Abstract
Background
Although both the alkylating agent temozolomide (TMZ) and oncolytic viruses hold promise for treating glioblastoma, which remains uniformly lethal, the effectiveness of combining the two treatments and the mechanism of their interaction on cancer stem cells are unknown.
Methods
We investigated the efficacy of combining TMZ and the oncolytic herpes simplex virus (oHSV) G47Δ in killing glioblastoma stem cells (GSCs), using Chou–Talalay combination index analysis, immunocytochemistry and fluorescence microscopy, and neutral comet assay. The role of treatment-induced DNA double-strand breaks, activation of DNA damage responses, and virus replication in the cytotoxic interaction between G47Δ and TMZ was examined with a panel of pharmacological inhibitors and short-hairpin RNA (shRNA)–mediated knockdown of DNA repair pathways. Comparisons of cell survival and virus replication were performed using a two-sided t test (unpaired). The survival of athymic mice (n = 6–8 mice per group) bearing GSC-derived glioblastoma tumors treated with the combination of G47Δ and TMZ was analyzed by the Kaplan–Meier method and evaluated with a two-sided log-rank test.
Results
The combination of G47Δ and TMZ acted synergistically in killing GSCs but not neurons, with associated robust induction of DNA damage. Pharmacological and shRNA-mediated knockdown studies suggested that activated ataxia telangiectasia mutated (ATM) is a crucial mediator of synergy. Activated ATM relocalized to HSV DNA replication compartments where it likely enhanced oHSV replication and could not participate in repairing TMZ-induced DNA damage. Sensitivity to TMZ and synergy with G47Δ decreased with O6-methylguanine-DNA-methyltransferase (MGMT) expression and MSH6 knockdown. Combined G47Δ and TMZ treatment extended survival of mice bearing GSC-derived intracranial tumors, achieving long-term remission in four of eight mice (median survival = 228 days; G47Δ alone vs G47Δ + TMZ, hazard ratio of survival = 7.1, 95% confidence interval = 1.9 to 26.1, P = .003) at TMZ doses attainable in patients.
Conclusions
The combination of G47Δ and TMZ acts synergistically in killing GSCs through oHSV-mediated manipulation of DNA damage responses. This strategy is highly efficacious in representative preclinical models and warrants clinical translation.
CONTEXTS AND CAVEATS
Prior knowledge
Glioblastoma multiforme (GBM) is the most common primary brain tumor in adults. The alkylating agent temozolomide (TMZ), which is part of the current standard of care, along with radiation therapy, extends survival by only a few months compared with radiation alone. Oncolytic herpes simplex viruses have been safely administered to patients with GBM, but the combination with TMZ is untested.
Study design
The combination of the oncolytic herpes simplex virus G47Δ with TMZ was tested in glioblastoma stem cells (GSCs), which were assessed for cell survival, virus replication, and DNA damage responses. The survival of athymic mice with GSC-derived glioblastoma tumors was also assessed after treatment with the G47Δ/TMZ combination.
Contribution
The combined treatment was effective in inducing a robust DNA damage response and killing GSCs, and the results suggest that the two agents act synergistically. The combination of the oncolytic virus with TMZ also statistically significantly extended the survival of mice with intracranial tumors compared with control mice and those treated with virus or TMZ alone.
Implication
The combination of the oncolytic virus G47Δ with TMZ may be a more potent treatment for GBM than either agent alone.
Limitations
The five GSCs that were examined differed in their sensitivity to TMZ, and therefore, the efficacy of the combined treatment will need to be tested in other GSC lines. Only immune-deficient mice were assessed, and therefore, the efficacy of the treatment in immune-competent models and patients may be different.
From the Editors
Glioblastoma multiforme (GBM), the most common primary brain tumor in adults, is invariably fatal despite the current optimal multimodal therapy, with the median survival (12–15 months) having barely improved since the 1980s (1). The alkylating agent temozolomide (TMZ) is part of the current standard of care, extending survival by a few months compared with radiation alone (2). The clinical benefits of TMZ are associated with epigenetic silencing of the O6-methylguanine-DNA-methyltransferase (MGMT) gene (3,4). Although the inactivating pseudosubstrates of MGMT, O6-benzylguanine (BG) and Lomeguatrib (LM), can inhibit MGMT activity (5), hematological toxic effects and lack of increased efficacy at tolerable doses have substantially limited their utility in the clinic (6).
GBM stem cells (GSCs), which have been recently isolated, form orthotopic tumors in mice, which closely resemble patients’ tumors genotypically and histopathologically, in contrast to GBM cell lines and primary serum-cultured glioma cells (7,8). Accumulating evidence suggests that GSCs are important in disease initiation, progression, recurrence, and resistance to radiation and chemotherapy (9–11). Therefore, targeting GSCs provides an important avenue for the development of much needed novel therapeutic strategies against GBM. It remains controversial whether GSCs are resistant to TMZ (12–14).
Oncolytic viruses are a new class of cancer therapeutics still awaiting successful incorporation into current standard therapies. Although oncolytic herpes simplex viruses (oHSVs) (eg, G207 [γ134.5−, ICP6−]) were among the first to be safely administered to patients with recurrent malignant glioma (15), evidence for clinical efficacy has been limited. Given the distinct mode of cell killing used by oHSV, combination with chemotherapy represents a promising and practical strategy to improve efficacy (16). Indeed, we previously reported that oHSV G207 synergizes with TMZ in killing GBM cell lines (17). However, we recently demonstrated that the γ134.5 deletion severely compromises oHSV replication in GSCs (18). A G207-derivative virus, G47Δ, exhibits enhanced antitumor activity while maintaining as favorable a safety profile as G207 (18,19), and it is now in clinical trial for progressive GBM (20). G47Δ replicates in and kills GSCs; however, its efficacy was transitory and unable to eradicate GSC-derived tumors in vivo (18), indicating the difficulty of achieving long-term remission by currently available oHSVs alone.
In this study, we combined oHSV-G47Δ and TMZ to target GSCs and to develop an effective and translatable strategy for GBM. We assessed treatment-induced DNA double-strand breaks (DSBs) and activation of DNA damage responses. We examined the role of DNA damage responses in the cytotoxic interaction between G47Δ and TMZ and evaluated the combinatorial strategy in vivo with mouse models of GSC-derived glioblastoma.
Materials and Methods
Cells
Human glioma (U87, U373, and T98) and Vero (African green monkey kidney) cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and were typically used within 15 passages from ATCC stock and verified by cell morphology in culture. U87/mp53 and U87/MGMT were isolated after transfection of U87 cells with pc53-SCX3 (human p53 cDNA with dominant Val-143 to Ala mutation) or pcDNA3-MGMT (MGMT cDNA cloned into pcDNA3) and selection for G418 resistance as described elsewhere (17). All cells except human neurons and GSCs were maintained in Dulbecco's modified Eagle medium (DMEM) with 4.5 g/L glucose, L-glutamine, and sodium pyruvate (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (Hyclone Thermo Scientific, Logan, UT). Human neurons (immature) were obtained from ScienCell (San Diego, CA) and maintained in neuronal medium (ScienCell). All cells were cultured at 37°C and 5% CO2. The isolation and characterization of GBM4, GBM6, GBM8, and BT74 GSCs were described previously, with GBM13 being isolated similarly (18). Briefly (with the exception of BT74), GBM surgical specimens, with Institutional Review Board approval, were mechanically minced, digested with trypsin and DNase I, strained and plated in EF20 medium composed of Neurobasal medium (Invitrogen, Carlsbad, CA) supplemented with 3 mM L-Glutamine (Mediatech), 1× B27 supplement (Invitrogen), 0.5× N2 supplement (Invitrogen), 2 μg/mL heparin (Sigma, St Louis, MO), 20 ng/mL recombinant human epidermal growth factor (R&D Systems, Minneapolis, MN), 20 ng/mL recombinant human fibroblast growth factor-2 (PeproTech, Rocky Hill, NJ), and 0.5× penicillin G/streptomycin sulfate/amphotericin B complex (Mediatech), and fed every third day with one-third of the volume of fresh medium. Spheres were dissociated using NeuroCult Chemical Dissociation kit (StemCell Technologies, Vancouver, BC, Canada). GBM# refers to a passaged GSC culture isolated from an individual patient. All passaged cells were confirmed to be mycoplasma-free (LookOut mycoplasma kit; Sigma).
Viruses
HSV-1 viruses, containing the indicated mutations/insertions and derived from wild-type strain F, G207 (γ34.5Δ, ICP6−, LacZ) (21), G47Δ (γ34.5Δ, ICP6−, ICP47/Us11proΔ, LacZ+) (19), G47Δ-BAC (γ34.5Δ, ICP6−, ICP47/Us11proΔ, eGFP+) (18), FΔ6 (ICP6−, LacZ) (18), or strain KOS, d120-BAC (ICP4Δ, ICP6−, eGFP+) (22), were grown (after low multiplicity of infection [MOI]), purified (benzonase treatment, low-speed centrifugation, filtration, and high-speed centrifugation), and titered on Vero cells as described previously (22,23).
Immunocytochemistry
Dissociated GSCs or glioma cell lines were plated onto fibronectin or laminin/poly-l-ornithine-coated coverslips. After overnight incubation, the plated cells were treated with TMZ in the presence or absence of BG, as indicated. Two days later, G47Δ infection was done at MOI = 1. For phosphorylated H2A histone family member X (γH2AX/H2AFX) detection, GSCs were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) (vol/vol), 24 hours after infection. For visualization of replication compartments, infection was done at MOI = 5, and GSCs were fixed 6 hours later. The fixed cells were then permeabilized with 0.1% Triton X, washed, and blocked with 10% goat or donkey serum before incubation with primary antibodies at 4°C overnight. The primary antibodies used were mouse anti-γH2AX (1:200) (Millipore, Temecula, CA), rabbit anti-HSV (1:500) (DAKO, Carpinteria, CA), mouse anti-phospho-ATM (pS1981) (1:200) (Rockland, Gilbertsville, PA), mouse anti-Nbs1 (1:200) (Novus Biologicals, Littleton, CO), and rabbit anti-ICP8 (1:400, 3-83, a generous gift from David M. Knipe, Harvard Medical School, Boston, MA) (24). Fluorescein isothiocyanate (FITC)–conjugated or cyanine dye (Cy3)–conjugated secondary antibodies (1:200) (Jackson ImmunoResearch, West Grove, PA) were applied at 4°C for 4 hours before counterstaining with 1.5 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) in Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Cells were observed under fluorescence or confocal microscopy.
