Abstract
One impediment to the use of TRAIL receptor targeted agents as anti-tumor drugs is the evolution of resistance, a common problem in cancer. On the other hand, many different kinds of drugs synergize with TRAIL in TRAIL-sensitive tumor cells, raising the question whether one can overcome resistance with the same drugs producing synergy. This is an important question, because recent clinical trials suggest that combination treatments with cytotoxic drugs and TRAIL receptor-targeted agents do not provide additional benefit compared with cytotoxic agents on their own. Such results might be expected if drug combinations that synergize in sensitive tumor cells but cannot overcome TRAIL resistance are used in patients whose tumors were not selected for retention of TRAIL sensitivity. We tested this idea by creating isogenic tumor cells with acquired TRAIL resistance or defined mechanisms of resistance that occur in human tumors then compared them to the TRAIL-sensitive parental cell line. Although diverse classes of anti-cancer drug were all able to synergize with TRAIL in sensitive cells, most agents were unable to overcome resistance and there was no relationship between the amount of synergy seen with a particular agent and its ability to overcome acquired resistance. An important exception was proteasome inhibitors, which were however able to overcome diverse resistance mechanisms. Our findings suggest that one should select drugs for TRAIL receptor agonist combination therapy based not just on their ability to synergize but rather on their ability to both overcome resistance as well as synergize.
Keywords: TRAIL, Six1, chemoresistance, lymphoma
Introduction
Cancer therapy is often hampered because the efficiency with which susceptible tumor cells are killed is too low and tumors evolve such that they either present with primary resistance or acquire resistance over time. To address the first issue it is useful to identify agents that synergize when they are combined. To address the second issue, it is necessary to find new treatments that avoid the resistance mechanisms or combine with agents that allow the resistance to be overcome. It is often assumed that if a drug synergizes with another drug, then the combination will provide a better way to treat cancer and that more synergy is better. However, obtaining synergy and overcoming resistance isn’t necessarily the same thing– just because a drug combination can synergize in susceptible tumor cells, it does not necessarily follow that the same combination will be able to overcome resistance.
Tumor Necrosis Factor-like Apoptosis Inducing Ligand (TRAIL) Receptor-targeted drugs are an interesting type of anti-cancer treatment with which to test these ideas because these drugs directly activate the apoptosis machinery, there are many different ways that tumor cells can become resistant to TRAIL R agonists, and there are a large number of other agents that can synergize with TRAIL in susceptible tumor cells (1, 2).
TRAIL R agonists bind to two receptors (DR4 and DR5, also known as TRAIL R1 and TRAIL R2, TNRSF10a and TNFRSF10b) to cause the recruitment of an adaptor protein called FADD, which in turn recruits caspase-8 to form a platform called the DISC that leads to the activation of caspase-8 (3). Active caspase-8 induces apoptosis by directly activating the effector caspase-3 or, more commonly, by cleavage of the BH3 protein Bid, which leads to release of cytochrome c and activation of the mitochondrial apoptosis machinery. Several TRAIL R activating drugs are in clinical trials and others are in pre-clinical development (1, 2), additionally some other agents may work indirectly through activation of DR4 and DR5.
Many tumor cell lines are TRAIL resistant (4) and resistance varies in primary human tumor cells. For example, it has been reported that primary colon cancer cells are usually sensitive to TRAIL (5) while primary astrocytoma cells (5) and B cell chronic lymphocytic leukemia cells (6) are not. Other tumor types, e.g. ovarian cancers, show more variable responses (7). Since TRAIL signaling is an important part of the host mechanisms to suppress tumor formation and metastasis (1, 2), it is to be expected that advanced cancers would often evolve TRAIL resistance. Diverse mechanisms that confer selective TRAIL resistance have been identified in human tumors (8). For example somatic mutations in TRAIL receptors, downregulation of DR4 or DR5 and overexpression of decoy receptors, DcR1 and DcR2 and expression of a tumor-related homeobox transcription factor called Six1 can all confer selective inhibition to TRAIL. Additionally more general anti-apoptotic mechanisms like increased expression of Bcl-2 can also cause TRAIL resistance.
A myriad of anticancer drugs including anti-metabolites, DNA damaging agents, microtubule-targeted drugs, protein kinase inhibitors, proteasome inhibitors, targeted toxins, deacetylase inhibitors, Bcl-2 antagonists and antibody-based therapeutics have been reported to synergize with TRAIL receptor targeted drugs (1, 2, 8, 9). This broad ability to synergize along with limited evidence of efficacy as single agents, has led to the view that optimal use of TRAIL R–targeted drugs will be in combination with cytotoxic chemotherapy or signal transduction pathway-targeted agents. Indeed, clinical trials with TRAIL or antibodies against DR4 or DR5 in combination with other drugs are already underway (1, 10–12). There has not been a consistent set of explanations for the ability of these diverse agents to synergize with TRAIL. Since a wide variety of different drugs synergize with TRAIL, it may be that rather than each different drug causing synergy by a specific molecular mechanism, much of the cooperation that has been seen with different drugs and TRAIL is due to a more general response to combinatorial stress—i.e. because tumor cells are primed to undergo apoptosis (13), the addition of any other apoptotic stimulus to TRAIL will significantly increase tumor cell killing and thus cause synergy. This begs the question of whether all these synergizing agents are equivalent when used in tumor cells that are resistant to TRAIL–i.e. do drugs that synergize with TRAIL also overcome resistance? This question is important because recent Phase II trials of TRAIL R-targeted drugs in combination with cytotoxic chemotherapy suggest that the addition of TRAIL R-targeted drug does not improve treatment compared with standard treatment alone (10–12). If overcoming TRAIL resistance and obtaining synergy are separable events, it might be expected that such combination treatments would fail unless the treatments are performed in selected patients whose tumors retain TRAIL sensitivity, or the combination is with an agent that can overcome TRAIL resistance as well as causing synergy in TRAIL-sensitive cells.
