacokinetic aspects generally defined as absorption, distribution, metabolism and excretion, which affect the disposition of a pharmaceutical compound in a cellular system potentially leading to ADRs; the drug, although having a conventional target, also affects the epigenome and thereby increases the probability of ADRs; and the drug has direct epigenetic targets and may thereby increase the risk for ADRs [36]. Although all three categories can be correlated with chromatin conformational changes and chromatin reorganization, this article focuses on the latter mode of action.
Cell-based assays in epigenetic drug discovery
Testing of epigenetic leads and compounds in the preclinical phases of drug development aim at determining drug action and efficacy as well as cytotoxicity and genetic toxicity using cell-based screening and in vitro assays. Currently, end points for measuring drug action and efficacy are: target abundance – in this case gDNA methylation and histone modifications or site-specific degree of promoter CG-dinucleotide methylation and locus-specific histone variation – and conversion rate/enzymatic activity of DNA methyltransferases and the so called chromatin modifiers. For that, there is a host of in vitro assays already commercially available [37]. Although concepts have been designed and are being pharmacologically pursued that encompass the entire known range of epigenetic targets and mechanisms, the epigenetic research and therapy field is being dominated by DNA methylation studies and manipulation. In light of this fact, the development of relevant quantitative assays towards holistic differential DNA methylation profiling was mainly pioneered by utilizing 2D gel electrophoresis. However, the era of epigenomics gained a strong momentum when microarray techniques – originally developed for gene-expression profiling and DNA copy number analysis – were recruited in the study of DNA methylation and histone modification patterns [38]. The current paradigm shift in sequencing technology has recently enabled single base-pair resolution of whole-genomic DNA methylation analysis in mammalian genomes [39–41], a principle that has begun to also significantly impact drug discovery and development. This shift became possible after the introduction of bisulfite treatment, which only converts unmethylated cytosine to uracil in extracted DNA for rapid and precise methylation detection by a plethora of downstream procedures; also in combination with chromatin immunoprecipitation, a method to determine the location of binding sites on the genome for chromatin-associated proteins of interest [42–44]. These techniques, despite currently being challenged by single-cell analysis, cost, time and labor can support the drug-discovery and -development pipeline to identify drug targets and mechanisms, as well as epigenetic side-effects. Developments toward nanoscaling of DNA sequencing and miniaturization of parallel biochemical processing of cell extracts may render this method economically more attractive for high-throughput drug testing in the future.
In preclinical in vitro drug testing adverse side effects are defined as cytotoxicity and genetic toxicity. Although in some therapeutic areas, such as cancer treatment, cytotoxicity still constitutes a primary strategy in eliminating fast growing tumor cells by inducing apoptosis and/or necrosis. Epigenetic treatment is shifting therapeutic objectives by reprogramming aberrant cells towards normal phenotypes; including controlled cell proliferation without mass cell eradication. Therefore, cytotoxicity is considered more of an unwanted effect in this context. Classically speaking, a compound or treatment is considered to be cytotoxic if it prevents cellular attachment, causes dramatic morphological changes, adversely affects replication rate or leads to a reduction in overall viability. Assessing cell membrane integrity is one of the most common ways to measure cell viability and cytotoxic effects using several classic vital dye inclusion, exclusion and lysosomal accumulation techniques. Cytotoxicity can also be monitored using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide or 3-(4,5-dimethylthi azol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium assay, which measures the reducing potential of the cell using a colorimetric reaction. Newer assays have been developed that are more sensitive and suitable for high-throughput screening. Among them are bioluminescence assays that use the cellular ATP content as a marker of viability [45]. On the other hand genetic toxicity (also termed genotoxicity) – the potential of drugs to damage DNA and become carcinogenic – remains a primary concern in drug development. The goal of genetic toxicology is to identify the carcinogenic potential of novel compounds by in vitro tests used in the early phases of drug development as surrogates for more expensive in vivo carcinogenesis testing conducted in animals. These liability tests usually consist of the so called Ames assay – the current gold standard for evaluating the mutagenic potential of chemicals – which allows for detecting reverse point mutations in a bacterial system; and the screening for chromosome breakage using either a micronucleus formation test or a mouse lymphoma test [46–48]. The micronucleus test can be performed with various cell types and applies a DNA intercalating dye to detect the existence of micronuclei in cells that form around chromosomal fragments due to breakage. As an alternative the more detailed chromosome aberration test can be performed, in which treated cells are arrested in mitosis followed by karyotyping of derived metaphase spreads. However, the method is tedious and not suitable for high-throughput screening, as it requires extensive technical skills in performance as well as outcome interpretation. The mouse lymphoma assay evaluates forward mutations in the tk locus of L5178Y mouse lymphoma cells, which are hemizygous for the tk locus. As a result, cells form colonies and the colony size correlates to the degree of aberration, ranging from small point mutations to larger chromosome rearrangements. Also in some instances a single-cell electrophoresis assay commonly known as the comet assay is performed [49]. It involves the encapsulation of cells in a low melting point agarose suspension, lysis of the cells and subsequent electrophoresis of the suspended lysed cells; followed by visual analysis and staining of DNA to determine the extent of DNA damage by means of fluorescence.
