Abstract
Objective
T cells from patients with systemic lupus erythematosus (SLE) display increased amounts of spleen tyrosine kinase (SYK) which is involved in the aberrant CD3/T cell receptor-mediated signaling process and increased amounts of cAMP response element modulator (CREM) α which suppresses the production of interleukin-2. Because SYK expression can be suppressed by CREM α we asked why CREM α fails to suppress SYK expression in SLE T cells.
Methods
Healthy T cells were overexpressed with CREM α expression vector and SYK expression and phosphorylation was measured. A newly identified CRE site on SYK promoter was characterized by ChIP and EMSA. The CREM α-mediated repression of SYK expression was further evaluated by analyzing SYK promoter activity. T cells from SLE patients and healthy individuals were subjected to ChIP to evaluate the CREM α binding and histone-H3 acetylation.
Results
We demonstrate that increased CREM α level can suppress SYK expression by direct binding on a CRE site of the SYK promoter in T cells from healthy individuals but failed to do so in SLE T cells. We demonstrate that failure of CREM α to suppress SYK expression in SLE T cells is due to weaker binding to the CRE site of the SYK promoter compared to healthy T cells because the promoter site is hypoacetylatylated and therefore of limited access to transcription factors.
Conclusions
Epigenetic alteration of the SYK promoter in SLE T cells results in inability of the transcriptional repressor CREM α to bind and suppress the expression of SYK resulting in aberrant T cell signaling.
INTRODUCTION
T cell receptor (TCR)-mediated signaling is subject to modulation through tyrosine phosphorylation of many effector molecules and via the activation of several families of protein tyrosine kinases (PTKs) (1). The Src family members of PTKs such as spleen tyrosine kinase (SYK) and zeta-associated protein (ZAP)-70, enable TCR-mediated signal transduction by phosphorylating immunoreceptor tyrosine-based activating motifs of the cytoplasmic region of immune receptors. SYK is one of the important non-receptor kinases isolated first from a porcine spleen cDNA library (2). SYK and ZAP-70 are members of PTKs that function as critical mediators of pre-TCR and TCR signaling, with ZAP-70 having a predominant role in mature T cells (3–5). Both kinases are activated after T cell receptor stimulation and share signaling pathways (6), but, while ZAP-70 requires Lck to be phosphorylated, SYK phosphorylation is Lck-independent (7, 8). T cells from patients with systemic lupus erythematosus (SLE) display decreased levels of CD3ζ chain. The function of the missing CD3ζ is carried out by the Fc receptor (FcR) γ chain (5, 9–11), which involves SYK rather than ZAP-70 causing a `rewired` TCR signaling (12). However, the regulation of SYK expression both in disease and health is largely unknown.
Of the cAMP response element (CRE)-binding proteins, cAMP response element-binding, cAMP response-element modulator protein (CREM) and activating transcription factor-1 belong to the superfamily of bZip proteins containing a basic leucine zipper domain, which binds to the 8-base pair palindrome DNA sequence of CRE (TGACGTCA). Isoforms of these three transcription factors can be activated by PKA and by the calcium calmodulin-dependent kinases such as calmodulin kinases (CaMK) II and IV (13, 14, 15). CREM α is a widely expressed transcriptional repressor important in the termination of T cell immune response (16, 17, 18). Increased levels of CREM α in SLE T cells have been linked to decreased IL-2 production. As PKA levels are decreased in SLE T cells (19), CaMKIV has been demonstrated to be involved in the phosphorylation of CREM α in SLE T cells (20) although the involvement of other kinases has not been studied.
In this communication we demonstrate that CREM α suppresses the expression of SYK by directly binding to the CRE motif on its promoter in normal T cells. Binding of CREM α to the SYK promoter in SLE T cells is limited and we propose that this accounts for a limited feedback suppression of SYK expression that occurs in normal T cells.