To answer this question we examined isogenic BJAB tumor cell lines. We tested if the parental cells, which are sensitive to all TRAIL receptor-targeted therapeutics, could synergize with anti-cancer drugs that work through different mechanisms. We developed an isogenic line with acquired TRAIL resistance through continued exposure to increasing doses of TRAIL Receptor-activating antibody and asked if these cells, or cells that were made resistant using defined molecular mechanisms that occur in human tumors, could be made sensitive to TRAIL by combining with drugs that synergize. Surprisingly, we found that most synergizing drugs could not overcome resistance and that more synergy does not make it more likely that the drug will overcome resistance. The most effective agents at overcoming resistance were proteasome inhibitors and this ability applied to different, but not all, TRAIL resistance mechanisms. These data demonstrate that it is possible to both obtain synergy and overcome TRAIL resistance by combining TRAIL receptor-targeted drugs with other agents. However, it is necessary to choose the correct agent for the combination because most synergizing drugs do not overcome acquired resistance and the most synergistic drugs are not necessarily the best ones to use in combination therapy if the objective is to try to both increase tumor killing in sensitive cells and overcome TRAIL resistance.
Materials and Methods
Cell Lines
Parental BJAB cells, which are an EBV negative B lymphoma cell line were provided previously by Marcus Peter (Univ. of Chicago). The cells were most recently fingerprinted using the ABI Identifiler kit in January 2010, and were distinct from other cells in the merged DSMZ, ATCC, JCRB, and Riken databases of DNA profiles of cell lines that is available at DSMZ website (14) and are therefore not contaminated with any other cell line. BJAB cells expressing mutant DR5 in place of the wildtype protein were described previously (15), the various resistant cells expressing DcR1, DcR2, FADD-DD and Six1 were made by stably expressing the respective cDNA in pcDNA3 puro (16). DR5 and XIAP knockdowns were achieved by stably expressing a lentivirus expressing DR5-targeted shRNA (Open Biosystems) while BJABLexR cells were made by gradually increasing Lexatumumab concentrations and selecting the cells capable of continued growth. All cell clones were derived from representative single clones isolated by limiting dilution. Cells were grown in RPMI 1640 with 10% FBS, Sodium Bicarbonate, and glucose in a 5% CO2 humidified atmosphere at 37°C.
MTS cell viability
Cells were plated in 96 well format at 40,000 cells per well. Lexatumumab (Human Genome Sciences) was cross-linked with anti-human IgG Fc (Sigma, St. Louis, MO) for 30 min before serial dilution. TRAIL (R&D Systems, Minneapolis, MN) and SuperFas Ligand (Enzo Life Sciences, Plymouth Meeting, PA) were prepared according to manufacturer’s instructions prior to addition to cells. Two hours prior to the end of the experiment, cells were treated with 20 ul of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethosxphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) (Promega, Madison, WI) according to manuacturer’s instructions. The plate was incubated at 37°C for 2 hrs. A Bio-Rad Benchmark Plus Microplate Spectrophotometer (Bio-Rad Laboratories, Hercules, CA) was used to measure absorbance of samples at 490 nm.
Immunoblotting
1 million cells were harvested and lysates were prepared by boiling in SDS buffer 5 min prior to gel electrophoresis. Lysates were resolved on 12% SDS-polyacrylamide gels. Proteins were transferred to Immobilon-P Transfer Membrane (Millipore Corporation, Bedford, MA). Blots were blocked with 5% milk in TBST and incubated with antibodies that recognize PARP, XIAP, Bid, Caspase 3, Caspase 8 (Cell Signaling Technologies, Danvers, MA), FADD (BD Biosciences, Franklin Lakes, NJ) and β-Actin (Sigma, St. Louis, MO). Blots were then incubated with anti-rabbit or anti-mouse horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technologies, Danvers, MA). Detection was performed using chemiluminescent ECL reagent (Millipore Corporation, Bedford, MA) and developed on Blue X-Ray film (Life Science Products, Inc., Frederick, CO).
Drug Synergy
Cells were plated in duplicate wells at a density of 40,000 cells per well in a 96 well plate and allowed to sit 24 hrs prior to treatment. TRAIL was used alone and in combination with the following drugs: MG132 (EMD Biosciences, Gibbstown, NJ), Doxorubicin hydrochloride, 5-Fluorouracil (5-FU), Etoposide, Oxamflatin, Staurosporine, and D-Sorbitol (Sigma, St. Louis, MO). Bortezomib (NSC #681239), Sorafenib (NSC # 747971) and Azacitidine (NSC# 102816) were obtained from The NCI/DTP Open Chemical Repository. MTS Assays were performed after 24 hrs incubation and CI values were obtained using Calcusyn, which determines synergy using the median effect principle comparing the sensitivity to each drug on its own and together with TRAIL. Synergy was scored as CI values 0.0–0.3 +++, 0.3–0.7 ++, and 0.7–1.0 +. For the data shown in Table 1, the doses used for each drug combination were: Bortezomib D1: 7.5 ng/ml TRAIL, 16 pM Bortez.; D2: 15 ng/ml TRAIL, 32 pM Bortez. MG132 D1: 2 ng/ml TRAIL, 1.05 uM MG132; D2: 4 ng/ml TRAIL, 2.1 uM MG132. Doxorubicin D1: 2 ng/ml TRAIL, 0.875 ug/ml Doxo; D2: 4 ng/ml TRAIL, 1.75 ug/ml Doxo. 5-FU D1: 2 ng/ml TRAIL, 25 nM 5-FU; D2: 8 ng/ml TRAIL, 100 nM 5-FU. Etoposide D1: 3.125 ng/ml TRAIL, 4 uM Etoposide; D2: 6.25 ng/ml TRAIL, 8 uM Etoposide. Oxamflatin D1: 4 ng/ml TRAIL, 1 ug/ml Oxam; D2: 8 ng/ml TRAIL, 2 ug/ml Oxam. Sorafenib D1:7.5 ng/ml TRAIL, 5 nM Sorafenib; D2: 15 ng/ml TRAIL, 10 nM Sorafenib. Azacitidine D1: 5 ng/ml TRAIL, 6.25 nM Aza.; D2: 10 ng/ml TRAIL, 12.5 nM Aza. Sorbitol D1: 4 ng/ml TRAIL, 50 mM Sorbitol; D2: 8 ng/ml TRAIL, 100 mM Sorbitol. Staurosporine D1: 2 ng/ml TRAIL, 0.375 uM Stauro; D2: 4 ng/ml TRAIL, 0.75 uM Stauro
Table 1. Many different drugs can synergize with TRAIL.