Although these tests are being used according to the International Conference on Harmonization guidance on genotoxicity testing [101], it is important to note that they do not reflect the wide range of conformational changes and genomic reorganizations that could be induced by epigenetic drugs with demethylating and chromatin remodeling potentials. Structural assays that assess the integrity of chromatin conformation and related nuclear genome distribution in direct response to epigenetic manipulations have been widely neglected in research, and more importantly, are currently not employed in drug testing. Investigations using imaging technologies have shown that epigenetic intervention can have direct implications on chromatin conformation and rearrangement [50–54]. These effects can occur without micronuclei formation. Therefore, these structural changes need to be appropriately addressed in an effort for better drug testing. In this context assessing the affected higher-order 3D genome organization may serve as a valuable entity for deriving multiple novel genotoxic end points in the evaluation of epigenetic drug actions.
Heterochromatin in cancer & epigenetic therapy
The global effect of these drugs on heterochromatin reasons for concern, as heterochromatin comprises a large portion (55%) of the human genome and is strongly involved in nuclear architecture, which functions as an infrastructure for a vast number of nuclear processes [55]. Furthermore, it is important to recognize that in cancer cells hypermethylation of single gene promoters – the actual focus of most epigenetic drug therapies – occurs against a strong background of gDNA hypomethylation including a decrease in the methylation load of heterochromatic regions of the genome. The loss of methyl groups is achieved mainly by hypomethylation of heterochromatin-residing repetitive DNA sequences, including transposable elements such as LINEs (especially LINE-1) and short interspersed elements (foremost Alu repeats) as part of the facultative heterochromatin and Sat2 DNA as part of the constitutive heterochromatin, when compared with normal tissue [18,56]. The extent of genome-wide hypomethylation converges closely with the degree of malignancy, in a tumor type-dependent manner [57]. In normal cells, the activity and interaction of these classes of DNA with neighboring chromatin regions (in the nuclear space) are strongly suppressed by methylation and compaction through histone modifications. Therefore, there are multiple risks associated with an increase in demethylation of repetitive elements [58], which can lead to their decondensation and related consequences: Sat2 DNA hypomethylation may favor pericentromeric instability, and LINE-1 elements may become transcriptionally active and relocate into other parts of the genome causing enhanced genome instability through mutational effects such as silencing of tumor-suppressor genes or the activation of oncogenes [17]. Hypomethylation of repetitive elements could lead to large-scale chromatin reorganization that can be made visible by cellular imaging. Therefore visualization of global nuclear DNA patterns that reflect these classes of DNA can be extremely supportive in drug testing [59].
3D quantitative DNA methylation imaging
-
■In this sense, 3D quantitative DNA methylation imaging (3D-qDMI) has been particularly developed as a tool towards the causal assessment of gDNA demethylation, heterochromatin decondensation and relevant genome reorganization in cell nuclei in response to environmental changes for directed differentiation of stem cells and in therapeutic reprogramming by demethylating agents [60–62]. The effects are measured through differential in situ analysis of the relevant nuclear structures represented by methylated CG-dinucleotides (MeC) and gDNA. The technology combines three steps:
-
■Visualization of cellular targets by nondistructive assays, such as immunofluorescence and FISH;
-
■High-resolution scanning microscopy realized through different imaging modalities;
-
■Computerized 3D image analysis utilizing advanced nuclear segmentation, signal extraction and image cytometry (cytomics).
-
■
Each step can be performed on currently existing platforms in research and is amenable to automation and scale-up for high-throughput clinical and industrial settings. Visualization of MeC is achieved by using a monoclonal antibody specific to the methylgroup bound to carbon 5 on the cytosine molecule, and gDNA is delineated by staining with the intercalating dye 4',6-diamidino-2-phenylindole (DAPI). The updated version of 3D-qDMI offers four modules for the characterization of cells and tissues based on gDNA methylation patterns: global nuclear load of MeC; MeC/gDNA codistribution displayed as a scatter plot; nuclear topology, i.e., spatial distribution of MeC and gDNA; and evaluation of similarity between cells in imaged populations based on the previous three features and the subsequent homogeneity assessment of these cell populations for statistical purposes [63]. The first three features are meant to provide an overall impression of changes in DNA methylation (differential MeC phenotypes) on a per-cell basis. The emphasis lies on monitoring the degree of demethylation caused by external influence (in this case drugs), and detection of critical changes in MeC and DAPI loads that indicate conformational changes ranging from local decondensation to massive relocations of heterochromatic areas in the nucleus [52,60,61]. The fourth module utilizes metrics frequently employed for similarity measurements in medical and systems biology applications including registration of image datasets, analysis of gene expression and determining DNA sequence homology. 3D-qDMI applies robust Kullback-Leibler divergence measurement [64,65] with proven high dynamic range in cell number variability to provide an estimate of feature homogeneity within a treated cell population. This module fits early phase high-throughput high-content screening practiced in pharmacology and toxicology, as the outcome can serve as a measure in determining demethylation efficacy [66]. Additionally, outlier cells can be identified, laser-captured and interrogated for possible mechanisms of drug resistance with downstream molecular methods.