Immunoblots
GSCs or glioma cell lines were treated for 2 days with effective dose (ED40) concentrations of TMZ, treated with G47Δ at MOI = 1 for 24 hours, and cell pellets were lysed in radioimmunoprecipitation (RIPA) buffer (Boston Bioproducts, Worcester, MA) with a cocktail of protease and phosphatase inhibitors (Roche, Indianapolis, IN). For MGMT and mismatch repair protein mutS homolog 6 (MSH6) detection, untreated GSCs were lysed in RIPA buffer. Protein (20μg) was separated by 6%–15% SDS–PAGE, and transferred to PVDF membranes by electroblotting. After blocking with 5% nonfat dry milk in TBST (20 mM Tris pH7.5, 150 mM NaCl, 0.1% Tween20) for 1 hour at room temperature, membranes were incubated with primary antibodies to MGMT (mouse monoclonal, 1:1000; Sigma), MSH6 (mouse monoclonal, 1:1000; BD Transduction Laboratories, San Jose, CA), γH2AX (mouse monoclonal, 1:1000; Millipore), ATM (rabbit monoclonal, 1:1000; Cell Signaling Technology, Danvers, MA), phospho-ATM (Ser1981) (mouse monoclonal; 1:1000, Cell Signaling Technology), phospho-ATR (Ser428) (rabbit polyclonal, 1:1000; Cell Signaling Technology), ATR (rabbit polyclonal, 1:10000; Bethyl Laboratories, Montgomery, TX), ATR-interacting protein (ATRIP) (goat polyclonal, 1:1000; Santa Cruz Biotechnology, Santa Cruz, CA), DNA-dependent protein kinase catalytic subunit (DNA-PKcs) (rabbit polyclonal, 1:5000; Santa Cruz Biotechnology), actin (rabbit polyclonal, 1:10 000; Sigma), or vinculin (mouse monoclonal, 1:10000; Thermo Scientific, Rockford, IL) at 4°C overnight. Membranes were then washed in TBST and incubated with appropriate peroxidase-conjugated secondary antibodies (Promega, Madison, WI) for 30 minutes at room temperature. Protein-antibody complexes were visualized using the enhanced chemiluminescense (ECL) kit (Amersham Bioscience, Piscataway, NJ).
MGMT Methylation-Specific Polymerase Chain Reaction (PCR)
Genomic DNA from untreated GSCs and corresponding primary glioma cells cultured in serum containing media was harvested using the Gentra Puregene kit (Qiagen, Hilden, Germany) per manufacturer's protocol. DNA quantification was performed using a Nanodrop ND-1000 UV-Vis spectrophotometer (Nanodrop Technologies, Wilmington, DE). Promoter methylation analysis of MGMT was accomplished by bisulfite conversion of 500 ng of genomic DNA using the EpiTect Bisulfite conversion kit (Qiagen). This was followed by methylation-specific PCR of the converted DNA with methylated- and unmethylated-specific PCR reactions using primers previously described and validated by Esteller et al. (25). Genomic DNA from Jurkat cell line methylated excessively by SSSI (New England Biolabs, Beverly, MA) and genomic DNA from normal male donors (Promega) were used as respective positive and negative controls. The PCR products were separated in 1.5% agarose gel and visualized under ultraviolet illumination.
Chemotherapeutic Drugs and Compounds
TMZ (Schering-Plough, Kenilworth, NJ) was dissolved in 5% dimethyl sulfoxide (DMSO) in PBS (vol/vol). MGMT inhibitor O6-benzylguanine (BG) (Sigma Chemicals Co, St Louis, MO) was dissolved in DMSO (Sigma) and further diluted in EF20 medium or DMEM for in vitro studies. Another MGMT inhibitor lomeguatrib (LM) was kindly provided by Geoffrey P. Margison (Paterson Institute for Cancer Research, Cancer Research-UK, Manchester, UK) and dissolved in DMSO. Ataxia telangiectasia mutated (ATM) kinase inhibitor KU55933 (Tocris Bioscience, Ellisville, MO), ATM/ ataxia telangiectasia and Rad3-related (ATR) kinase inhibitor CGK733 (Calbiochem, San Diego, CA), cyclin-dependent kinase inhibitor UCN-01 (Sigma), checkpoint kinase 2 (Chk2) inhibitor II (Sigma), DNA-dependent protein kinase inhibitor NU7026 (Tocris Bioscience), poly-(ADP-ribose) polymerase (PARP) inhibitor XIV (Calbiochem), PARP inhibitor lithocholic acid (Sigma), and Akt inhibitor V (triciribine) (Sigma) were dissolved in DMSO. Autophagy inhibitor 3-methyladenine (3-MA) (Sigma) was dissolved in water.
Cell Susceptibility Assays and Chou–Talalay Analysis
GSCs were dissociated and seeded into 96-well plates at 5000 cells per well. Two hours later, the cells were treated with either TMZ or virus at indicated doses. For GBM13 and BT74, BG was added 2 hours before TMZ, as indicated. 72 hours after infection, MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) cell viability assays (Promega) were performed according to the manufacturer's instructions. For glioma cell lines (U87, U373), the cells were plated the day before addition of agents. For Chou–Talalay analysis, threefold serial dilutions were used, with the doses of TMZ ranging from 0.07 to 3000 μM and the doses of HSV ranging from MOI = 0.0004 to 10. Dose–response curves and effective dose (ED) values were obtained and compared at day 5. For MGMT-positive cells (GBM13, BT74, U87/MGMT, and T98), experiments were also done in the presence of BG or LM, as indicated. The experiments were repeated at least three times for each of the conditions, each time in triplicate. After the ED50 values of both agents were obtained for each cell line, both agents were then added to the cells in combination, in a ratio equal to the ratio of their ED50 values. Combined dose–response curves were fitted to Chou–Talalay lines, which are derived from the law of mass action and described by the equation log (fa/fu) = mlogD − mlogDm, in which fa is the fraction affected, fu is the fraction unaffected, D is the dose, Dm is the median-effect dose, and m is the coefficient signifying the shape of the dose–response curve. The combination indices (CIs) were calculated using the equation CI = (D1/Dx1) + (D2/Dx2) + (D1)(D2)/[(Dx1)(Dx2)], where Dx1 and Dx2 are the TMZ and HSV doses, respectively, that are required to achieve a particular fa, and D1 and D2 are the doses of the two agents (combined treatment) required for achieving the same fa (26). Fixed ratios of drug and virus concentrations and mutually exclusive equations were used to determine CIs. Values of CI equal to 1, greater than 1, and less than1 indicate additive, antagonistic, and synergistic interactions, respectively.
Viral Replication, Infectivity, and Spread Assays
GSCs were dissociated into single cell suspensions and seeded into 24-well plates at 2 × 104 cells per well in 500 μL of EF20 medium. Glioma cell lines (U87 or U373) were plated in 500 μl of DMEM–10% fetal bovine serum (FBS). TMZ was applied at the indicated concentrations 24 hours before virus infection. GSCs or glioma cell lines (U87, U373) were infected with virus at a MOI of 1.5 in the presence or absence of TMZ, and various inhibitors as indicated, in EF20 (GSCs) or DMEM–10% FBS (glioma cell lines) and harvested with supernatant at the indicated time points in triplicate. After three freeze/thaw cycles and sonication, the titers of infectious virus were determined by plaque assay on Vero cells. For virus infectivity assays, glioma cells were plated at 4 × 104 cells per well in 12-well plates, infected at a MOI of 1.5 in 500 μL of medium, and 60 minutes later, anti-HSV human immunoglobulin G was added at a final concentration of 0.2% (vol/vol) and further incubated for 15 hours. The infected cells were then fixed and stained with X-gal. For virus spread assays, GSCs were infected with G47Δ-BAC at MOI of 0.1, and green fluorescent protein intensity was measured by plate reader (Synergy HT; BioTek Instruments, Winooski, VT).
Neutral Comet Assay to Assess DSBs
GSCs (GBM4, GBM8, BT74) and U87 cells were pretreated with indicated doses of TMZ or TMZ + BG for 36 hours and then infected with G47Δ at MOI of 1. Twenty-four hours later, the treated cells were suspended in low-melting agarose (Trevigen, Gaithersburg, MD), placed onto comet slides (Trevigen), lysed when the agarose solidified, and electrophoresed according to the company's protocol for the neutral comet assay. Nuclei were labeled with SYBR green dye (Trevigen), cells were photographed using fluorescent microscopy, and tail moments (product of DNA amount in tail and distance of tail migration) from more than 100 cells per slide were calculated with CometScore software (v1.5; TriTek Corporation, Sumerduck, VA). Experiments were performed in triplicate.