Drugs were combined with TRAIL in different dose combinations (see methods section) and synergy determined using the Calcusyn program. All drugs displayed synergy, however some drugs were significantly more synergistic than others.
| Drug | Type of Drug | Doses | CI | Synergy |
|---|---|---|---|---|
| Bortezomib | Proteasome inhibitor | 1 | 0.049 | +++ |
| 2 | 0.123 | +++ | ||
| MG132 | Proteasome inhibitor | 1 | 0.313 | ++ |
| 2 | 0.299 | +++ | ||
| Doxorubicin | Topoisomerase inhibitor | 1 | 0.087 | +++ |
| 2 | 0.092 | +++ | ||
| 5-FU | Inhibitor of thymidine synthesis | 1 | 0.463 | ++ |
| 2 | 0.366 | ++ | ||
| Etoposide | Topoisomerase II inhibitor | 1 | 0.065 | +++ |
| 2 | 0.068 | +++ | ||
| Oxamflatin | HDAC inhibitor | 1 | 0.195 | +++ |
| 2 | 0.287 | +++ | ||
| Sorafenib | Selective kinase inhibitor | 1 | 0.795 | + |
| 2 | 0.316 | ++ | ||
| Azacitidine | DNA/RNA Methyltransferase inhibitor | 1 | 0.324 | ++ |
| 2 | 0.189 | +++ | ||
| Sorbitol | Hyperosmotic agent | 1 | 0.089 | +++ |
| 2 | 0.002 | +++ | ||
| Staurosporine | General kinase inhibitor | 1 | 0.307 | ++ |
| 2 | 0.175 | +++ |
Cell Death Assays
Cells were plated at 40,000 cells per well in triplicate in a 96 well plate and allowed to sit 24 hrs prior to treatment. Drug was added at the indicated values and an MTS Assay was performed after 24 hrs. For sensitization experiments, cells were plated at 40,000 cells per well in triplicate in a 96 well plate and allowed to sit for 4 hrs. The sensitization drug was added to all wells to a concentration to kill less than 35% of the cells on its own. 24 hrs later, TRAIL was added in a dose dependent manner. MTS cell viability assay was performed 24 hrs later.
DISC IP
1 × 107 cells were suspended in 10 ml of culture medium, incubated with lexatumumab at 1 μg/ml at 4 °C for 30 min, transferred to 37 °C for another 1 h, washed in phosphate-buffered saline three times, and then lysed in IP buffer (150 mm NaCl, 20 mm Tris·Cl, pH 7.5/1% Triton X-100) supplemented with complete protease inhibitors (Roche Applied Science). After the lysates were centrifuged (15 min at 13,000 rpm), antibodies were precipitated at 4 °C overnight. The beads were washed three times with IP buffer supplemented with 0.5 m NaCl, and samples were subjected to Western blotting analysis.
DNA Fragmentation Assay
Cells were pretreated with MG132 (0.35 uM) for 24 hrs. 5 × 106 cells were suspended in 10 mls of culture medium and treated with 150 ng/ml TRAIL for 4 hrs. After treatment cells were washed with ice cold PBS and lysis buffer (20 mM EDTA, 10mM Tris pH 8.0, 200 mM NaCl, 0.2% Triton-X 100, RNase A 0.5 mg/ml) was added for 2 hrs @ 37 °C. Proteinase K (5 mg/ml) was added and incubated at 56 °C overnight. DNA was precipitated and run on 1.6% agarose gel for 3 hrs at 75 V.
Results
TRAIL synergizes with multiple agents in TRAIL-sensitive BJAB cells
Although many different kinds of drugs have been reported to synergize with TRAIL (1, 2, 9), there are few reports where the same tumor cells have been treated with different classes of anti-cancer drug along with TRAIL to compare the extent of synergy between different kinds of agents. To do this, we compared the sensitivity of BJAB cells to TRAIL and different kinds of anti-cancer drug alone and in combination. To determine the extent of synergy, we used different dose combinations and calculated synergy using the Calcusyn program, which assesses synergy using the median effect principal to calculate the Combination Index (CI) where CI=1 indicates additivity, CI<1 indicates synergy and CI>1 indicates antagonism. Table 1 shows CI values for different dose combinations for various anti-cancer drugs including; proteasome inhibitors, DNA damaging agents, anti-metabolites, deactylase inhibitors, a selective protein kinase inhibitor and an inhibitor of methyltransferases. Additionally, we compared two general apoptotic stimuli, sorbitol, which induces hyperosmolar stress and staurosporine, which is a broad kinase inhibitor. All these agents caused synergy with TRAIL as indicated by CI values < 1. For example the topoisomerase inhibitor etoposide led to very high synergy (CI<0.1) while the proteasome inhibitor MG132 produced strong synergy (CI~ 0.3) and the kinase inhibitor sorafinib, was somewhat less effective (CI 0.3–0.7). These data indicate that in the same tumor cells, different anti-cancer agents, and even general pro-apoptotic stimuli all synergize with TRAIL although to different extents.