Investigations using 3D-qDMI disclosed significant changes in the nuclear patterns of MeC and heterochromatin-derived signals in AtT20 mouse pituitary tumor cells upon treatment with AZA (Figure 1) [53], an agent that alters molecular methylation patterns on a genomic scale, in gene promoters and repetitive elements [67,68]. Furthermore, the methodology was able to monitor the dual effect of demethylating agents in human cancer cells: a decrease in the number of MeCs in gDNA, and the subsequent reorganization of highly compact heterochromatic regions of the genome as reflected by a significant decrease of DAPI intensity in the relevant nuclear regions. The effects resulted in low-intensity MeC and low-intensity DAPI sites, whose distributions can be mapped within cell nuclei [62]. Thereby, more nimble changes in gDNA methylation load/distribution can be detected as experienced with more gently demethylating drugs such as zebularine in human cells, which to begin with, also exhibit a finer granulation of heterochromatin compared with rodent cells [13]. The information of low-intensity MeC and low-intensity DAPI topology can therefore be applied in the assessment of risks associated with genome-wide demethylation, specifically leading to hypomethylation of repetitive elements causing an adverse reorganization of the genome. In summary, 3D-qDMI supports profiling whole populations at single-cell level, and provides a rapid landscape of cell-specific DNA methylation (MeC) phenotypes across drug-targeted cells [59].
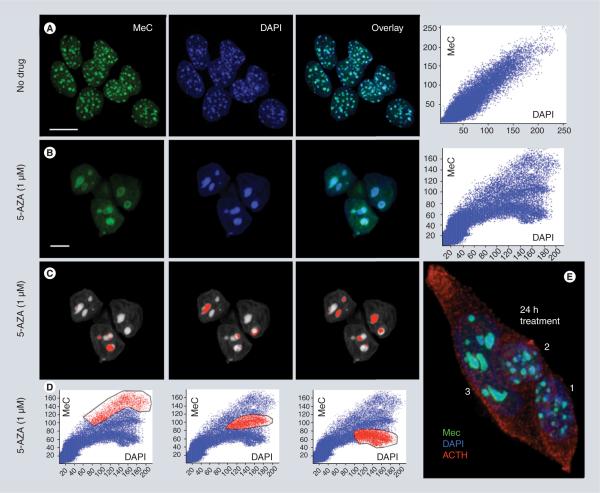
Figure 1. Effect of 5-azacytidine on the heterochromatin of AtT20 pituitary tumor cells in culture.
5-AZA causes both, demethylation of gDNA including DAPI-positive heterochromatic regions and significant reorganization of the chromatin as inferred by maximum intensity projections of three cell nuclei imaged by confocal microscopy, and quantitative 3D image ana lysis (correlated scatterplots). Untreated cells show numerous small (diameter ~1 μm) MeC foci (so-called chromocenters that consist of centromeric and pericentromeric repetitive DNA) – close to the number of different chromosomes – which display an even MeC distribution (in green), and almost fully overlap with the nuclear DAPI signals (in red) (A); in contrast, cells treated for 48 h with 5-AZA show only a few (2–5) giant MeC foci (diameter ~3–5 μm), which mostly exhibit a drastically different MeC distribution. A hypomethylated heterochromatic core surrounded by a hypermethylated ring (B). This phenomenon is represented by changes in the distribution of the MeC and DAPI signals displayed as respective 2D scatter plots (A & B). The reverse mapping of selected groups of plotted signals (red labeled in [D]) explains that corresponding heterochromatic areas of the genome (red labeled in [C]) have experienced different degrees of demethylation in lieu of drug exposure; including loci that stay hypermethylated (left column), loci that have become less demethylated (middle column) and more strongly hypomethylated (right column). Also, drug-treated cells appear larger and flatter than their naive counterparts. (E) The cluster of three ACTH-producing AtT20 cells in this subfigure shows the more heterogeneous drug response of the cells after only 24 h of exposure. The normal-sized nuclei 1 and 2 present foci with unchanged and slightly changed morphology and heterochromatin organization, whereas cell 3 already presents full-blown demethylation effects, including significantly altered morphology and merged chromocenters (scale bars are 10 μm).
5-AZA: 5-azacytidine; ACTH: Adrenocorticotropic hormone; DAPI: 4′,6-diamidino-2-phenylindole; MeC: Methylcytosine.