Short-hairpin RNA (shRNA)–Mediated Knockdown of ATM, ATR, ATM/ATR, or MSH6
Plasmid constructs containing shRNA sequences against ATM mRNA (TRCN0000039951, TRCN0000194861) and ATR mRNA (TRCN0000196538, TRCN0000219647) or non-targeting shRNA (SHC002; should not target human or mouse genes but will engage the RNA-induced silencing complex) were obtained from Sigma. The construct against MSH6 mRNA (TRCN0000078543) was originally obtained from the Massachusetts General Hospital shRNA core (Dr Toshi Shioda, Massachusetts General Hospital, Boston, MA). Generation of lentiviral constructs for shRNA-mediated knockdown were as described in the protocol listed at the RNAi Consortium/Broad Institute website (http://www.broadinstitute.org/rnai/public/static/protocols/TRC_Protocols_Section_II_Viral_Production_100809.pdf). Briefly, 293T cells were transfected with shRNA-containing plasmids, packaging plasmid (pCMV-dR8.9), and envelope plasmid (pCMV-VSV-G) using Lipofectamine and Plus reagent (Invitrogen). Harvested medium was centrifuged to remove packaging cells, and lentivirus was pelleted by high-speed centrifugation and resuspended in EF20 medium for infection. GSCs (GBM4, GBM8, BT74) or U87 cells were infected with concentrated lentivirus or supernatants, which were removed after overnight incubation, and the cells were allowed to grow in EF20 (GSCs) or in DMEM with 10% FBS (U87) for 36 hours before selection with Puromycin (Sigma). Protein levels of target genes were assessed by Western blot analysis. The experiments were repeated at least three times for each of the conditions, each time in triplicate.
In Vivo Experiments
Female athymic mice of 6–8 weeks of age with median weight of 20 g were obtained from the National Cancer Institute (Frederick, MD). To generate intracranial xenograft models, 1 × 105 dissociated GSCs in 4 μL were stereotactically implanted into the right striatum (2.5-mm lateral from Bregma and 2.5-mm deep). For GBM8, mice were randomly assigned into four groups (n = 8 per group). On days 7, 8, and 9 after tumor implantation, mice were treated with 5 mg/kg of TMZ (groups 1, 2) or PBS (groups 3, 4) by intraperitoneal injection. On day 8, mice were treated by intratumoral injection of 2 × 106 PFU of G47Δ in 3 μL of virus buffer (150 mM NaCl, 20 mM Tris, pH 7.5) (groups 2, 3), or virus buffer (Mock, groups 1, 4) using the same burr hole and coordinates from which tumor cells were implanted. For BT74, mice were randomly assigned into six groups (n = 6 per group). On days 7, 8, and 9 after tumor implantation, mice were treated with 50 mg/kg of TMZ (Groups 1–4) or PBS (Groups 5, 6). Two groups of mice were given 0.3 mg of BG dissolved in 40% polyethylene glycol-400 (Sigma) in saline (vol/vol) (27) (groups 2, 3) or solvent alone (groups 1, 4) by intraperitoneal injection 1 hour before TMZ. On day 8, mice were treated with G47Δ (groups 3–5) or PBS (groups 1, 2, 6) as for GBM8. Mice were monitored and killed by overdose anesthesia when they became moribund, lethargic, anorexic, or developed marked neurologic symptoms. The presence of intracranial tumors was macroscopically or microscopically confirmed postmortem in all mice. All in vivo procedures were approved by the Subcommittee on Research Animal Care at Massachusetts General Hospital. Animals were housed in isolator cages with 4–5 mice per cage. For detection of γH2AX in vivo, mice (n = 3 per each group) were treated with TMZ (5 mg/kg/d for GBM8 and 50 mg/kg/d with BG [0.3 mg/d] for BT74) for 3 consecutive days starting 23 days after GSC implantation as above. G47Δ (2 × 106 PFU) was intratumorally injected on the second day of chemotherapy, and mice were killed 10 hours after the last chemotherapy. Brains were fixed with 4% PFA, and frozen sections (8 μm thickness) were doubly immunostained with anti-γH2AX (1:100) (Millipore) and anti-HSV (1:200) (DAKO), followed by FITC and Cy3-conjugated secondary antibodies (Jackson ImmunoResearch). Sections were mounted with Vectashield with DAPI (Vector Laboratories).
Statistical Analysis
Comparisons of data in cell survival, viral replication, and comet assays were performed using a two-sided t test (unpaired). The ratios of γH2AX-positive cells were compared by one-way analysis of variance using Bonferroni Multiple Comparison Test. Survival analyses were performed by Kaplan–Meier curves, and their comparison was evaluated by a two-sided log-rank (Mantel–Cox) test. P values less than .05 were considered statistically significant. Prism (GraphPad Software Inc, San Diego, CA) and Statcel2 (OMS Publishing Inc, Saitama, Japan) software packages were used for analysis.
Results
Sensitivity of Glioblastoma Stem Cells to TMZ
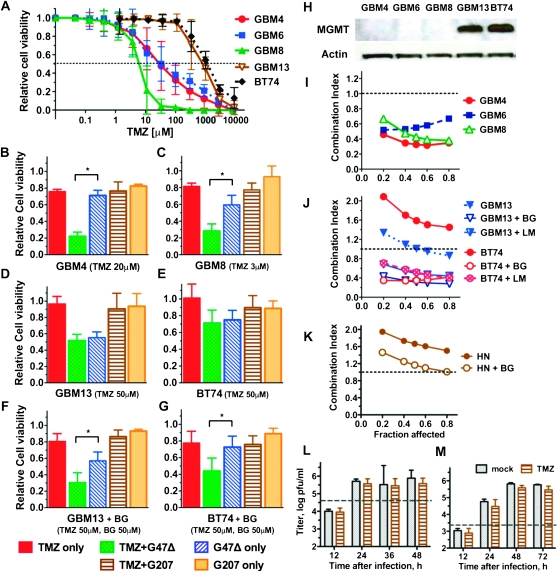
We previously isolated GSCs from patients with primary GBM to obtain a representative model of GBM (18). In tests of sensitivity to TMZ, the five GSC lines demonstrated variable sensitivities (Figure 1, A), with one GSC (GBM8) being highly sensitive (ED50 < 10 μM), two GSCs (GBM4, GBM6) being sensitive (ED50 < 50 μM), and two GSCs (GBM13, BT74) being highly resistant (ED50 > 900 μM) (Table 1). As expected, TMZ-resistant cells expressed MGMT, whereas sensitive cells did not (Figure 1, H), and MGMT promoter methylation patterns were associated with protein levels (Supplementary Figure 1, A, available online). When MGMT-positive GSCs (GBM13 and BT74) were treated with BG or LM to inhibit MGMT activity, their sensitivity to TMZ more than doubled, although TMZ was still above clinically relevant doses (Table 1, Supplementary Figure 1, B, available online).
Figure 1.
Effect of G47Δ and TMZ and their combination on GSCs in vitro. Five GSC lines (MGMT positive and negative) were tested. A) TMZ dose–response curves. GSCs were cultured in triplicate wells in vitro with various concentrations of TMZ, and cell viability was measured by MTS assay 5 days after adding TMZ. Error bars represent 95% confidence intervals. Dotted line indicates 50% cell viability relative to mock-treated cells. (B–G) Effect of TMZ and G47Δ combination on GSC killing in vitro. Five thousand cells were seeded in 96-well plates, treated with TMZ B) GBM4: TMZ, 20 μM, C) GBM8: TMZ, 3 μM, D) GBM13: TMZ, 50 μM or phosphate-buffered saline (PBS) and 24 hours later infected with G47Δ or G207 at MOI = 0.1, or mock (TMZ-only control). Cell viability was measured 3 days later by MTS assay and plotted as a percent of PBS + mock infection. F, G) MGMT-positive GSCs (GBM13, BT74) were also treated with BG (50 μM) 1 hour before TMZ. Asterisks denote statistically significant differences (B, P < .001; C, P < .001; F, P = .002; G, P = .004) (unpaired t test, two-sided). Error bars represent 95% confidence intervals. H) MGMT expression in GSCs was examined by western blot. Mouse anti-MGMT antibody (1:1000), rabbit anti-actin antibody (1:10 000), and secondary antibodies, Horseradish peroxidase (HRP)–conjugated anti-mouse immunoglobulin G (IgG) and HRP-conjugated anti-rabbit IgG (1:10 000; Promega). (I–K) The interaction between TMZ and G47Δ on cell killing was examined by the median-effect method of Chou–Talalay. Data are plotted as Fraction affected versus Combination Index (CI). I) MGMT-negative GSCs. J) MGMT-positive GSCs were also analyzed in the presence of BG (50 μM) or LM (20 μM). K) Human neurons. CI < 1, CI = 1, and CI > 1 represent synergistic, additive, and antagonistic interactions, respectively. L) Virus yield (titer) in GBM8 GSCs treated with TMZ (5 μM) or PBS (mock) after G47Δ infection at MOI = 1.5 and M) MOI = 0.1. Dashed lines indicate the infecting dose of G47Δ. No statistically significant difference between mock- and TMZ-treated cells (unpaired t test, two-sided). The experiments were repeated at least three times for each of the conditions, each time in triplicate. BG = O6-benzylguanine; GSC = glioblastoma stem cell; HN = human neurons; LM = lomeguatrib; MGMT = O6-methylguanine-DNA-methyltransferase; MOI = multiplicity of infection; MTS = (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium); pfu = plaque forming unit; TMZ = temozolomide.
Table 1.
Median-effect doses of TMZ and G47Δ in vitro*
| GSC line | ED50: drug or virus alone | |
| TMZ, μM | G47Δ, MOI | |
| GBM4 | 32 | 0.21 |
| GBM6 | 36 | 0.31 |
| GBM8 | 5.8 | 0.1 |
| GBM13 | 928 | 0.05 |
| GBM13 + BG | 325 | 0.05 |
| GBM13 + LM | 457 | 0.05 |
| BT74 | 1561 | 0.23 |
| BT74 + BG | 415 | 0.23 |
| BT74 + LM | 628 | 0.22 |
| HN | 1629 | 0.38 |
| HN +BG | 815 | 0.41 |
Sensitivity to TMZ was determined 5 days after adding TMZ. O6-methylguanine methyltransferase (MGMT)-positive GSCs (GBM13, BT74) and HN were also treated with O6-benzylguanine (BG) (50 μM) or lomeguatrib (LM) (20 μM) 2 hours before TMZ. Sensitivity to G47Δ was determined 4.5 days after infection. ED50 or Dm (dose required for 50% effect) values were determined from dose–response curves. GSC = glioblastoma stem cells; G47Δ = oncolytic herpes simplex virus; HN = human neurons; MOI = multiplicity of infection; TMZ = temozolomide.