Acquired TRAIL resistance in BJAB cells is overcome by only some synergizing agents
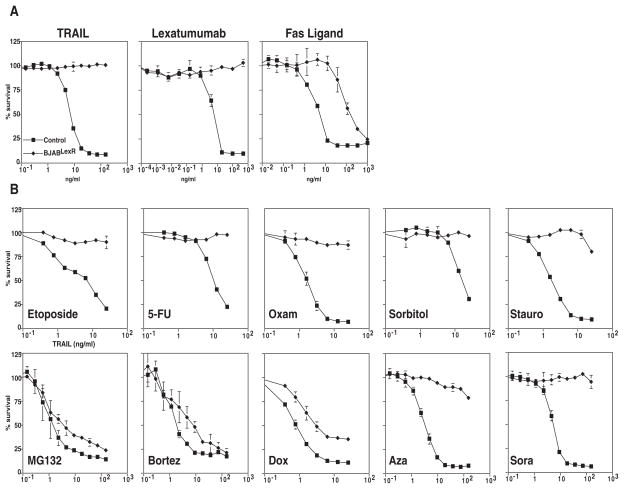
To test if acquired TRAIL resistance could be overcome by combining with the other agents, we selected BJAB cells for resistance to TRAIL-targeted therapeutics by treating with increasing doses of the TRAIL R2/DR5 agonistic antibody Lexatumumab. Fig. 1A shows that these cells (named BJABLexR) displayed essentially complete resistance to TRAIL and Lexatumumab and were also partially resistant to Fas Ligand, which works through a different receptor but a similar downstream pathway. To test if drugs that synergize with TRAIL could overcome this acquired resistance, we treated with increasing doses of TRAIL in the presence of a low dose of each of the synergizing agents. To ensure that we examined the effect on TRAIL sensitivity, we normalized each dose response curve in the absence of the sensitizing drug compared with sensitizing doses of each of the other treatments that resulted in no more than 35% cell death on its own in parental BJAB cells. The parental BJAB and BJABLexR cells displayed similar sensitivity to all the cytotoxic agents used (Supp. Fig. 1). Fig. 1B shows that most of the synergistic drugs were unable to sensitize the BJABLexR cells to TRAIL-induced death. Two types of drugs were however able to cause TRAIL sensitivity in the BJABLexR cells; doxorubicin caused some increase in TRAIL sensitivity and the proteasome inhibitors MG132 and Bortezomib both caused the resistant cells to display dose-dependent TRAIL sensitivity close to that observed with wildtype parental BJAB cells. Despite the fact that some agents (e.g. etoposide) induced stronger synergy with TRAIL than the proteasome inhibitors in parental BJABs, resistance was not overcome by the more synergistic drug. Because etoposide was significantly more synergistic than proteasome inhibitor, but proteasome inhibitors were effective at overcoming acquired resistance and etoposide was not, further experiments focused on these two treatments.
Figure 1. Only some synergizing agents can overcome acquired TRAIL resistance.
Panel a, MTS Assay quantifying cell viability of BJABLexR cells in response to TRAIL, Lexatumumab, and Fas Ligand indicating that LexR cells display complete resistance to TRAIL and Lexatumumab and partial resistance to Fas ligand. 1b MTS Assay quantifying cell viability of BJABLexR cells after treatment with sensitizing drugs. Cells were pretreated with drugs Etoposide (1.5 μM), 5-FU (100 nM), Oxamflatin (1μg/ml), Sorbitol (5 mM), Staurosporine (0.1 μM), MG132 (0.5 μM), Bortezomib (10 pM), Doxorubicin (2.5 ng/ml), Azacitadine (5 nM), or Sorafenib (7 nM) for 24 hrs prior to TRAIL treatment. Cell viability was determined using MTS assays 24 hours later indicating that although all the agents caused synergy with TRAIL in parental cells, most do not overcome TRAIL resistance in LexR cells.
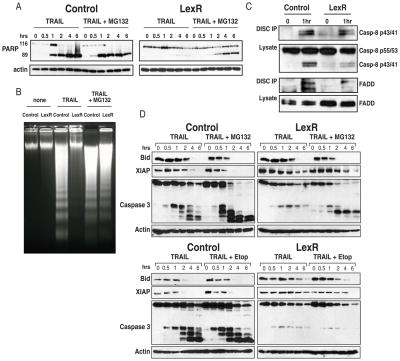
To determine if the sensitivity caused by proteasome inhibition was associated with altered signaling downstream of the TRAIL receptors, we analyzed proximal events in TRAIL R signaling in matched wildtype and BJABLexR cells. Parental BJAB cells were similarly sensitive to TRAIL whether or not Bid was knocked down (Supp. Fig. 2) indicating that these cells are Type I cells that do not require amplification through the mitochondrial pathway; however Bid cleavage (Fig. 2D) and cytochrome c release (data not shown) occurs after TRAIL R activation in the parental cells meaning that Bid cleavage is a useful way to monitor activation of the caspase-8. Figure 2A shows PARP cleavage in control parental cells and the BJABLexR cells after treatment with TRAIL with or without MG132. In the BJABLexR cells, PARP cleavage only occurred when MG132 was present but was reduced compared with parental cells. Similarly apoptotic cleavage of DNA was only observed in the BJABLexR cells when MG132 was added (Fig. 2B). TRAIL R stimulation leads to DISC formation, caspase-8 activation and cleavage leading to Bid cleavage, XIAP degradation and caspase-3 cleavage. We therefore examined the kinetics of these responses in parental BJABs and the BJABLexR cells with and without MG132 (Fig. 2D upper panel) or etoposide (Fig. 2D lower panel). In the resistant cells with MG132 or etoposide, TRAIL R signaling leads to similar cleavage of full-length Bid and XIAP with relatively small differences in the extent or kinetics of these proximal signaling events in the presence or absence of MG132 or etoposide. As expected, there was a marked difference in caspase-3 cleavage with efficient cleavage in control cells but only MG132-treated BJABLexR cells displaying extensive caspase-3 cleavage. Additionally, both BJAB and BJABLexR cells displayed similar DISC formation as determined by recruitment of FADD and caspase-8 to precipitated DR5 (Fig. 2C). These data indicate that the proximal events in TRAIL R signaling are intact in the resistant BJABLexR cells and the mechanism by which proteasome inhibition overcomes resistance is downstream of DISC formation and initial signaling events but upstream of caspase-3 cleavage.