MATERIALS AND METHODS
Antibodies and reagents
Mouse monoclonal antibodies against SYK (clone 4D10), goat anti-rabbit HRP and goat anti-mouse HRP were procured from Santa Cruz Biotechnology (Santa Cruz, CA). To generate antibody against human CREM α, a peptide encoding the N-terminal of CREM was used to immunize rabbits. The resulting serum which detected a band of approximately 26 kDa was used in this project (20). An antibody against phosphorylated SYK (Phospho-SYK –Tyr348) was collected from BD Pharmigen (San Jose, CA). Anti-human CD3 antibody clone OKT3 was purchased from BioXcell (West Lebanon, NH) and anti-human CD28 was procured from BioLegend (San Diego, CA). Anti-acetyl-histone H3 antibody was collected from Millipore Corporation (Billercia, MA).
Expression vectors
A SYK promoter luciferase reporter construct (product identifier 108157-CHR9-P0393 R1(SYK) was procured from Switch Gear Genomics (Menlo Park, CA) and cloned in pSGG vector (modified pGL4). The promoter region of the construct length is −782 from the transcription initiation site. The CREM α expression plasmid (gift of Dr. Sassone Corsi, Strasbourg, France) has been described before (21).
Isolation of T cells from peripheral blood of healthy donors and SLE patients
T cells were isolated from human peripheral blood of healthy donors and SLE patients using the RosetteSep Kit (Stemcell Technologies Inc., Vancouver, Canada) following the manufacturer’s instruction. T cells were negatively separated from labeled cell complexes by Lymphoprep gradient centrifugation (purity ≥ 98%) (Fresenius Kabi Norge AS, Oslo, Norway). Freshly isolated T cells were cultured in RPMI 1640 in 5% CO2 supplemented with 10% fetal bovine serum for 2h and used for further experiments. Patients fulfilling the American College of Rheumatology-established criteria for the diagnosis of SLE were included. The study was approved by the Institutional Review Board of Beth Israel Deaconess Medical Center.
Western blot
Total protein was extracted from T cells by radio-immunoprecipitation assay (RIPA) buffer (Boston Bioproducts, MA) supplemented with protease and phosphatase inhibitors (5 mM sodium fluoride, 4 mM sodium vanadate, aprotinin, leupeptin, 1 mM dithiothreitol and 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride). Equal amount of proteins were resolved on a 4–12% Bis-Tris NuPAGE precast gel at 200 V for 1h and transferred to polyvinylidene difluoride (PVDF) membrane at 30 V for 2h, blocked in 5% non-fat milk in Tris-buffered saline with Tween 20 (TBS-T) for 3h, incubated with primary antibody overnight, washed three times with TBS-T, incubated with HRP-conjugated goat anti-mouse or goat anti-rabbit secondary antibody for 2–3h, washed three times with TBS-T buffer, incubated for 5–10 min with ECL reagent (GE Healthcare, Piscataway, NJ) and visualized by Fuji LAS-3000 scanner.
Real-time quantitative reverse transcriptase-polymerase chain reaction
Total RNA was isolated from T cells (transfected or non-transfected controls) by RNeasy Mini Kit (Qiagen, Valencia, CA). Reverse transcription was performed at 37 °C for 120 min from 100 ng total RNA using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). Quantitative real-time PCR for SYK was performed (LightCycler 480, Roche, Indianapolis, IN) with 40 cycles at 94 °C for 12 sec and 60 °C for 60 sec using Taqman gene expression assay (Applied Biosystems). All PCR reactions were run in triplicates with a control reaction containing no RT enzyme. The comparative Ct method was used to quantify transcripts relative to the endogenous control gene 36B4.
Transient transfection
Serial concentrations of CREM α (0.5, 1 and 2 µg) expression constructs or empty vector were transfected separately into T cells using the Amaxa nucleofector system. In some experiments transfected cells were treated with 1 µg anti-CD3/anti-CD28 antibodies for 1h. Time- and dose-curve of anti-CD3/anti-CD28 treatment were optimized in preliminary experiments (data not shown). After 24h of incubation, transfected cells were washed with PBS and lysed with RIPA buffer supplemented with protease and phosphatase inhibitors. Equal amount of proteins were subjected to Western blot using antibodies against total SYK, phosphorylated SYK, CREM α and β-actin.