Interaction of TMZ With oHSV in Glioma Cells In Vitro
GSCs were sensitive to killing by G47Δ (Table 1). In MGMT-negative GSCs, the combination of TMZ with G47Δ statistically significantly decreased cell viability compared with TMZ or G47Δ alone (relative cell viability, for GBM4, TMZ only vs TMZ + G47Δ: difference = 0.54, 95% CI = 0.50 to 0.57, P < .001; TMZ + G47Δ vs G47Δ only: difference = −0.49, 95% CI = −0.54 to −0.44, P < .001; Figure 1, B; for GBM8, TMZ only vs TMZ + G47Δ: difference = 0.53, 95% CI = 0.47 to 0.59, P < .001; TMZ + G47Δ vs G47Δ only: difference = −0.31, 95% CI = −0.40 to −0.22, P < .001; Figure 1, C). In MGMT-positive GSCs, the combination with TMZ did not alter viability at a TMZ dose of 50 μM (Figure 1, D and E), the peak TMZ plasma concentration reached in patients (28). BG only modestly increased TMZ sensitivity in these cells (Table 1) but statistically significantly enhanced the combination activity (relative cell viability, for GBM13 + BG, TMZ only vs TMZ + G47Δ: difference = 0.50, 95% CI = 0.40 to 0.60, P < .001; TMZ + G47Δ vs G47Δ only: difference = −0.26, 95% CI = −0.37 to −0.16, P = .002, Figure 1, F; for BT74 + BG, TMZ only vs TMZ + G47Δ: difference = 0.34, 95% CI = 0.20 to 0.47, P = .002; TMZ + G47Δ vs G47Δ only: difference = −0.29, 95% CI = −0.42 to −0.15, P = .004; Figure 1, G). GSCs are not permissive for G207 replication (18, 29), and combination with TMZ did not enhance G207 oncolytic activity (Figure 1, B–G).
To evaluate the interaction between TMZ and oHSV, we used the median-effect method of Chou and Talalay (26). TMZ and G47Δ acted synergistically in reducing the viability of MGMT-negative GSCs (Figure 1, I). FΔ6, an ICP6-mutated oHSV, was also synergistic with TMZ in GSCs, whereas G207 and replication-deficient d120-BAC were not (Supplementary Figure 1, C, available online), suggesting that oHSV replication is necessary for synergy with TMZ. The combination of G47Δ and TMZ was also synergistic in MGMT-negative glioma cell lines, including p53 wild-type U87 and mutated U373 and U87mp53 (Supplementary Figure 1, D, available online). No synergy was observed between TMZ and G47Δ in MGMT-positive GSCs (Figure 1, J) or glioma cell lines (Supplementary Figure 1, F, available online). However, in the presence of MGMT inhibitors BG or LM, synergy was observed, including in p53-mutated BT74 (Figure 1, J and Supplementary Figure 1, E and F, available online). The absence of synergy in MGMT-positive glioma cells is likely a direct effect of MGMT because MGMT-transfected U87 cells acquire this phenotype (Supplementary Figure 1, F, available online). Importantly, no synergistic killing was observed in human neurons, with or without BG (Figure 1, K).
The DNA Damage Response in Glioma Cells After TMZ and G47Δ Treatment
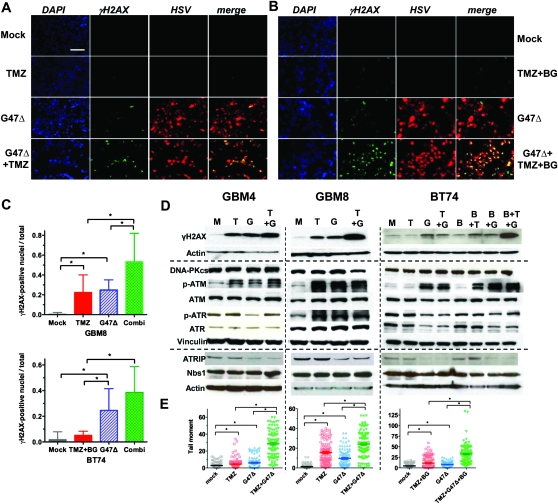
Synergy in GSCs and glioma cell lines is not attributable to increased G47Δ infectivity, replication, or spread because TMZ did not alter these properties in MGMT-negative or MGMT-positive GSCs or glioma cells, with or without BG (Figure 1, L and M and Supplementary Figure 2, available online). Both TMZ and HSV induce DNA damage responses (DDR) that are associated with phosphorylation of H2AX (γH2AX) (30–32), a sensitive marker of DSBs (33). Therefore, we sought to determine whether TMZ and G47Δ were interacting through DDRs and effects on DNA repair. TMZ alone induced γH2AX in MGMT-negative GSCs (γH2AX-positive nuclei fraction, GBM8: mock vs TMZ alone, difference = −0.220, 95% CI = −0.33 to −0.11, P = .006; Figure 2, A and C) and glioma cell lines (Supplementary Figure 3, available online), and only in the presence of BG in MGMT-positive BT74 (not statistically significant, P = .06; Figure 2, B and C), GBM13 (Supplementary Figure 4, A, available online), and T98 (Supplementary Figure 3, available online). G47Δ infection alone induced γH2AX in all GSCs and glioma cell lines examined (γH2AX-positive nuclei fraction, GBM8: mock vs G47Δ alone, difference = −0.24, 95% CI = −0.31 to −0.18, P < .001; BT74: mock vs G47Δ alone, difference = −0.23, 95% CI = −0.34 to −0.11, P = .003; Figure 2, A–D, and Supplementary Figure 3, available online). Combination treatment further increased γH2AX protein levels and positive cells (for GBM8, G47Δ vs combination: difference = −0.29, 95% CI = −0.48 to −0.09, P = .01, and TMZ vs combination: difference = −0.31, 95% CI = −0.53 to −0.09, P = .02; for BT74, G47Δ vs combination: difference = −0.14, 95% CI = −0.31 to −0.03, P = .04, and TMZ + BG vs combination: difference = −0.33, 95% CI = −0.47 to −0.20, P = .001; Figure 2, A–D, and Supplementary Figures 3 and 4, A and C, available online). We used a neutral comet assay to directly assess DSBs (could include virus DNA), which were increased by single agents alone and further increased by combination treatment in all GSCs (for GBM4 [Mock vs TMZ: difference = −1.5, 95% CI = −2.8 to −0.29, P = .012; Mock vs G47Δ: difference = −3.2, 95% CI = −4.6 to −1.8, P < .001; TMZ vs TMZ + G47Δ: difference = −24, 95% CI = −27 to −21, P < .001; G47Δ vs TMZ + G47Δ: difference = −23, 95% CI = −26 to −19, P < .001], for GBM8 [Mock vs TMZ: difference = −14, 95% CI = −16 to −12, P <.001; Mock vs G47Δ: difference = −8.3, 95% CI = −10 to -6.5, P < .001; TMZ vs TMZ + G47Δ: difference = −8.6, 95% CI = −12 to −5.3, P <.001; G47Δ vs TMZ + G47Δ: difference = −15, 95% CI = −18 to −11, P < .001], and for BT74 (Mock vs TMZ + BG: difference = −6.7, 95% CI = −9.6 to −3.8, P < .001; Mock vs G47Δ: difference = −3.2, 95% CI = −4.8 to −1.6, P < .001; TMZ + BG vs TMZ + BG + G47Δ: difference = −21, 95% CI = −26 to −17, P < .001; G47Δ vs TMZ + BG + G47Δ: difference = −25, 95% CI = −29 to −21, P <.001) (unpaired t test, two-sided); Figure 2, E), and U87 (Supplementary Figure 3, B and D, available online).
Figure 2.
DNA damage and DNA damage responses after treatment with TMZ and/or G47Δ in GSCs in vitro. A) γH2AX induction in GBM8 cells (MGMT negative). Cells were mock treated or treated with TMZ (3 μM) for 36 hours and then mock infected or infected with G47Δ (MOI = 1). Twenty-four hours later, cells were fixed and processed for immunocytochemistry (DAPI, blue; anti-γH2AX, green; anti-HSV, red). B) γH2AX induction in BT74 cells (MGMT positive). Cells were mock treated or treated with TMZ (200 μM) + BG (50 μM) for 36 hours and then mock infected or infected with G47Δ (MOI = 1) in the presence of BG (50 μM). Twenty-four hours later, cells were processed as in (A). Scale bar = 100 μm in (A) and (B). C) The ratio of γH2AX-positive nuclei/total was determined in three randomly selected fields with GBM8 (upper panel) and BT74 with BG (50μM) (lower panel). Combi = TMZ + G47Δ (GBM8); TMZ + BG + G47Δ (BT74). Groups are significantly different in GBM8 and BT74 (P < .001, one-way ANOVA). Asterisks indicate statistically significant differences between indicated pairs (P < .05, Bonferroni Multiple Comparison Test). Error bars represent 95% confidence intervals. D) GSCs were treated as in (A), harvested 24 hours later, and processed for western blotting. Left panels: GBM4 (TMZ 20 μM), Middle panels: GBM8 (TMZ 3 μM), Right panels: BT74 (TMZ 200 μM, BG 50 μM). M: mock, T = TMZ, G = G47Δ, B: BG. E) DSBs induced by TMZ (+BG), G47Δ, or the combination, were assessed by neutral comet assay and plotted as individual tail moments, with the dark horizontal bars representing the means and 95% confidence intervals. Groups are significantly different in GBM4, GBM8, and BT74 (P < .001, one-way ANOVA). Asterisks indicate statistically significant differences between indicated pairs (P < .001, except for GBM4 Mock vs TMZ, P = .012, unpaired t test, two-sided). Experiments were performed in triplicate. ANOVA = analysis of variance; ATM = ataxia telangiectasia mutated; ATR = ataxia telangiectasia and Rad3-related; ATRIP = ATR interacting protein; BG = O6-benzylguanine; DAPI = 4′, 6-diamidino-2-phenylindole; DNA-PKcs = DNA-dependent protein kinase catalytic subunit; DSB = DNA double-strand break; GSC = glioblastoma stem cell; HSV = herpes simplex virus; MGMT = O6-methylguanine-DNA-methyltransferase; MOI = multiplicity of infection; p-ATM = phosphorylated ATM (Ser1981); p-ATR = phosphorylated ATR (Ser428); TMZ = temozolomide.