Figure 2. LexR cells have functional DISC formation and proximal signaling events.
PARP Western blot analysis of parental BJAB cells and BJABLexR cells treated with TRAIL after pre-treatment with MG132 (0.35μM) (panel A). Panel B, DNA Fragmentation Assay of parental BJAB cells and BJABLexR cells. TRAIL was added at a concentration of 150 ng/ml after pre-treatment with MG132 (0.35μM) and treated for 4 hrs to assess apoptosis. Panel C, DISC IP using Lexatumumab in BJAB and BJABLexR cells; the western blot was probed with anti-FADD and anti-caspase-8 indicating no apparent difference in FADD and caspase-8 recruitment. Panel D, TRAIL R signaling events in parental BJAB cells and BJABLexR cells treated with TRAIL after pre-treatment with MG132 (0.35μM) or etoposide (0.75μM). TRAIL was added at 150 ng/ml for the indicated times before harvesting in RIPA buffer. These data show that proximal TRAIL R signaling events are functional in the LexR cells with acquired TRAIL resistance and that the block in signaling lies between caspase-8 and caspase-3.
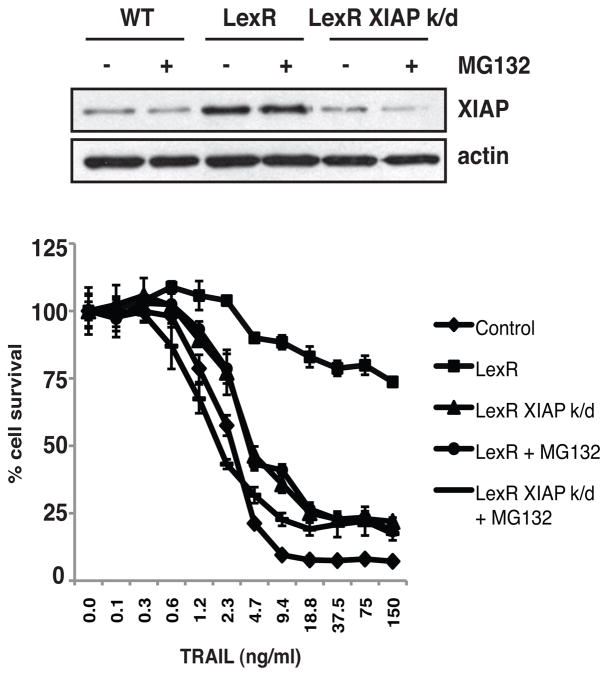
One potential mechanism by which the BJABLexR cells could become resistant to TRAIL is through increased levels of XIAP, which were higher in the BJABLexR cells as shown by comparison of the 0hr time point in Fig. 2D, and which was degraded more slowly in the BJABLexR cells. XIAP can determine both the kinetics and extent of apoptosis after TRAIL R signaling (17) and which has been identified as a potential mechanism through which proteosome inhibitors can sensitize to TRAIL (18). The combination of bortezimib and TRAIL R antibodies has also been shown to be more effective in vivo through a mechanism that was reported to involve increased activity of caspase-8 and caspase-3 but not necessarily more mitochondrial depolarization (19). Figure 3 shows that BJABLexR cells had higher levels of XIAP than the parental BJAB cells. Treatment with MG132 caused a small reduction in the amount of XIAP, but this was still above that in the parental cells. Knockdown of XIAP in the BJABLexR cells to levels equal to that in the parental line led to increased sensitivity to TRAIL, that was further increased if MG132 was added as well. These data indicate that XIAP elevation may contribute to but does not fully explain the resistance, and that the ability to overcome resistance by the proteasome inhibitor involves but is not fully explained by reduction in XIAP.
Figure 3. XIAP contributes but does not fully explain TRAIL resistance.
Western blot analysis of parental BJAB cells and BJABLexR cells treated with TRAIL after pre-treatment with MG132 (0.35μM). XIAP was knocked down in BJABLexR cells using pGIPZ Lentiviral shRNA. MTS Assay quantifying cell viability of BJAB parental and BJABLexR cells after treatment with TRAIL.