Chromatin immunoprecipitation
3×106 T cells from healthy individuals were crosslinked with 1% formaldehyde and incubated for 10 minutes at 37 °C. Cells were washed twice with ice cold PBS containing protease inhibitors at 2000 rpm at 4 °C for 5 min. The cell pellet was re-suspended in appropriate volume of SDS lysis buffer to obtain the appropriate number of cells/150 µl and incubated for 10 min on ice. DNA was sheared to lengths between 200 and 1000 bp by sonication. Sonicated DNA was transferred to another tube and diluted with ChIP dilution buffer. To reduce non-specific background, chromatin solution were pre-cleared with 30 µl salmon sperm DNA/Protein A agarose-50% slurry for 2h at 4 °C with agitation. Pre-cleared chromatin was precipitated by anti-CREM α antibody overnight. Immunoprecipitated DNA was collected by 50 µl salmon sperm DNA/Protein A agarose slurry with rotation for 6h followed by gentle centrifugation (1000 rpm, 5 min). Supernatant contained unbound, non-specific DNA was removed. Protein A agarose/antibody/DNA complex was washed sequentially by low salt, high salt, LiCl immune complex and Tris-EDTA washing buffer for 5 min on a rotating platform. Freshly prepared elution buffer (1% SDS, 0.1 M NaHCO3) was used to elute the DNA protein complex. Elution was repeated twice. DNA was purified from combined elutes by using QIAamp DNA purification kit collected from Qiagen (Valencia, CA) and analyzed by PCR with the sense-GGTATCTCAAAACCATTCTTAG and antisense-TGGGCAACTTCCTTAACG primer pair. The PCR reaction was repeated with a second set of primers (sense-TAACTCGAGTCCAGCACAGTAGCTTGGAGCTT and antisense-CGCAAGCTTTAGAACAAGAAAGGGCATGAAAGCA). The experiment was repeated with T cells from 5 SLE patients and 5 age- and sex-matched controls. In a separate experiment, T cells isolated from 7 SLE patients and 7 age- and sex-matched controls were immunoprecipitated with anti-acetyl-histone H3 antibody as stated above using equal number of cells. Equal amount of DNA was used in PCR reactions to compare the relative enrichment of acetyl groups between the SLE and control samples.
Electrophoretic mobility shift assay
5 pmol double-stranded synthetic oligonucleotides were annealed and labeled with [γ32P]ATP using T4 polynucleotide kinase. CRE-sense-GGTTGTGGACGTCAGAGCCGT and CRE-antisense-ACGGCTCTGACGTCCACAACC primer pair was used as wild-type probe, and MuCRE-sense-GGTTGTAGACGACTGAGCCGT and MuCRE-antisense-ACGGCTCAGTCGTCTACAACC primer pair was used as a mutated probe to analyze the complex formed by CRE site on SYK promoter with T cell nuclear extract. These probes were designed to encompass one potential CRE site on SYK promoter. T cell nuclear extracts were prepared following the procedure described before (22). 5 µg of T cell nuclear extracted total protein were incubated with labeled probe in presence of binding buffer and 1 µg of poly(dI-dC) for 1h at 20°C. In a separate binding reaction 2 µg antibody against CREM α was used to verify the specificity of binding. Additional binding reaction was performed with a probe where CRE site was disrupted by three bases alongside the wild-type probe. The DNA-protein complex was isolated on a 6% DNA retardation gel. Gels were fixed in 10% acetic acid/10% methanol for 10 min, dried, exposed overnight to phospho-screen and scanned using a Phospho-Imager scanner (GE Healthcare).