The persistence of DSBs after therapy indicates impaired repair pathways. We next examined DSB repair signaling (34–37). Only ATM was activated (p-Ser 1981) by both TMZ, alone in MGMT-negative cells or with BG in MGMT-positive cells, and G47Δ in GSCs (Figure 2, D) and glioma cell lines (Supplementary Figure 3, F, available online). The activation of ATR (p-ATR) was variable, being induced in GBM8 by both TMZ and G47Δ and reduced by G47Δ in GBM4 and BT74 (Figure 2, D). However, ATRIP, a regulatory binding partner of ATR (38), was reduced after G47Δ infection in all GSCs, including GBM8, and glioma cell lines examined (Figure 2, D, and Supplementary Figure 3, F, available online), suggesting that the ATR pathway is being impaired by infection. NBS1 interacts with and recruits ATM to DSBs (38), and there was no change in its level (Figure 2, D, and Supplementary Figure 3, F, available online). HSV infection has been reported to reduce DNA-PKcs levels because of ICP0 expression (32, 39), which might decrease DSB repair and increase γH2AX levels. However, we found no evidence for decreased DNA-PKcs levels after G47Δ (ICP0+) infection, except in U373 cells (Figure 2, D and Supplementary Figure 3, F, available online).
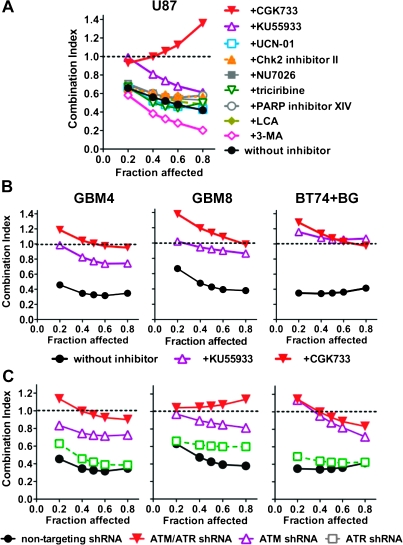
Pharmacological Inhibition of ATM/ATR
To investigate the possible involvement of cellular DDR pathways in synergistic GBM killing by combination treatment, we initially screened U87 with a panel of chemical inhibitors at nontoxic concentrations (Supplementary Table 1, available online). Only inhibitors of ATM impaired TMZ + G47Δ synergy, with CGK733, an ATM/ATR inhibitor (40), reducing the interaction to antagonistic-additive, and KU55933, a selective ATM inhibitor (41), decreasing the interaction to additive-weak synergy (Figure 3, A and B, and Supplementary Figure 5, A, available online). The effect of the ATM/ATR inhibitors on TMZ or G47Δ sensitivity was modest in GSCs and glioma cell lines, with a slight increase for TMZ and decrease for G47Δ (Supplementary Table 2, available online). Inhibition of DNA-PKcs, involved in the nonhomologous end joining (NHEJ) pathway of DSB repair, or Akt, activated in an ATR-dependent fashion after TMZ (42), had no effect on synergy in glioma cell lines (Figure 3, A and Supplementary Figure 5, A, available online). Inhibition of Chk1 or Chk2, downstream substrates for ATR and ATM, respectively, which induce cell cycle arrest, did not affect synergy (Figure 3, A).
Figure 3.
Effect of inhibition of DNA damage response pathways on synergy. A) U87 cells were treated with pharmacological inhibitors of DDR pathways at the concentrations indicated in Supplementary Table 1 (available online), and the interactions between TMZ and G47Δ analyzed. Data are shown as Fraction affected–Combination Index (CI) plot. B) Interaction between G47Δ and TMZ in GSCs (GBM4, GBM8, and BT74 [pretreated with 50 μM BG]) treated with ATM inhibitors CGK733 (5 μM) or KU55933 (20 μM). C) Interaction between G47Δ and TMZ in GSCs (GBM4, GBM8, and BT74 [pretreated with 50 μM BG]) lentivirally transduced with shRNA targeting ATM, ATR, ATM + ATR, or nontargeting shRNA. CI < 1, CI = 1, and CI > 1 represent synergistic, additive, and antagonistic interactions, respectively. The experiments were repeated at least three times for each of the conditions, each time in triplicate. ATM = ataxia telangiectasia mutated; ATR = ataxia telangiectasia and Rad3-related; BG = O6-benzylguanine; CGK733 = ATM/ ataxia telangiectasia and Rad3-related (ATR) kinase inhibitor; Chk2 = checkpoint kinase 2; DDR = DNA damage response; GSCs = glioblastoma stem cells; KU55933 = activated ATM kinase inhibitor; LCA = lithocholic acid; NU7026 = DNA-dependent protein kinase inhibitor; PARP = poly-(ADP-ribose) polymerase; shRNA = short-hairpin RNA; TMZ = temozolomide; UCN-01 = cyclin-dependent kinase inhibitor; 3-MA = autophagy inhibitor 3-methyladenine.
shRNA-Mediated Knockdown of ATM or ATM/ATR and TMZ/G47Δ Synergy in Glioma Cell Killing
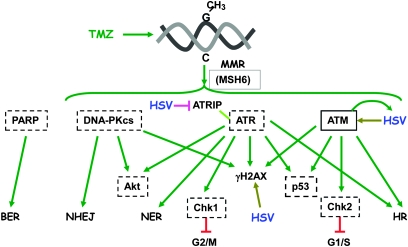
Because no ATR-specific inhibitors were available to distinguish ATM- and ATR-mediated effects, we assessed the contribution of ATM- vs ATR-mediated responses using shRNA. After transduction with lentivirus vectors expressing shRNA, there was a large decrease in the targeted steady state protein levels compared with nontarget shRNA, except for ATR in BT74 cells (Supplementary Figure 5, B, available online). In all cases, ATM knockdown greatly reduced or abrogated synergistic interactions, consistent with the pharmacological inhibitors, whereas ATR knockdown did not, and the ATM + ATR combination further reduced the interaction, except for BT74 (Figure 3, C), suggesting only a minor role for ATR in synergy in GSCs in the absence of ATM. In U87 cells, ATR shRNA reduced synergy (Supplementary Figure 5, C and D, available online); this was the only cell line in which G47Δ increased total ATR levels (Supplementary Figure 3, F, available online). ATM shRNA modestly increased TMZ and decreased G47Δ sensitivities (Supplementary Table 2, available online), similar to the inhibitors. Taken together, these results indicate that among the different DDR pathways, ATM makes a substantial contribution to synergy (Figure 4).
Figure 4.
Diagram of DNA damage response pathways. Hatched-line boxes indicate proteins whose inhibition does not affect synergy between TMZ and G47Δ. Solid-line box indicates protein whose inhibition abrogates synergy. Arrows indicate activation/phosphorylation. Inverted T indicates inhibition. ATM = ataxia telangiectasia mutated; ATR = ataxia telangiectasia and Rad3-related; ATRIP = ATR interacting protein; BER = base excision repair; Chk1 = checkpoint kinase 1; Chk2 = checkpoint kinase 2; DNA-PKcs = DNA-dependent protein kinase catalytic subunit; HR = homologous recombination; HSV = herpes simplex virus; MMR = mismatch repair; MSH6 = mutS homolog 6; NER = nucleotide excision repair; NHEJ = non-homologous end joining; PARP = poly(ADP-ribose) polymerase; TMZ = temozolomide.
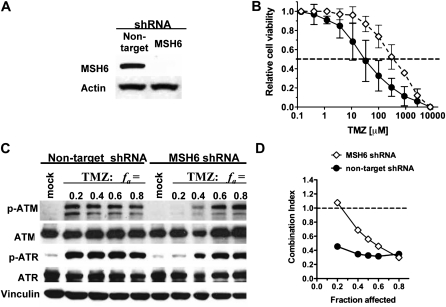
MSH6 Knockdown, TMZ Resistance in MGMT-Negative GSCs, and Cytotoxic Interaction Between TMZ and G47Δ
Because MSH6 mutations and loss are an alternate mechanism for TMZ resistance (43,44), we examined MSH6 loss on synergy. MSH6 knockdown with shRNA (Figure 5, A) increased the ED50 for TMZ by almost 10-fold (compared with nontarget shRNA), to a dose that would confer TMZ resistance (ED50 = 408 μM) (Figure 5, B). MSH6 knockdown had minimal effect on G47Δ sensitivity (Supplementary Table 2, available online), suggesting that the mismatch repair pathway is not used by HSV. MSH6 shRNA impaired synergy at low but not high fa doses (Figure 5, D). ATM is only fully activated at higher TMZ doses (fa = 0.6) in MSH6 shRNA cells compared with fa = 0.2 in nontarget shRNA GSCs, in which synergy occurs (Figure 5, C) that further implicates activated ATM as critical for TMZ + G47Δ synergy.
Figure 5.
Effects of MSH6 knockdown on GSC sensitivity to TMZ, ATM activation, and synergy. A) GBM4 cells were lentivirally transduced with MSH6 or nontargeting shRNA, with associated decrease in MSH6 protein by western blot. B) Dose–response curves of TMZ sensitivity after 5-day incubation. Error bars indicate 95% confidence intervals. Dotted line indicates 50% cell viability relative to mock-treated cells. C) Nontargeting (left) or MSH6 (right) shRNA-expressing cells were treated with indicated doses of TMZ [determined from dose–response curves in (B)] or PBS (mock) and 2.5 days later harvested and processed for western blotting. D) Effect of MSH6 shRNA on TMZ and G47Δ interaction. Data are shown as Fraction affected–Combination Index (CI) plot. CI < 1, CI = 1, CI > 1 represent synergistic, additive, and antagonistic interactions, respectively. The experiments were repeated at least three times for each of the conditions, each time in triplicate. ATM = ataxia telangiectasia mutated; ATR = ataxia telangiectasia and Rad3-related; GSC = glioblastoma stem cell; MSH6 = mutS homolog 6; PBS = phosphate buffered saline; p-ATM = phosphorylated ATM (Ser1981); p-ATR = phosphorylated ATR (Ser428); TMZ = temozolomide.