Different methods of TRAIL resistance can be overcome by proteasome inhibition
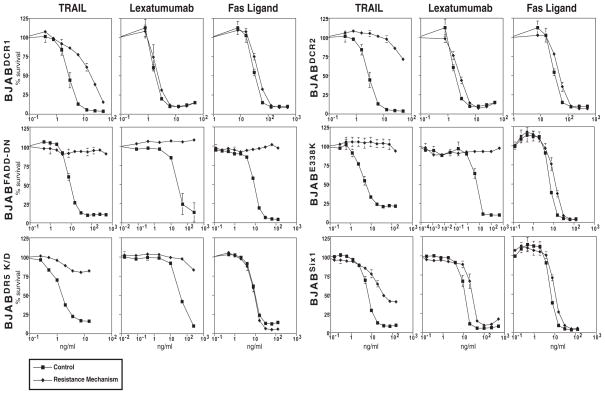
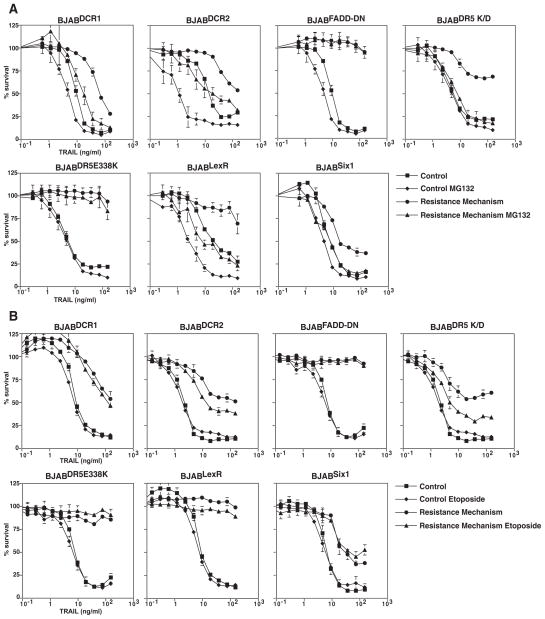
Since reduction of the elevated XIAP levels only partly explains the ability of proteasome inhibitors to overcome resistance in the BJABLexR cells, we hypothesized that by targeting the proteasome, it may be possible to overcome mechanistically distinct ways of conferring TRAIL resistance. To test this hypothesis, we tested various different TRAIL resistance mechanisms that have been found in human tumors (8). We tested a variety of different mechanisms including an inactivating mutation in DR5, E338K, which acts as a dominant negative mutant, increased expression of two TRAIL decoy receptors, DcR1 and DcR2, reduced expression of DR5 and expression of the transcription factor Six1. We also expressed a dominant negative FADD molecule (FADD-DD) that blocks all TRAIL receptor signaling. Figure 4 shows that each of these manipulations caused TRAIL resistance (albeit to different extents). To test if the various resistance mechanisms could be overcome, we compared proteasome inhibition and etoposide treatment. MG132 treatment caused TRAIL sensitivity in the resistant lines equivalent to at least that seen with the parental BJAB cells for all the resistance mechanisms except FADD-DD and the mutant DR5 receptor. However, etoposide treatment did not. The failure of proteasome inhibitors to overcome resistance by FADD-DD is expected because this mutant prevents recruitment of endogenous FADD to the DISC and thus blocks all signaling, additionally it has been reported that dominant negative FADD expressing cells are resistant to proteasome inhibitors in the absence of exogenous TRAIL (20). Failure to overcome resistance in the BJABDR5E338K cells is also expected because we previously showed that this mutant inhibits TRAIL signaling by DR4 through competition for TRAIL binding (15). Decoy receptor-induced resistance was overcome, presumably because despite their name, these receptors work not just by competing for TRAIL binding but instead by forming heteromeric complexes with the signaling receptors and altering their downstream signaling capacity (21). These data show that distinct mechanisms of TRAIL resistance can be overcome by proteasome inhibition but not etoposide treatment and that this extends to mechanisms that act at the receptor and those that activate downstream of the DISC.
Figure 4. Different mechanisms can confer TRAIL resistance.
Matched cell lines BJABDCR1, BJABDCR2, BJABFADD-DN, BJABDR5E338K, BJABDR5 K/D, BJABSix1 were treated with TRAIL, Lexatumumab, or Fas Ligand for 24 hrs and viability determined by MTS assay. All the resistance mechanisms conferred TRAIL resistance to various degrees.
Discussion
Our experiments and data from other groups show that TRAIL can synergize with various different kinds of anti-cancer treatment when tested in tumor cells that are TRAIL sensitive. Indeed we find strong synergy even with general apoptotic stimuli such as increased hyperosmolar stress. The ability of a wide variety of agents to synergize with TRAIL suggests that there is no common underlying mechanism such as increased receptor expression– instead these data are more consistent with synergy being caused by the addition of distinct apoptotic stimuli moving the tumor cells closer to their apoptotic threshold thus making it easier for the added signal coming from TRAIL receptor activation to push the cell over the threshold.
However, many tumor cells display resistance to TRAIL and, as shown here, susceptible tumor cells can easily acquire resistance. Thus given the wide variety of agents that can synergize with TRAIL in sensitive cells, the question arises whether these agents can also work effectively with TRAIL in resistant cells. Our data show that achieving synergy in susceptible tumor cells and sensitizing resistant tumor cells are not the same and it may be better to design combination therapies with agents like the proteasome inhibitors that are able to do both rather than develop combinations with drugs like etoposide that while very good at synergizing with TRAIL are not good at overcoming resistance. Proteasome inhibitors may be able to do this because in addition to moving the tumor cells closer to their apoptotic threshold, inhibition of the proteasome affects multiple apoptosis regulators and thus increases the likelihood that any specific resistance mechanism can be bypassed. The fact that proteasome inhibition can overcome resistance in matched cells that occurs both downstream of the DISC (e.g. with BJABSix1 and BJABLexR cells) and at the DISC (e.g. with BJABDCR1, BJABDCR2 cells) supports this idea. Additional support comes from the fact that other studies of the ability of proteasome inhibitors to synergize with TRAIL have identified different alterations in the TRAIL signaling pathway associated with proteasome inhibition including reduced degradation of effector caspases (17), altered Bcl-xL levels (22), increased expression of receptors (23), and increased caspase activity (19). It has also been reported that a group of primary glioma cells from different patients can all be sensitized irrespective of the heterogeneity of the tumors (5). Together these results from diverse tumor types along with our experiments in matched cells suggest that the reason proteasome inhibitors are not only good at synergizing but are also good at overcoming TRAIL resistance is due to their ability to target different resistance mechanisms at the same time, thus making it more likely that any given mechanism will be affected by the drug combination.