Conclusion & future perspective
Today's possibilities to use more advanced imaging approaches in an automated high-throughput fashion – including confocal laser scanning and two-photon excitation microscopy, as well as high-content cell imaging, and digital tissue scanning – have rendered high-resolution optical imaging an essential tool for testing new chemical substances in the pharmaceutical pipeline by using nondisruptive cell-based assays [69]. In contrast to pure biochemical analytics, imaging and cytomics provide the ability to measure the spatial and temporal distribution of molecules and cellular components within their native environment [70]. This helps to understand drug activity at the cell systemic level. Respectively, improvements in image cytometry towards a hybrid platform, that tries to combine the diagnostic speed of flow cytometry and the capability of high-resolution scanners for intracellular target mapping, have been very supportive [71]. In this context, the impact of 3D-qDMI can be considered as twofold: it introduces spatial DNA methylation patterns as a potential surrogate pharmacodynamic biomarker of demethylating drug action; beyond that, it may serve as an encouraging model system to a field of future assays that utilize chromatin structure genotoxicity relationships as an integral component in drug and compound characterization in the pre-clinical phases of drug development. If necessary, 3D-qDMI could be combined with more elaborate (however not with cell-by-cell) molecular methods such as chromosome conformation capture (3C) methodology and its derivatives, 5C and Hi-C, which map the 3D architecture of the genome by proximity-based ligation and subsequent next-generation sequencing at the resolution of ~1 Mbp [72,73]. It is conceivable that decondensation/reorganization of larger portions of heterochromatic regions (due to demethylation of repetitive elements) could skew the genomic interaction maps of treated cells generated by said methods compared with untreated cells. The introduction of in situ and in vitro chromatin conformational assays for genotoxicity testing would also leverage the use of cell models, and improve efficient data generation and interpretation in this very critical phase of drug development (Figure 2). This opens possibilities with regard to personalized medicine that focuses on utilizing a patient's own cells in order to consider interindividual variability in drug response (pharmacoepigenomics) for the design of tailored drug-based therapies [74]. Importantly, these approaches – which aim at manipulating chromatin-associated epigenetic mechanisms – require optimization of dosing schedule and sequences for improved prediction of responses and a better risk assessment of immediate to long-term genotoxic side effects, in an effort to minimize patients' risk [75].
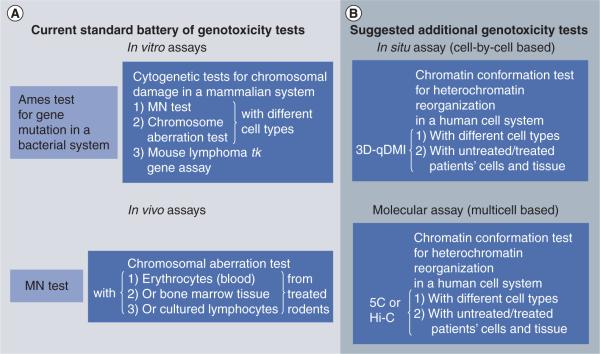
Figure 2. Currently applied and suggested future genotoxicity tests.
(A) The current battery of recommended tests in accordance with the International Conference on Harmonization S2 guidance. The combination of the Ames test, and one in vitro and one in vivo assay is accepted for being sufficient to demonstrate absence or presence of genotoxicity for a an agent. Thereby, all three cytogenetic in vitro tests are recognized as equivalent and are therefore interchangeable. The guidance allows for an additional test. (B) This article suggests additional in situ (3D-qDMI) and molecular chromatin conformational tests to assess unwanted drug effects on the 3D genome architecture, not detectable by the presented conventional methods. 3D-qDMI is a true cell-by-cell assay that can be combined with chromosome conformation capture methods (5C or Hi-C), which have a lower cellular resolution but probe at a higher molecular resolution. These novel approaches can be adapted for genotoxicity testing to provide information regarding possible decondensation and/or reorganization of heterochromatic areas of the genome as a result of drug application. Such effects may bear risks for unwanted interactions of these normally suppressed and compacted regions with other sites within the genome and mutations caused by unleashed transposition events.
3d-qDMI: 3D-quantitative DNA methylation imaging.
With the existing advanced tools that are at hand, drug testing clearly needs to integrate additional assays and complementary informatics for detecting unwanted genomic-scale adverse effects such as heterochromatin reorganization, mentioned in this paper. These topologic end points cannot simply be covered by the micronucleus assay or tedious karyotyping. The implementation of such tests in pharmacology and pharmacovigilance will ultimately not only benefit patient safety but also significantly contribute to reducing attrition in the very costly drug-development pipeline. Furthermore, since epigenetic mechanisms seem to be either directly or indirectly involved in many cellular processes and pathologies, this novel type of genotoxicity assessment could be practically expanded to any agent beyond epigenetic drugs, for which we have no current knowledge of epigenetic side effects: including drugs that are already in practice, and perhaps also in environmental toxicology, under the new umbrella task coined toxicoepigenomics [75] and its subdivision toxicomethylomics [35].
Executive summary.