Transient transfection of T cells and measurement of luciferase activity
T cells were transfected using Amaxa nucleofector system. Briefly, 5×106 T cells were resuspended in T cell nucleofector solution (Lonza Walkerrsville Inc., Walkersville, MD) and 2 µg of SYK reporter plasmids were transfected along with 0.5 µg CREM α DNA or empty vector. 50 ng of Renilla luciferase gene driven by the cytomegalovirus (CMV) promoter (pRL-CMV; Promega, Madison, WI) was used as an internal control in all transfection experiments. After 24h incubation, cells were washed twice with PBS (Invitrogen, Carlsbad, CA) and lysed with passive Lysis Buffer (Promega). Luciferase activities were measured by injecting luciferase assay reagent (Promega) into cell extracts and recording luminescence (a 15-sec light output) in a D-20/20 Luminometer (Turner Biosystems, Mountain View, CA). The Renilla luciferase activity as internal control was measured with the Dual Luciferase Reporter Assay System (Promega).
Mutation of CRE site on SYK promoter
The QuikChange site-directed mutagenesis method was performed using PfuTurbo DNA polymerase in temperature cycler. The basic procedure utilizes a supercoiled double-stranded DNA vector with an insert of interest and two synthetic oligonucleotide primers containing the desired mutation. The primers were used to mutate CRE site are M-CRE-sense-TTTATTTGGTTGTGGATACGAGAGCCGTCATGG and M-CRE-antisense-CCATGACGGCTCTCGTATCCACAACCAAATAAA. Primers are extended during temperature cycling by PfuTurbo DNA polymerase. Incorporation of the primers generates a mutated plasmid containing staggered nicks. Following temperature cycling, the product is treated with DpnI. The DpnI endonuclease is specific for methylated and hemimethylated DNA and is used to digest the parental DNA template and to select for mutation-containing synthesized DNA. The nicked vector DNA containing the desired mutation is then transformed into OneShot TOP10 supercompetent cells (Invitrogen). The final clone was sequenced and used in transfection. Luciferase activity was measured following the procedure as discussed before.
Silencing RNA transfection
Mixture of 3 different human CREM α-specific siRNAs and a scrambled negative control siRNA were collected from Origene Technologies (Rockville, MD). 500 nM control siRNA or 100 or 500 nM CREM α-specific siRNA were transfected into healthy T cells by Amaxa nucleofector system. After 72h post-transfection some groups were stimulated with anti-CD3/anti-CD28 antibodies for 1h, washed with PBS and lysed with RIPA buffer supplemented with protease and phosphatase inhibitors. Total proteins were subjected to Western blot with total SYK, phosphorylated SYK, CREM α and β-actin antibodies.
Statistical analysis
Statistical analyses were performed using Student t-test and ANOVA. * p≤0.05, ** p≤0.01, ***p≤0.001.