Inhibition or Knockdown of ATM, G47Δ Replication in Glioma Cells, and Localization of Activated ATM and HSV Replication Compartments
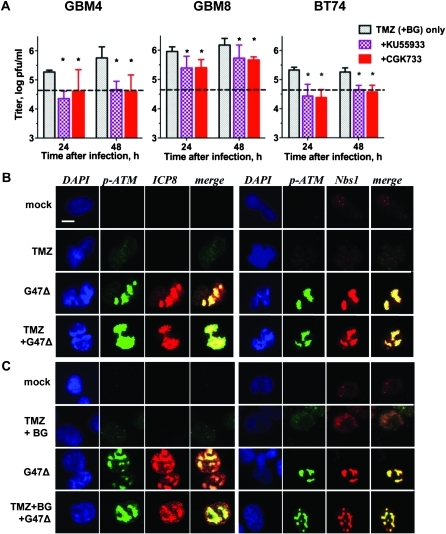
Because ATM inhibition or knockdown decreased G47Δ cytotoxic activity and synergy with TMZ in glioma cells (Figure 3 and Supplementary Table 2, available online), we examined whether this was attributable to inhibition of virus replication (Figure 6, A). G47Δ replication was reduced approximately fivefold by the addition of KU55933 after TMZ treatment (TMZ + BG for MGMT+ BT74) (virus yield, log pfu/mL: for GBM4 [24-hour TMZ vs TMZ + KU55933: difference = 0.91, 95% CI = 0.74 to 1.08, P < .001; 48-hour TMZ vs TMZ + KU55933: difference = 1.09, 95% CI = 0.78 to 1.39, P < .001]; for GBM8 [24-hour TMZ vs TMZ + KU55933: difference = 0.563, 95% CI = 0.28 to 0.84, P = .005; 48-hour TMZ vs TMZ + KU55933: difference = 0.441, 95% CI = 0.12 to 0.76, P = .02]; and for BT74 [24-hour TMZ + BG vs TMZ + BG + KU55933: difference = 0.892, 95% CI = 0.62 to 1.16, P < .001; 48-hour TMZ + BG vs TMZ + KU55933: difference = 0.607, 95% CI = 0.47 to 0.74, P < .001]; Figure 6, A and Supplementary Figure 6, A, available online).
Figure 6.
Interaction of activated ATM with HSV replication. A) G47Δ replication in the presence of ATM inhibitors KU55933 and CGK733. GSCs were pretreated with TMZ for 12 hours in the presence of KU55933 or CGK733 and virus yields (pfu/mL) determined in triplicate. Left panel: GBM4 (TMZ 20 μM), Middle panel: GBM8 (TMZ 3 μM), Right panel: BT74 (TMZ 50 μM, BG 50 μM). Asterisks indicate statistically significant differences in G47Δ titers from those without inhibitors. Dashed line indicates the dose of G47Δ used for infection. Error bars represent 95% confidence intervals, unpaired t test, two-sided. B, C) Accumulation of activated ATM (p-ATM) and Nbs1 (MRN complex) at G47Δ replication compartments. GBM8 (B; TMZ 3 μM) and BT74 (C; TMZ 200 μM, BG 50 μM) were fixed 6 hours after infection and examined for immunofluorescence (DAPI, blue; p-ATM, green; ICP8 and Nbs1, red; merge, yellow). Scale bar = 10μm. ATM = ataxia telangiectasia mutated; BG = O6-benzylguanine; DAPI = 4′, 6-diamidino-2-phenylindole; GSCs = glioblastoma stem cells; Nbs1 = Nijmegen breakage syndrome 1; pfu = plaque forming unit; p-ATM = phosphorylated ATM (Ser1981); TMZ = temozolomide.
A similar effect was seen with shRNA to ATM, but not ATR (Supplementary Figure 6, B, available online). In glioma cell lines, reduction of G47Δ replication by ATM inhibition was independent of TMZ (Supplementary Figure 6, A, available online). There was no additional decrease in virus yield in cells with ATM + ATR shRNA or the ATM/ATR inhibitor CGK733 (Figure 6, A and Supplementary Figure 6, A, available online). Thus, ATM contributes to oHSV replication and virus yield in glioma cells. In light of previous reports demonstrating that activated ATM and associated DNA repair factors colocalize with HSV replication compartments (31, 32, 45), we examined possible colocalization in GSCs. There was a large increase in the size of activated ATM–MRN complexes (p-ATM, Nbs1) in G47Δ-infected GSCs compared with uninfected cells treated with TMZ (±BG), and these complexes colocalized with HSV replication compartments (ICP8) (Figure 6, B and C, and Supplementary Figure 4, B, available online). The combination of G47Δ with TMZ (±BG) did not alter this colocalization. Thus, G47Δ infection of GSCs sequestered activated ATM-containing repair complexes or foci at viral replication compartments where they likely contribute to efficient virus replication.
Effect of Combination Therapy on Efficacy in GSC-Derived Intracerebral Tumors and Increased DDR
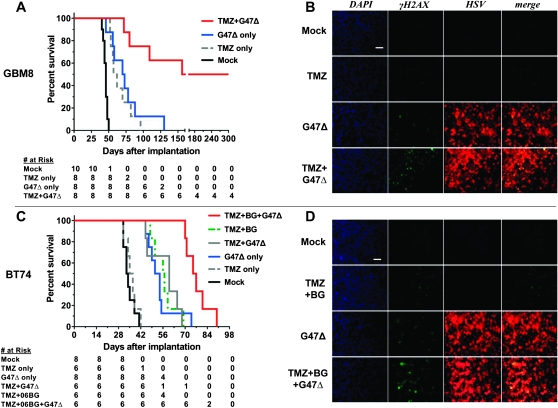
We evaluated the in vivo efficacy of the combination strategy in two GSC orthotopic xenograft mouse models (GBM8 and BT74) with different histology and MGMT status. Both GSCs form aggressive tumors, with all mice succumbing to tumor growth within 50 days, except when treated with G47Δ (Figure 7, A and C). In the MGMT-negative GBM8 model, 5 mg/kg of TMZ for 3 days was effective in prolonging survival (median survival = 59.5 days; TMZ vs mock: hazard ratio [HR] of survival = 18.7, 95% CI = 4.7 to 73.7, P < .001) to a similar extent as G47Δ alone (median survival = 71.5 days; G47Δ vs mock, HR of survival = 12.1, 95% CI = 3.2 to 45.2, P < .001). The combination of TMZ with G47Δ further prolonged survival, achieving long-term remission in four of eight mice (median survival = 228 days; G47Δ vs G47Δ + TMZ, HR of survival = 7.1, 95% CI = 1.9 to 26.1, P = .003; Figure 7, A). In the MGMT-positive BT74 model, 50 mg/kg of TMZ for 3 days had no effect, and the combination with G47Δ was only slightly, but not statistically significantly, better than G47Δ alone (median survival = 60 days vs 52.5 days for G47Δ alone; Figure 7, C). As in vitro, when MGMT was inhibited by BG, survival was extended (median survival = 56.5 days vs 36 days for TMZ alone; TMZ + BG vs TMZ, HR of survival = 21.9, 95% CI = 3.8 to 124.6, P < .001), and now the combination with G47Δ further extended survival (median survival = 76 days; G47Δ vs TMZ + BG + G47Δ, HR of survival = 8.7, 95% CI = 2.2 to 34.3, P = .002; Figure 7, C). In both GSC tumor models, G47Δ or TMZ (±BG) alone induced some γH2AX, but this was greatly increased after combination treatment, particularly in virus-positive tumor regions (Figure 7, B and D). Thus, in vivo efficacy was associated with increased γH2AX, consistent with the mechanism of action determined in vitro.
Figure 7.
Combination treatment of mice intracerebrally implanted with GSCs. A) Athymic mice were stereotactically implanted with GBM8 on day 0 and PBS or TMZ (5 mg/kg/d) administered intraperitoneally on days 7–9. Mock (PBS) or 2 × 106 pfu of G47Δ were intratumorally injected on day 8 (n = 8 per group). Combination vs G47Δ only, HR of survival = 7.1, 95% CI = 1.9 to 26.1, P = .003; combination vs TMZ only, TMZ vs Mock, and G47Δ alone vs Mock, P < .001 (two-sided log-rank test). B) γH2AX induction by the combination treatment in vivo. Mice were treated for 3 consecutive days with TMZ (5 mg/kg/d) starting 23 days after GSC (GBM8) implantation (1 × 105 per mouse). G47Δ (2 × 106 pfu) was intratumorally injected on the second day of chemotherapy, and mice were killed 10 hours after the last chemotherapy. Brains were fixed, sectioned, and examined for immunofluorescence (DAPI, blue; γH2AX, green; HSV, red; merge, yellow). Scale bar = 100 μm. C) Athymic mice were stereotactically implanted with BT74 on day 0, and treated as in (A), except for TMZ dose (50 mg/kg/d) and BG (0.3 mg/d) administration on days 7–9 (Mock: n = 8, TMZ only: n = 6, TMZ + BG: n = 6, G47Δ only: n = 8, TMZ + G47Δ: n = 6, TMZ + BG + G47Δ: n = 6). P < .001 (G47Δ only vs Mock, TMZ + BG vs Mock, TMZ + BG + G47Δ vs TMZ + BG), P = .002 (TMZ + BG + G47Δ vs G47Δ only) (two-sided log-rank test). D) γH2AX induction by the combination treatment in vivo (BT74 model: MGMT positive). Mice were treated for 3 consecutive days with TMZ (50 mg/kg/d) + BG (0.3 mg/kg/d) starting 23 days after GSC (BT74) implantation (1 × 105 per mouse). G47Δ (2 × 106 pfu) was intratumorally injected on the second day of chemotherapy, and mice were treated as in (B). Scale bar = 100 μm. BG = O6-benzylguanine; DAPI = 4’,6-diamidino-2-phenylindole; GSC = GBM stem cell; HSV = herpes simplex virus; MGMT = O6-methylguanine methyltransferase; PBS = phosphate buffered saline; TMZ = temozolomide.