Therapeutic targeting of chromatin
-
■
Epigenetic processes such as DNA methylation, histone modifications and the metabolism of ncRNAs are involved in the dynamic structure and function of chromatin.
-
■
Pharmacologic targeting of chromatin and its basic components has become one of the major strategies in the treatment of complex traits.
-
■
The boost in epigenetic drug development calls for appropriate high-throughput assays to be implemented into the early phases of drug discovery for assessing the agents' genotoxic side effects on chromatin architecture and the correlated risk of genome instability in targeted cells and tissues in order to improve the agents' clinical utility.
Cell-based assays in epigenetic drug discovery
-
■
Preclinical phases of drug development focus on determining drug action and efficacy as well as cytotoxicity and genetic toxicity using cell-based screening.
-
■
Cytotoxicity is evaluated by multiplexed fluorescent assays with DNA intercalating dyes and labeled reporters of apoptosis in conjunction with flow cytometry to determine the percentage of apoptotic and necrotic cells.
-
■
Genotoxicity, which refers to drug-induced DNA damage is currently obtained by two to three different in vitro tests: the Ames assay as the gold standard for detection of point mutations in a bacterial system; the micronuclei assay for detection of chromosome breakage; and the mouse lymphoma assay for detection of smaller mutations up to chromosome rearrangements.
-
■
Alternative tests include single-cell karyotyping and gel electrophoresis (comet test).
-
■
These tests do not reflect the wide range of conformational changes and genomic reorganizations that could be induced by epigenetic drugs with demethylating and chromatin remodeling potentials.
Heterochromatin in cancer & epigenetic therapy
-
■
Heterochromatin (55% of the human genome) is strongly involved in genome architecture and nuclear processes.
-
■
Potential hazard of demethylating agents in cancer cells: heterochromatin-residing repetitive elements such as long interspersed nuclear element-1 and Sat-2 are often hypomethylated in cancer cells, therefore further demethylation would increase the chances of heterochromatin reorganization and genome instability.
3D quantitative DNA methylation imaging
-
■
3D quantitative DNA methylation imaging (3D-qDMI) combines immunofluorescence, high-resolution imaging, and 3D image analysis, and has been developed to assess gDNA demethylation, heterochromatin decondensation, and relevant genome reorganization induced by environmental changes in thousands of single cells in parallel.
-
■
The effects are measured through differential in situ analysis of the relevant nuclear structures represented by methylated CG-dinucleotides and gDNA.
-
■
3D-qDMI measures end points such as methylated CG-dinucleotides and 4′,6-diamidino-2-phenylindole loads, colocalization, and differential distribution, as well as cell population heterogeneity based on these values towards drug efficacy.
-
■
The technology is flexible and adaptable to automation for high-content and high-throughput lead screening, and compound analysis in academic research and industrial routine.
-
■
3D-qDMI has proven itself as a method that could reconcile the reported effects of the drugs obtained by more elaborate molecular analyses with a rapid imaging-based approach at single-cell resolution.
Conclusion & future perspective
-
■
The design of better drug-based therapies that manipulate chromatin-associated epigenetic mechanisms requires improved prediction of responses to these and a better risk assessment of immediate to long-term genotoxic side effects such as heterochromatin reorganization.
-
■
Drug testing needs to integrate additional in situ assays and complementary informatics that assess chromatin structure–phenotype relationships for detecting unwanted genome-scale adverse effects.
-
■
3D-qDMI introduces spatial DNA methylation patterns as a potential surrogate pharmacodynamic biomarker of demethylating drug action, plus serves as a model for future genotoxicity assays that utilize chromatin structure and discerned patterns in drug testing.
-
■
Universal implementation of such assays beyond epigenetic drug testing should lead to better patient safety and reduction of attrition in drug development.
Footnotes
Financial & competing interests disclosure The Department of Surgery at the Cedars-Sinai Medical Center and the National Institutes of Health are acknowledged for funding epigenetics projects that led to the consideration of the issues discussed in this manuscript. The author is a shareholder of Epilumina Inc. The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
■■ of considerable interest
- 1.Misteli T. Higher-order genome organization in human disease. Cold Spring Harb. Perspect. Biol. 2010;2(8):a000794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deal RB, Henikoff S. Capturing the dynamic epigenome. Genome Biol. 2010;11(10):218. doi: 10.1186/gb-2010-11-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 4.Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science. 1974;184(139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- 5.Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr. Opin. Genet. Dev. 2011;21(2):175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crutchley JL, Wang XQ, Ferraiuolo MA, Dostie J. Chromatin conformation signatures: ideal human disease biomarkers? Biomark. Med. 2010;4(4):611–629. doi: 10.2217/bmm.10.68. [DOI] [PubMed] [Google Scholar]
- 7.Brock MV, Herman JG, Baylin SB. Cancer as a manifestation of aberrant chromatin structure. Cancer J. 2007;13(1):3–8. doi: 10.1097/PPO.0b013e31803c5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447(7143):433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 9.Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu. Rev. Pharmacol. Toxicol. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- 10.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349(21):2041–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 11.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 13.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat. Rev. Drug Discov. 2006;5(2):37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 14.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J. Natl Cancer Inst. 2005;97(20):1498–1506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]
- 15.Luebbert M. DNA methylation inhibitors in the treatment of leukemias, myelodysplastic syndromes and hemoglobinopathies: clinical results and possible mechanismas in action. Curr. Top. Microbiol. Immunol. 2000;249:135–164. doi: 10.1007/978-3-642-59696-4_9. [DOI] [PubMed] [Google Scholar]
- 16.Issa JP, Kantarjian HM. Targeting DNA methylation. Clin. Cancer Res. 2009;15(12):3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Momarler RL. Epigenetic therapy of cancer with 5-aza-2'-deoxycytidine (decitabine) Semin. Oncol. 2005;32(5):443–451. doi: 10.1053/j.seminoncol.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1(2):239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■■ Provides a comprehensive overview of the risks in connection with demethylating tumor suppressor genes and heterochromatic regions of the genome.