Recent results of randomized Phase II trials of TRAIL R-targeted drugs reported at the 2010 ASCO meeting were not encouraging; combinations with cytotoxic chemotherapy with or without bevacizumab did not demonstrate additional benefit from the TRAIL R targeted drug in non-small cell lung cancer (NSCLC) patients (10–12). However, these combinations were not chosen because the other drugs were able to overcome TRAIL resistance, rather the origination of these treatments was that (in cells that are susceptible to TRAIL) the cytotoxic agents could synergize and the drugs were already used to treat NSCLC. Our data therefore demonstrate the need to understand the underlying reason for resistance if one wants to use “targeted” agents like TRAIL R agonists. For example, one resistance mechanism that has been found in human tumors (somatic mutation in DR5) was not overcome with the proteasome inhibitors, however we previously showed that the inhibitory effect of the DR5 mutation could be bypassed through the use of a TRAIL receptor agonist (Mapatumumab) that is targeted only to DR4 (15). However, the practical importance of this kind of approach may be limited because such TRAIL Receptor mutations are rare in human tumors (24–27). On the other hand, another resistance mechanism (Six1 overexpression) that can be overcome by combination with proteasome inhibitors is common, being found in more than 60% of metastatic ovarian cancers (28) and up to 90% of metastatic breast cancers (29, 30) and associated with poor clinical outcomes in lymphoma and other cancer types (31). These data suggest that optimal use of TRAIL R targeted drugs will require that we identify resistance mechanisms in a person’s tumor and use strategies like selective targeting of DR4 in DR5 mutant tumors when they are justified, but that in more common cases (such as with Six1 overexpressing tumors), we maximize both the amount of tumor killing and the ability to overcome resistance by combining TRAIL R-targeted drugs with agents that not only synergize but are also able to overcome resistance and, especially, resistance that is driven by as more than one molecular mechanism if possible. Proteasome inhibitors may be a good place to start with this kind of approach.
Supplementary Material
Figure 5. Sensitization experiments with TRAIL resistant cell lines.
Parental BJAB cells and matched cell lines with different mechanisms of TRAIL resistance were pretreated with MG132 or etoposide 24 hrs prior to TRAIL treatment, then treated with increasing doses of TRAIL and viability determined by MTS assay after a further 24 hours. MG132 caused TRAIL sensitivity equivalent to the parental BJAB cells for all resistant cells except the BJABFADD-DN, BJABDR5E338K lines. Etoposide treatment failed to overcome TRAIL resistance for any of the resistant cell lines.
Acknowledgments
Supported by NIH grants CA111421 (A.T.) and CA124545 (A.T., K.B. & H.L.F.). and Flow cytometry and Tissue Culture shared resources supported by the University of Colorado Cancer Center Support Grant P30 CA46934.
References
- 1.Ashkenazi A, Herbst RS. To kill a tumor cell: the potential of proapoptotic receptor agonists. J Clin Invest. 2008;118:1979–90. doi: 10.1172/JCI34359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer. 2008;8:782–98. doi: 10.1038/nrc2465. [DOI] [PubMed] [Google Scholar]
- 3.Thorburn A. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) pathway signaling. J Thorac Oncol. 2007;2:461–5. doi: 10.1097/JTO.0b013e31805fea64. [DOI] [PubMed] [Google Scholar]
- 4.Walczak H, Miller RE, Ariail K, et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med. 1999;5:157–63. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 5.Koschny R, Holland H, Sykora J, et al. Bortezomib sensitizes primary human astrocytoma cells of WHO grades I to IV for tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Clin Cancer Res. 2007;13:3403–12. doi: 10.1158/1078-0432.CCR-07-0251. [DOI] [PubMed] [Google Scholar]
- 6.MacFarlane M, Harper N, Snowden RT, et al. Mechanisms of resistance to TRAIL-induced apoptosis in primary B cell chronic lymphocytic leukaemia. Oncogene. 2002;21:6809–18. doi: 10.1038/sj.onc.1205853. [DOI] [PubMed] [Google Scholar]
- 7.Lane D, Cartier A, L’Esperance S, Cote M, Rancourt C, Piche A. Differential induction of apoptosis by tumor necrosis factor-related apoptosis-inducing ligand in human ovarian carcinoma cells. Gynecol Oncol. 2004;93:594–604. doi: 10.1016/j.ygyno.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 8.Thorburn A, Behbakht K, Ford H. TRAIL receptor-targeted therapeutics: Resistance mechanisms and strategies to avoid them. Drug Resist Updat. 2008;11:17–24. doi: 10.1016/j.drup.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shankar S, Srivastava RK. Enhancement of therapeutic potential of TRAIL by cancer chemotherapy and irradiation: mechanisms and clinical implications. Drug Resist Updat. 2004;7:139–56. doi: 10.1016/j.drup.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Blackhall FH, Mark Z, Zatloukal P, et al. A randomized phase II study of paclitaxel (P) and carboplatin (C) +/− bevacizumab (B) +/− dulanermin (D) in non-small cell lung cancer (NSCLC) J Clin Oncol. 2010;28:7s. doi: 10.1200/JCO.2011.37.2623. (suppl: abstr 7534) [DOI] [PubMed] [Google Scholar]