RESULTS
CREM α overexpression downregulates SYK expression and SYK phosporylation
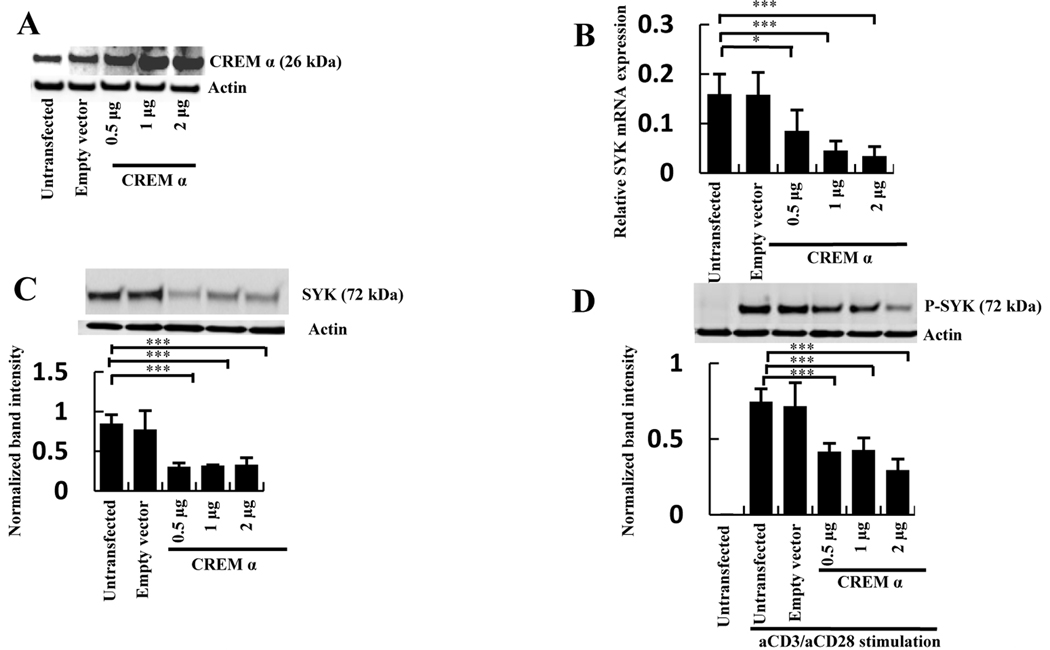
To determine whether CREM α is involved in the regulation of SYK expression, CREM α cDNA was overexpressed in normal human T cells. Transfection of T cells with a CREM α DNA expression construct (Figure 1A) resulted in significant suppression of SYK mRNA (Figure 1B) and protein levels (Figure 1C). Phosphorylation of SYK induced by anti-CD3/anti-CD28 antibodies was also significantly reduced following CREM α overexpression (Figure 1D). These results demonstrate that CREM α is able to suppress SYK mRNA and protein expression as well as the level of SYK phosphorylation in T cells.
Figure 1. Overexpression of CREM α downregulates SYK expression.
A, T cells were transfected with empty vector or with 0.5, 1 and 2 µg of CREM α expression vector. After 24h post-transfection Western blot was performed with antibodies against CREM α or β-actin. B, T cells were transfected with different concentrations of CREM α expression vector or with an empty vector. SYK mRNA level was detected by quantitative PCR. Relative mRNA expression normalized to 36B4 is shown as mean ± SEM from one of 3 independent experiments. C, T cells were transfected with different concentrations of CREM α expression vector or with an empty vector. After 24h incubation total SYK protein or β-actin were detected by Western blot and represented as mean ± SEM of densitometry scan of 5 different blots. D, T cells were transfected with different concentrations of CREM α expression vector or with an empty vector. The transfected cells were treated with 1 µg anti-CD3/anti-CD28 antibodies and the levels of phosphorylated SYK and β-actin were measured by Western blot. The data presented as mean ± SEM of densitometry scan of 5 independent blots.
Silencing of endogenous CREM α enhances SYK protein expression and phosphorylation
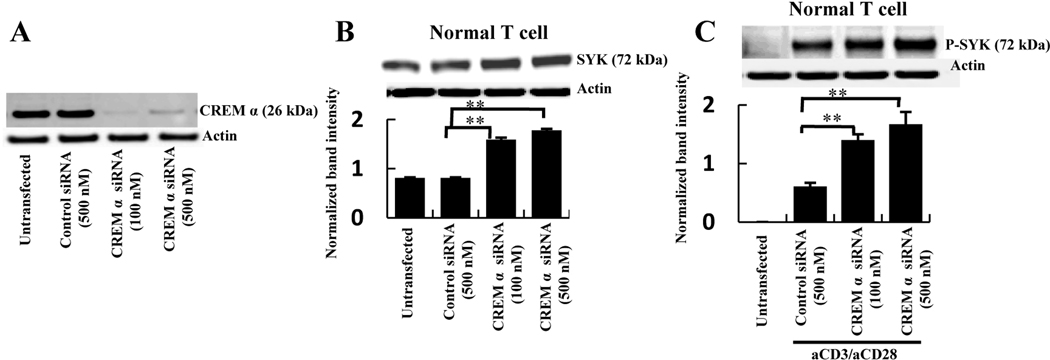
To verify the functional importance of the endogenous CREM α in the regulation of SYK expression in normal cells CREM α was silenced using the mixture of 3 specific siRNAs. Using two different concentrations of CREM α-specific siRNAs (Figure 2A) total SYK protein expression (Figure 2B) as well as the phosphorylation of SYK protein (Figure 2C) were significantly elevated compared to the control siRNA-treated samples in normal T cells.