- 19.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187(4173):226–232. [PubMed] [Google Scholar]
- 20.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 21.Bestor TH, Tycko B. Creation of genomic methylation patterns. Nat. Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 22.Ehrlich M. DNA methylation: normal development, inherited disease and cancer. J. Clin. Ligand Assay. 2000;23:144–146. [Google Scholar]
- 23.Vera E, Canela A, Fraga MF, Esteller M, Blasco MA. Epigenetic regulation of telomeres in human cancer. Oncogene. 2008;27(54):6817–6833. doi: 10.1038/onc.2008.289. [DOI] [PubMed] [Google Scholar]
- 24.Mund C, Lyko F. Epigenetic cancer therapy: proof of concept and remaining challenges. Bioessays. 2010;32(11):949–957. doi: 10.1002/bies.201000061. [DOI] [PubMed] [Google Scholar]
- 25.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai HC, Baylin SB. Cancer epigenetics: linking basic biology to clinical medicine. Cell Res. 2011;21(3):502–517. doi: 10.1038/cr.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat. Med. 2011;17(3):330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 28.Gore SD, Baylin S, Sugar E, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66(12):6361–6369. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, et al. Phase 1/2 study of the combination of 5-aza-2'-deoxycytidine with valproic acid in patients with leukemia. Blood. 2006;108(10):3271–3279. doi: 10.1182/blood-2006-03-009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurtubise A, Momparler RL. Effect of histone deacetylase inhibitor LAQ824 on antineoplastic action of 5-aza-2′-deoxycytidine (decitabine) on human breast carcinoma cells. Cancer Chemother. Pharmacol. 2006;58(5):618–625. doi: 10.1007/s00280-006-0225-6. [DOI] [PubMed] [Google Scholar]
- 31.D'Alessio AC, Szyf M. Epigenetic tête-à-tête: the bilateral relationship between chromatin modifications and DNA methylation. Biochem. Cell Biol. 2006;84(4):463–476. doi: 10.1139/o06-090. [DOI] [PubMed] [Google Scholar]
- 32.Detich N, Hamm S, Just G, Knox JD, Szyf M. Valproate induces replication-independent active DNA demethylation. J. Biol. Chem. 2003;278(30):27586–27592. doi: 10.1074/jbc.M303740200. [DOI] [PubMed] [Google Scholar]
- 33.Ou JN, Torrisani J, Unterberger A, et al. Histone deacetylase inhibitor Trichostatin A induces global and gene-specific DNA demethylation in human cancer cell lines. Biochem. Pharmacol. 2007;73(9):1297–1307. doi: 10.1016/j.bcp.2006.12.032. [DOI] [PubMed] [Google Scholar]
- 34.Milutinovic S, D'Alessio AC, Detich N, Szyf M. Valproate induces widespread epigenetic reprogramming which involves demethylation of specific genes. Carcinogenesis. 2007;28(3):560–571. doi: 10.1093/carcin/bgl167. [DOI] [PubMed] [Google Scholar]
- 35.Szyf M. The implications of DNA methylation for toxicology: toward toxicomethylomics, the toxicology of DNA methylation. Toxicol. Sci. 2011;120(2):235–255. doi: 10.1093/toxsci/kfr024. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■■ Discusses the importance of and provides a rationale for considering DNA demethylation in drug toxicology.