Discussion
In this study, we demonstrated synergy between TMZ and oHSV-G47Δ in treating GBM including the GSC subpopulation, an important therapeutic target of GBM. Synergy occurs primarily through oHSV-mediated manipulation of DNA damage responses, a universal mechanism of cancer resistance to radiation and chemotherapy. To our knowledge, this study is the first to show that combination of an oncolytic virus and chemotherapeutic agent exploits specific DDR pathways to promote killing of cancer cells including cancer stem cells. Since the initial description of genetically engineered oHSV, many oncolytic viruses with different alterations have been described and tested in clinical trials (15). Because of tumor selectivity, oncolytic viruses typically have a large therapeutic index with limited toxic effects and are not known to allow cancer cells to develop resistance (46). In clinical trials, they have shown some promise when used as single agents (47,48). However, like many cancer therapies, combination with other therapeutic modalities might improve efficacy. The effects of chemotherapies are not specific to cancer cells, leading to dose-limiting toxic effects, and are coupled with the emergence of resistance. Therefore, identification of a mechanistic rationale for combining the two modalities could have a profound impact on advancing the management of refractory cancers including GBM.
Controversy remains about whether GSCs are inherently chemoresistant (12,13,49). An analysis of 20 GSC lines found that about half were TMZ resistant, which was associated with MGMT expression (14). In patients, TMZ dosing varies from 75–150 mg/m2 daily, with different “on–off” schedules, which result in peak tumor concentrations of approximately 15–35 μM (50), corresponding to a dose of 0.4–1.2 mg TMZ in mice. We administered 0.125 and 1.25 mg TMZ for sensitive and resistant tumors, respectively, doses comparable to those used in the clinic and well below the reported LD50 dose (5 mg) in athymic mice (51). At these clinically relevant doses, TMZ statistically significantly increased median survival in mice bearing MGMT-negative GBM8 tumors and had no effect on MGMT-positive BT74 tumors. Inhibition of MGMT by BG or LM sensitized MGMT-positive GSCs to TMZ (approximately two- to threefold), although not to equivalent levels as those for MGMT-negative GSCs. In the MGMT-positive BT74 intracerebral tumor model, BG (15 mg/kg) extended median survival after TMZ from 36 to 56 days.
TMZ interacted synergistically with G47Δ in vitro in killing GSCs and glioma lines tested in the absence of MGMT. Importantly, there was no synergy in human neurons, indicative of glioma specificity. Combination therapy resulted in a greater than fivefold decrease in the TMZ and G47Δ doses necessary for a similar level of GSC killing. This effect is likely to be more important for MGMT-positive GSCs, in which the TMZ dose with BG could be reduced to about 20 and 80 μM for GBM13 and BT74, respectively, within a range obtainable in patients. TMZ treatment did not affect G47Δ entry, replication, or spread, suggesting that the combination sensitized GBM cells to killing by TMZ and/or G47Δ. MSH6 mutation or loss frequently occurs in recurrent GBM after TMZ treatment (43,44). Knockdown of MSH6 in GBM4 decreased the sensitivity to TMZ killing, as previously shown in glioma cell lines (44), and also increased the cytotoxic TMZ damage required to activate the ATM and ATR pathways. This decrease in ATM/ATR activation may make MSH6-deficient recurrent GBMs more sensitive to DNA damage from other chemotherapeutics in the presence of TMZ.
As reported by others for wild-type HSV (52), γH2AX levels were increased after oHSV-G47Δ infection, even in vivo, as well as induction of cellular DSBs. HSV DNA replication occurs in discrete compartments in the nucleus that assemble as prereplicative sites with viral DNA and DNA-binding replication proteins (ie, ICP8), followed by recruitment of HSV DNA polymerase and cellular factors to form replication compartments (53). HSV-1 replication is decreased and delayed in cells unable to activate ATM but is not affected by expression of dominant-negative ATR (32). Using oHSV-G47Δ, we found that ATM inhibitors KU55933 or CGK733 reduced virus yields. Similarly, ATM shRNA, but not ATR shRNA, inhibited G47Δ replication. This decrease occurred in the presence or absence of TMZ, indicating that HSV-mediated ATM activation alone is sufficient for efficient replication. It is unclear whether the normal activity and localization of ATM are detrimental to HSV or whether the relocalization of ATM is beneficial; however, the effects on virus replication suggest that it has been co-opted by HSV, possibly to enhance recombinational repair of branched concatameric replication products.
In response to DNA damage, DDR pathways are immediately activated by PI3K-related kinases, ATM, ATR, and DNA-PK. Repair of DSBs occurs primarily through NHEJ or homologous recombination, with the latter, not NHEJ, required for protection from TMZ-mediated cytotoxicity (54). HSV induces DDR early after infection, activating the ATM pathway and inactivating NHEJ and ATR signaling (32,39,52,55,56). As we demonstrated using oHSV-G47Δ, activated ATM is sequestered to replication compartments, thereby becoming unavailable for productive DNA repair of the host genome. These intrinsic properties of HSV provide a unique opportunity to effectively target cancer stem cells by combining oHSV with current cytotoxic therapeutics because these DNA damage repair pathways contribute to survival and maintenance of cancer stem cells after radiation therapy and chemotherapy (9). Increased activation of DDR pathways including ATM and subsequent checkpoint kinases renders CD133+ GSCs resistant to radiation-induced toxic effects (57). We also demonstrate that ATM/ATR pathways play a role in protecting GSCs from TMZ-mediated cytotoxicity.
This study also had some limitations. We examined only five GSCs, and these varied considerably in their sensitivity to TMZ, their xenograft histopathology (18), and their genotype (58). While the expression of MGMT correlated with TMZ resistance, many more GSCs will need to be tested to draw conclusions about how GSC phenotype and/or genotype impacts TMZ sensitivity. Our in vivo studies clearly demonstrate that the combination of TMZ-induced damage with G47Δ greatly enhances survival; however, there were differences between the two GSC models. For example, the combination was curative in GBM8 but not in BT74, for reasons that we do not understand. Treatment of other GSC-derived tumors will be necessary to determine whether these two GSC-derived tumors are representative of other GSCs. Because we used immune-deficient mice for the GBM xenograft models, it remains to be determined how the immune system, as in immunocompetent models and patients, might affect combination therapy, be it beneficial or detrimental. Finally, only clinical translation will indicate whether these GSC models are predictive of patient responses.
In summary, we demonstrated synergy between TMZ and G47Δ in killing human GSCs, which occurs via a mechanism in which HSV manipulates DDRs, leading to increased cell death. Other therapeutics that induce DDRs may also have similar interactions with oHSV. Many viruses, such as adenovirus, perturb cellular DDR (55), which might be similarly harnessed to induce favorable interactions between other oncolytic viruses and chemotherapy/radiation therapy. Importantly, synergy was observed in both TMZ-sensitive and TMZ-resistant GSCs, as long as MGMT-mediated repair was inoperative, and to some extent in MSH6 knock-down GSCs, representative of recurrent GBM, making this combination attractive for translation to clinical trials for this otherwise fatal tumor.
Funding
This work was supported by the National Institutes of Health (NS-032677 to R.L.M. and CA57683 to D.N.L.), and the Department of Defense (W81XWH-07-1-0359 to S.D.R.). S.Y. was supported by a Clinician Investigator Fellowship from the Royal College of Physicians and Surgeons of Canada and BrainCare BC.
Supplementary Material
Footnotes
We thank Dr David Knipe and Dr Lynne Chang (Harvard Medical School) for anti-ICP8 antibodies and instructions, and Dr Geoffrey P. Margison (Cancer Research-UK Carcinogenesis Group, University of Manchester, Paterson Institute for Cancer Research, Manchester, UK) for providing Lomeguatrib. The construct against MSH6 mRNA (TRCN0000078543) was originally obtained from the Massachusetts General Hospital shRNA core (Dr Toshi Shioda, Massachusetts General Hospital, Boston, MA).