- 11.Von Pawel J, Harvey JH, Spigel DR, et al. A randomized phase II trial of mapatumumab, a TRAIL-R1 agonist monoclonal antibody, in combination with carboplatin and paclitaxel in patienstbwith advanced NSCLC. J Clin Oncol. 2010;28:7s. (suppl; abstract LBA7501) [Google Scholar]
- 12.Karapetis CS, Clingan PR, Leighl NB, Durbin-Johnson B, O’Neill V, Spigel DR. Phase II study of PRO95780 plus paclitaxel, carboplatin, and bevacizumab (PCB) in non-small cell lung cancer (NSCLC) J Clin Oncol. 2010;28:7s. (suppl; abstr 7535) [Google Scholar]
- 13.Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19–30. doi: 10.1016/s1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 14.DSMZ cell line data base. Available from http://www.dsmz.de/
- 15.Bin L, Thorburn J, Thomas LR, Clark PE, Humphreys R, Thorburn A. Tumor-derived mutations in the TRAIL receptor DR5 inhibit TRAIL signaling through the DR4 receptor by competing for ligand binding. J Biol Chem. 2007;282:28189–94. doi: 10.1074/jbc.M704210200. [DOI] [PubMed] [Google Scholar]
- 16.Thomas LR, Bender LM, Morgan MJ, Thorburn A. Extensive regions of the FADD death domain are required for binding to the TRAIL receptor DR5. Cell Death Differ. 2006;13:160–2. doi: 10.1038/sj.cdd.4401714. [DOI] [PubMed] [Google Scholar]
- 17.Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leverkus M, Sprick MR, Wachter T, et al. Proteasome inhibition results in TRAIL sensitization of primary keratinocytes by removing the resistance-mediating block of effector caspase maturation. Mol Cell Biol. 2003;23:777–90. doi: 10.1128/MCB.23.3.777-790.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shanker A, Brooks AD, Tristan CA, et al. Treating metastatic solid tumors with bortezomib and a tumor necrosis factor-related apoptosis-inducing ligand receptor agonist antibody. J Natl Cancer Inst. 2008;100:649–62. doi: 10.1093/jnci/djn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller CP, Ban K, Dujka ME, et al. NPI-0052, a novel proteasome inhibitor, induces caspase-8 and ROS-dependent apoptosis alone and in combination with HDAC inhibitors in leukemia cells. Blood. 2007;110:267–77. doi: 10.1182/blood-2006-03-013128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merino D, Lalaoui N, Morizot A, Schneider P, Solary E, Micheau O. Differential inhibition of TRAIL-mediated DR5-DISC formation by decoy receptors 1 and 2. Mol Cell Biol. 2006;26:7046–55. doi: 10.1128/MCB.00520-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson TR, Stone K, Nikrad M, et al. The proteasome inhibitor PS-341 overcomes TRAIL resistance in Bax and caspase 9-negative or Bcl-xL overexpressing cells. Oncogene. 2003;22:4953–63. doi: 10.1038/sj.onc.1206656. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Yue P, Chen S, et al. The proteasome inhibitor PS-341 (bortezomib) up-regulates DR5 expression leading to induction of apoptosis and enhancement of TRAIL-induced apoptosis despite up-regulation of c-FLIP and survivin expression in human NSCLC cells. Cancer Res. 2007;67:4981–8. doi: 10.1158/0008-5472.CAN-06-4274. [DOI] [PubMed] [Google Scholar]
- 24.Lee SH, Shin MS, Kim HS, et al. Somatic mutations of TRAIL-receptor 1 and TRAIL-receptor 2 genes in non-Hodgkin’s lymphoma. Oncogene. 2001;20:399–403. doi: 10.1038/sj.onc.1204103. [DOI] [PubMed] [Google Scholar]
- 25.Pai SI, Wu GS, Ozoren N, et al. Rare loss-of-function mutation of a death receptor gene in head and neck cancer. Cancer Res. 1998;58:3513–8. [PubMed] [Google Scholar]
- 26.Seitz S, Wassmuth P, Fischer J, et al. Mutation analysis and mRNA expression of trail-receptors in human breast cancer. Int J Cancer. 2002;102:117–28. doi: 10.1002/ijc.10694. [DOI] [PubMed] [Google Scholar]
- 27.Wu WG, Soria JC, Wang L, Kemp BL, Mao L. TRAIL-R2 is not correlated with p53 status and is rarely mutated in non-small cell lung cancer. Anticancer Res. 2000;20:4525–9. [PubMed] [Google Scholar]
- 28.Behbakht K, Qamar L, Aldridge CS, et al. Six1 Overexpression in Ovarian Carcinoma Causes Resistance to TRAIL-Mediated Apoptosis and Is Associated with Poor Survival. Cancer Res. 2007;67:3036–42. doi: 10.1158/0008-5472.CAN-06-3755. [DOI] [PubMed] [Google Scholar]
- 29.Coletta RD, Christensen K, Reichenberger KJ, et al. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci U S A. 2004;101:6478–83. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reichenberger KJ, Coletta RD, Schulte AP, Varella-Garcia M, Ford HL. Gene amplification is a mechanism of Six1 overexpression in breast cancer. Cancer Res. 2005;65:2668–75. doi: 10.1158/0008-5472.CAN-04-4286. [DOI] [PubMed] [Google Scholar]
- 31.Micalizzi DS, Christensen KL, Jedlicka P, et al. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-beta signaling. J Clin Invest. 2009;119:2678–90. doi: 10.1172/JCI37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.