- 36.Kacevsca M, Ivanov M, Ingelman-Sundberg M. Perspectives on epigenetics and its relevance to adverse drug reactions. Clin. Pharmacol. Ther. 2011;89(6):902–907. doi: 10.1038/clpt.2011.21. [DOI] [PubMed] [Google Scholar]
- 37.Comley J. Epigenetics: an emerging target class for drug screening, Spring 2011. Drug Discov. World. 2011:40–55. [Google Scholar]
- 38.Laird PW. Principles and challenges of genome-wide DNA methylation ana lysis. Nat. Rev. Genet. 2010;11(3):191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 39.Meissner A, Mikkelsen TS, Gu H, et al. Genomescale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng J, Shoemaker R, Xie B, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 2009;27(4):353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ammerpohl O, Martin-Subero JI, Richter J, Vater I, Siebert R. Hunting for the 5th base: techniques for analyzing DNA methylation. Biochim. Biophys. Acta. 2009;1790:847–862. doi: 10.1016/j.bbagen.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Lister R, Ecker JR. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19(6):959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith ZD, Gu H, Bock C, Gnirke A, Meissner A. High-throughput bisulfite sequencing in mammalian genomes. Methods. 2009;48(3):226–232. doi: 10.1016/j.ymeth.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niles AL, Moravec RA, Riss TL. Update on in vitro cytotoxicity assays for drug development. Expert Opin. Drug Discov. 2008;3(6):655–669. doi: 10.1517/17460441.3.6.655. [DOI] [PubMed] [Google Scholar]
- 46.Ames BN, Lee FD, Durston WE. An improved bacterial test system for the detection and classification of mutagens and carcinogens. Proc. Natl Acad. Sci. USA. 1973;70(3):782–786. doi: 10.1073/pnas.70.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmid W. The micronucleus test. Mutat. Res. 1975;31(1):9–15. doi: 10.1016/0165-1161(75)90058-8. [DOI] [PubMed] [Google Scholar]
- 48.Pfuhler S, Albertini S, Fautz R, et al. Genetic toxicity assessment: employing the best science for human safety evaluation part IV. Toxicol. Sci. 2007;97(2):237–240. doi: 10.1093/toxsci/kfm019. [DOI] [PubMed] [Google Scholar]
- 49.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA danage in individual cells. Exp. Cell Res. 1988;175(1):184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 50.Haaf T. The effects of 5-azacytidine and 5-azadeoxycytidine on chromosome structure and function: implications for methylation-associated cellular processes. Pharmacol. Ther. 1995;65(1):19–46. doi: 10.1016/0163-7258(94)00053-6. [DOI] [PubMed] [Google Scholar]; ■■ Emphasizes the effects of the demethylating agent azacytidine on chromatin structure, chromosome organization and nuclear architecture.
- 51.de Capoa A, Menendez F, Poggesi I, et al. Cytological evidence for 5-azacytidine-induced demethylation of the heterochromatic regions of human chromosomes. Chromosome Res. 1996;4(4):271–276. doi: 10.1007/BF02263676. [DOI] [PubMed] [Google Scholar]; ■■ Emphasizes the effects of the demethylating agent azacytidine on chromatin structure, chromosome organization and nuclear architecture.
- 52.Gilbert N, Thomson I, Boyle S, Allan J, Ramsahoye B, Bickmore WA. DNA methylation affects nuclear organization, histone modifications, and linker histone binding but not chromatin compaction. J. Cell Biol. 2007;177(3):401–411. doi: 10.1083/jcb.200607133. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■■ Emphasizes the effects of the demethylating agent azacytidine on chromatin structure, chromosome organization and nuclear architecture.
- 53.Tajbakhsh J, Wawrowsky KA, Gertych A, et al. Characterization of tumor cells and stem cells by differential nuclear methylation imaging. In: Farkas DL, Nicolau DV, Leif RC, editors. Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues. San Jose, CA, USA: 2008. Proceedings of the SPIE 6856. 6859F1-10. [Google Scholar]; ■■ Marks the first-time ana lysis of differential DNA methylation patterns in a mouse cancer cell model and differentiating human stem cells based on high-resolution confocal microscopy. Also emphasizes the effects of the demethylating agent azacytidine on chromatin structure, chromosome organization and nuclear architecture.
- 54.Shvachko LP. Alterations of constitutive pericentromeric heterochromatin in lymphocytes of cancer patients and lymphocytes exposed to 5-azacytidine is associated with hypomethylation. Exp. Oncol. 2008;30(3):230–234. [PubMed] [Google Scholar]; ■■ Emphasizes the effects of the demethylating agent azacytidine on chromatin structure, chromosome organization and nuclear architecture.
- 55.Espada J, Esteller M. Epigenetic control of nuclear architecture. Cell Mol. Life Sci. 2007;64(4):449–457. doi: 10.1007/s00018-007-6358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■■ Emphasizes the effects of the demethylating agent azacytidine on chromatin structure, chromosome organization and nuclear architecture.
- 56.Choi SH, Worswick S, Byun HM, et al. Changes in DNA methylation of tandem DNA repeats are different from interspersed repeats in cancer. Int. J. Cancer. 2009;125(3):723–729. doi: 10.1002/ijc.24384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costello JF, Plass C. Methylation matters. J. Med. Genet. 2001;38(5):285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300(5618):489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]; ■■ Demonstrates the first-time engineered induction of global demethylation through DNA methyltransferase manipulation in a mouse model and the evidence of cancerogenesis as a consequence.