References
- 1.Barnholtz-Sloan JS, Sloan AE, Schwartz AG. Relative survival rates and patterns of diagnosis analyzed by time period for individuals with primary malignant brain tumor, 1973-1997. J Neurosurg. 2003;99(3):458–466. doi: 10.3171/jns.2003.99.3.0458. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Hegi ME, Liu L, Herman JG, et al. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26(25):4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 4.Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27(34):5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 5.Watson AJ, Middleton MR, McGown G, et al. O(6)-methylguanine-DNA methyltransferase depletion and DNA damage in patients with melanoma treated with temozolomide alone or with lomeguatrib. Br J Cancer. 2009;100(8):1250–1256. doi: 10.1038/sj.bjc.6605015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinn JA, Jiang SX, Reardon DA, et al. Phase II trial of temozolomide plus o6-benzylguanine in adults with recurrent, temozolomide-resistant malignant glioma. J Clin Oncol. 2009;27(8):1262–1267. doi: 10.1200/JCO.2008.18.8417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 8.Li A, Walling J, Kotliarov Y, et al. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res. 2008;6(1):21–30. doi: 10.1158/1541-7786.MCR-07-0280. [DOI] [PubMed] [Google Scholar]
- 9.Eyler CE, Rich JN. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. 2008;26(17):2839–2845. doi: 10.1200/JCO.2007.15.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alcantara Llaguno S, Chen J, Kwon CH, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15(1):45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Yang J, Zheng H, et al. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell. 2009;15(6):514–526. doi: 10.1016/j.ccr.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beier D, Rohrl S, Pillai DR, et al. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer Res. 2008;68(14):5706–5715. doi: 10.1158/0008-5472.CAN-07-6878. [DOI] [PubMed] [Google Scholar]
- 14.Blough MD, Westgate MR, Beauchamp D, et al. Sensitivity to temozolomide in brain tumor initiating cells. Neuro Oncol. 2010;12(7):756–760. doi: 10.1093/neuonc/noq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aghi M, Martuza RL. Oncolytic viral therapies—the clinical experience. Oncogene. 2005;24(52):7802–7816. doi: 10.1038/sj.onc.1209037. [DOI] [PubMed] [Google Scholar]
- 16.Kanai R, Wakimoto H, Cheema T, Rabkin SD. Oncolytic herpes simplex virus vectors and chemotherapy: are combinatorial strategies more effective for cancer? Future Oncol. 2010;6(4):619–634. doi: 10.2217/fon.10.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aghi M, Rabkin S, Martuza RL. Effect of chemotherapy-induced DNA repair on oncolytic herpes simplex viral replication. J Natl Cancer Inst. 2006;98(1):38–50. doi: 10.1093/jnci/djj003. [DOI] [PubMed] [Google Scholar]
- 18.Wakimoto H, Kesari S, Farrell CJ, et al. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009;69(8):3472–3481. doi: 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Todo T, Martuza RL, Rabkin SD, Johnson PA. Oncolytic herpes simplex virus vector with enhanced MHC class I presentation and tumor cell killing. Proc Natl Acad Sci U S A. 2001;98(11):6396–6401. doi: 10.1073/pnas.101136398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ino Y. A clinical study of a replication-competent, recombinant herpes simplex virus type 1 (G47delta) in patients with progressive glioblastoma. WHO International Clinical Trials Registry. 2009 http://apps.who.int/trialsearch/trial.aspx?trialid=JPRN-UMIN000002661. Accessed November 15, 2011. [Google Scholar]
- 21.Mineta T, Rabkin SD, Yazaki T, Hunter WD, Martuza RL. Attenuated multi-mutated herpes simplex virus-1 for the treatment of malignant gliomas. Nat Med. 1995;1(9):938–943. doi: 10.1038/nm0995-938. [DOI] [PubMed] [Google Scholar]
- 22.Todo T, Feigenbaum F, Rabkin SD, et al. Viral shedding and biodistribution of G207, a multimutated, conditionally replicating herpes simplex virus type 1, after intracerebral inoculation in aotus. Mol Ther. 2000;2(6):588–595. doi: 10.1006/mthe.2000.0200. [DOI] [PubMed] [Google Scholar]
- 23.Kuroda T, Martuza RL, Todo T, Rabkin SD. Flip-Flop HSV-BAC: bacterial artificial chromosome based system for rapid generation of recombinant herpes simplex virus vectors using two independent site-specific recombinases. BMC Biotechnol. 2006;6:40. doi: 10.1186/1472-6750-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knipe DM, Senechek D, Rice SA, Smith JL. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J Virol. 1987;61(2):276–284. doi: 10.1128/jvi.61.2.276-284.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esteller M, Toyota M, Sanchez-Cespedes M, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60(9):2368–2371. [PubMed] [Google Scholar]
- 26.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 27.Friedman HS, Keir S, Pegg AE, et al. O6-benzylguanine-mediated enhancement of chemotherapy. Mol Cancer Ther. 2002;1(11):943–948. [PubMed] [Google Scholar]
- 28.Ostermann S, Csajka C, Buclin T, et al. Plasma and cerebrospinal fluid population pharmacokinetics of temozolomide in malignant glioma patients. Clin Cancer Res. 2004;10(11):3728–3736. doi: 10.1158/1078-0432.CCR-03-0807. [DOI] [PubMed] [Google Scholar]
- 29.Kanai R, Wakimoto H, Martuza RL, Rabkin SD. A novel oncolytic herpes simplex virus that synergizes with phosphoinositide 3-kinase/Akt pathway inhibitors to target glioblastoma stem cells. Clin Cancer Res. 2011;17(11):3686–3696. doi: 10.1158/1078-0432.CCR-10-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirzoeva OK, Kawaguchi T, Pieper RO. The Mre11/Rad50/Nbs1 complex interacts with the mismatch repair system and contributes to temozolomide-induced G2 arrest and cytotoxicity. Mol Cancer Ther. 2006;5(11):2757–2766. doi: 10.1158/1535-7163.MCT-06-0183. [DOI] [PubMed] [Google Scholar]
- 31.Wilkinson DE, Weller SK. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J Virol. 2004;78(9):4783–4796. doi: 10.1128/JVI.78.9.4783-4796.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lilley CE, Carson CT, Muotri AR, Gage FH, Weitzman MD. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc Natl Acad Sci U S A. 2005;102(16):5844–5849. doi: 10.1073/pnas.0501916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24(4):679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 34.Ohnishi T, Mori E, Takahashi A. DNA double-strand breaks: their production, recognition, and repair in eukaryotes. Mutat Res. 2009;669(1–2):8–12. doi: 10.1016/j.mrfmmm.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 35.Riches LC, Lynch AM, Gooderham NJ. Early events in the mammalian response to DNA double-strand breaks. Mutagenesis. 2008;23(5):331–339. doi: 10.1093/mutage/gen039. [DOI] [PubMed] [Google Scholar]
- 36.Bolderson E, Richard DJ, Zhou BB, Khanna KK. Recent advances in cancer therapy targeting proteins involved in DNA double-strand break repair. Clin Cancer Res. 2009;15(20):6314–6320. doi: 10.1158/1078-0432.CCR-09-0096. [DOI] [PubMed] [Google Scholar]
- 37.Bonner WM, Redon CE, Dickey JS, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8(12):957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434(7033):605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 39.Lees-Miller SP, Long MC, Kilvert MA, Lam V, Rice SA, Spencer CA. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70(11):7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Won J, Kim M, Kim N, et al. Small molecule-based reversible reprogramming of cellular lifespan. Nat Chem Biol. 2006;2(7):369–374. doi: 10.1038/nchembio800. [DOI] [PubMed] [Google Scholar]
- 41.Hickson I, Zhao Y, Richardson CJ, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64(24):9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 42.Caporali S, Levati L, Starace G, et al. AKT is activated in an ataxia-telangiectasia and Rad3-related-dependent manner in response to temozolomide and confers protection against drug-induced cell growth inhibition. Mol Pharmacol. 2008;74(1):173–183. doi: 10.1124/mol.107.044743. [DOI] [PubMed] [Google Scholar]
- 43.Cahill DP, Levine KK, Betensky RA, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13(7):2038–2045. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yip S, Miao J, Cahill DP, et al. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 2009;15(14):4622–4629. doi: 10.1158/1078-0432.CCR-08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shirata N, Kudoh A, Daikoku T, et al. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J Biol Chem. 2005;280(34):30336–30341. doi: 10.1074/jbc.M500976200. [DOI] [PubMed] [Google Scholar]
- 46.Russell SJ, Peng KW. Viruses as anticancer drugs. Trends Pharmacol Sci. 2007;28(7):326–333. doi: 10.1016/j.tips.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Senzer NN, Kaufman HL, Amatruda T, et al. Phase II clinical trial of a granulocyte-macrophage colony-stimulating factor-encoding, second-generation oncolytic herpesvirus in patients with unresectable metastatic melanoma. J Clin Oncol. 2009;27(34):5763–5771. doi: 10.1200/JCO.2009.24.3675. [DOI] [PubMed] [Google Scholar]
- 48.Geevarghese SK, Geller DA, de Haan HA, et al. Phase I/II study of oncolytic herpes simplex virus NV1020 in patients with extensively pretreated refractory colorectal cancer metastatic to the liver. Hum Gene Ther. 2010;21(9):1119–1128. doi: 10.1089/hum.2010.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frosina G. DNA repair and resistance of gliomas to chemotherapy and radiotherapy. Mol Cancer Res. 2009;7(7):989–999. doi: 10.1158/1541-7786.MCR-09-0030. [DOI] [PubMed] [Google Scholar]
- 50.Rosso L, Brock CS, Gallo JM, et al. A new model for prediction of drug distribution in tumor and normal tissues: pharmacokinetics of temozolomide in glioma patients. Cancer Res. 2009;69(1):120–127. doi: 10.1158/0008-5472.CAN-08-2356. [DOI] [PubMed] [Google Scholar]
- 51.Kokkinakis DM, Bocangel DB, Schold SC, Moschel RC, Pegg AE. Thresholds of O6-alkylguanine-DNA alkyltransferase which confer significant resistance of human glial tumor xenografts to treatment with 1,3-bis(2-chloroethyl)-1-nitrosourea or temozolomide. Clin Cancer Res. 2001;7(2):421–428. [PubMed] [Google Scholar]
- 52.Wilkinson DE, Weller SK. Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J Cell Sci. 2006;119(pt 13):2695–2703. doi: 10.1242/jcs.02981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quinlan MP, Chen LB, Knipe DM. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984;36(4):857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- 54.Roos WP, Nikolova T, Quiros S, et al. Brca2/Xrcc2 dependent HR, but not NHEJ, is required for protection against O(6)-methylguanine triggered apoptosis, DSBs and chromosomal aberrations by a process leading to SCEs. DNA Repair (Amst). 2009;8(1):72–86. doi: 10.1016/j.dnarep.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 55.Lilley CE, Schwartz RA, Weitzman MD. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 2007;15(3):119–126. doi: 10.1016/j.tim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 56.Mohni KN, Livingston CM, Cortez D, Weller SK. ATR and ATRIP are recruited to herpes simplex virus type 1 replication compartments even though ATR signaling is disabled. J Virol. 2010;84(23):12152–12164. doi: 10.1128/JVI.01643-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 58.Wakimoto H, Mohapatra G, Kanai R, et al. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells [published online ahead of print November 7, 2011] Neuro Oncol. 2011 doi: 10.1093/neuonc/nor195. doi:10.1093/neuonc/NOR195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.