- 59.Tajbakhsh J, Gertych A, Farkas DL. Utilizing 3D nuclear DNA methylation patterns in cell-based assays for epigenetic drug screening. Drug Discovery World. 2010 Spring;:27–35. 2010. [Google Scholar]
- 60.Tajbakhsh J, Gertych A, Fagg WS, Hatada S, Fair JH. Early In vitro differentiation of mouse definitive endoderm is not correlated with progressive maturation of nuclear DNA methylation patterns. PLoS ONE. 2011;6(7):e21861. doi: 10.1371/journal.pone.0021861. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■■ Reconciles molecular results made in early differentiation of mouse embryonic stem cells and provides the first cell-by-cell impression of changes in gDNA methylation patterns, with striking evidence for the importance of single-cell ana lysis in heterogeneous populations such as stem cells.
- 61.Gertych A, Wawrowsky KA, Lindsley E, Vishnevsky E, Farkas DL, Tajbakhsh J. Automated quantification of DNA demethylation effects in cells via 3D mapping of nuclear signatures and population homogeneity assessment. Cytometry A. 2009;75(7):569–583. doi: 10.1002/cyto.a.20740. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■■ Introduces the first-time application of 3D quantitative DNA methylation imaging with a novel and dedicated algorithm, to demonstrate the effect of demethylating drugs on DNA methylation patterns and chromatin organization.
- 62.Gertych A, Farkas DL, Tajbakhsh J. Measuring topology of low-intensity DNA methylation sites for high-throughput assessment of epigenetic drug-induced effects in cancer cells. Exp. Cell Res. 316(19):3150–3160. doi: 10.1016/j.yexcr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tajbakhsh J, Gertych A. 3-D quantitative DNA methylation imaging for chromatin texture ana lysis in pharmacoepigenomics and toxicoepigenomics. In: Appasani K, editor. From Chromatin Biology to Therapeutics. Cambridge University Press; Cambridge, UK: In press. [Google Scholar]
- 64.Kullback S, Leibler R. On information and sufficiency. Ann. Math. Stat. 1951;22(1):79–86. [Google Scholar]
- 65.Kullback S. Information Theory and Statistics. Dover Publications; New York, NY, USA: 1997. p. 8. [Google Scholar]
- 66.Gertych A, Tajbakhsh J. Homogeneity assessment of cell populations for high-content screening platforms. In: Pietka E, Kawa J, editors. Information Technology in Biomedicine. Advances in intelligent and soft computing. volume 69. Springer Verlag; Heidelberg, Germany: 2010. pp. 309–319. [Google Scholar]
- 67.Soriano AO, Yang H, Faderl S, et al. Safety and clinical activity of the combination of 5-azacytidine, valproic acid, and all-trans retinoic acid in acute myeloid leukemia and myelodysplastic syndrome. Blood. 2007;110(7):2302–2308. doi: 10.1182/blood-2007-03-078576. [DOI] [PubMed] [Google Scholar]
- 68.Hagemann S, Heil O, Lyko F, Brueckner B. Azacytidine and decitabine induce gene-specific and non-random DNA demethylation in human cancer cell lines. PLoS ONE. 2011;6(3):e17388. doi: 10.1371/journal.pone.0017388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lang P, Yeow K, Nichols A, Scheer A. Cellular imaging in drug discovery. Nat. Rev. Drug Discov. 2006;5(4):343–356. doi: 10.1038/nrd2008. [DOI] [PubMed] [Google Scholar]
- 70.Tárnok A. Cytomics for discovering drugs. Cytometry A. 2010;77(1):1–2. doi: 10.1002/cyto.a.20845. [DOI] [PubMed] [Google Scholar]
- 71.Telford WG. Imaging cytometry: an expanding role in biomedical image ana lysis. Int. Drug Discov. 2010;5(5):54–58. [Google Scholar]
- 72.Dostie J, Richmond TA, Arnaout RA, et al. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16(10):1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ingelman-Sundberg M, Gomez A. The past, present and future of pharmacoepigenomics. Pharmacogenomics. 2010;11(5):625–627. doi: 10.2217/pgs.10.59. [DOI] [PubMed] [Google Scholar]
- 75.Csoka AB, Szyf M. Epigenetic side-effects of common pharmaceuticals: a potential new field in medicine and pharmacology. Med. Hypotheses. 2009;73(5):770–780. doi: 10.1016/j.mehy.2008.10.039. [DOI] [PubMed] [Google Scholar]; ■■ Avant-garde review article that introduces a visionary scope on considering potential side effects of agents with epigenetic modes of action, specifically in chemicals that manipulate the methylome of cells.
Website
- 101.International Conference on Harmonization Guidance on Genotoxicity Testing and Data Interpretation for Pharamceuticals Intended for Human Use S2(R1) Draft. 2009 www.ich.org. [PubMed]