Figure 2. Silencing of endogenous CREM α upregulates SYK protein expression and phosphorylation.
A, T cells were transfected with 500 nM control siRNA or 100 and 500 nM CREM α-specific siRNA. At 72h post-transfection cells were lysed with RIPA buffer and Western blotted with antibody against CREM α. B, Normal T cells were transfected with 500 nM of control siRNA or 100 and 500 nM CREM α-specific siRNA. Cells were collected after 72h post-transfection in RIPA buffer and Western blotted with antibody against total SYK and β-actin. Data represented as mean ± SEM of densitometry scan of 5 different blots. C, Normal T cells were transfected with 500 nM control siRNA and 100 and 500 nM CREM α-specific siRNA. 1 µg of anti-CD3/anti-CD28 stimulation was given 1h before cell harvesting. Cells were collected after 72h of post-transfection in RIPA buffer and Western blotted with antibody against phosphorylated SYK and β-actin. Data represented as mean ± SEM of densitometry scan of 5 different blots.
In sum, endogenous level of CREM α effectively regulates the SYK expression and phosphorylation in normal T cells.
Binding of CREM α on SYK promoter
In considering possible trans sites on the SYK promoter that would enable CREM binding , we identified a CRE motif (Figure 3A) and we showed, using chromatin immunoprecipitation assays, that CREM α bound to the SYK promoter through this element (Figure 3B). To ascertain that CREM α indeed binds to this CRE site on the SYK promoter EMSA was performed using a [γ32P]-labeled oligonucleotide harboring the CRE sequence on the SYK promoter and nuclear proteins extracted from normal T cells. Specific protein-DNA complexes were observed with the oligonucleotides containing the CRE site that could be displaced by a cold CRE site whereas a mutated oligonucleotide as competitor failed to displace the specific complex (Figure 3C). The presence of CREM α protein in these protein-DNA complexes was confirmed with a specific antibody against CREM α which was able to displace the protein-DNA complex (Figure 3D). An oligonucletide spanning the CRE site but altered by three bases failed to form any specific complexes with the T cell nuclear extract in EMSA (Figure 3E). Taken together these data demonstrate that CREM α binds to the CRE site of the SYK promoter.
Figure 3. CREM α binds on SYK promoter.
A, The SYK gene promoter sequence from initiator element to −782. The transcription start site is marked as red; the CRE site on SYK promoter as blue; one AP1 site marked as green; Ets core binding sites as pink. B, Sonicated chromatin from normal T cells was immunoprecipitated by CREM α antibody or a control IgG. Amplified SYK promoter was detected by PCR. C, An oligonucleotude spanning the region of CRE site on SYK promoter was labeled with [γ32P] and incubated with 5 µg T cell nuclear extract. Protein-DNA complexes were separated on a 6% DNA retardation gel and visualized by phosphoimage analyzer. Unlabeled cold competitors (20 to 500 molar excess) were used to displace complexes formed by labeled oligonucleotides. A cold mutated oligonucleotide was used as non specific competitor. One representative experiment out of 3 is shown. D, 2 µg of specific antibody against CREM α or a non-specific IgG were incubated with labeled oligonucleotides and EMSA was performed. E, Mutated CRE site-bearing oligonucleotide was labeled with [γ32P] and EMSA was performed. In a separate reaction a wild-type probe was used. One representative experiment out of 3 is shown.
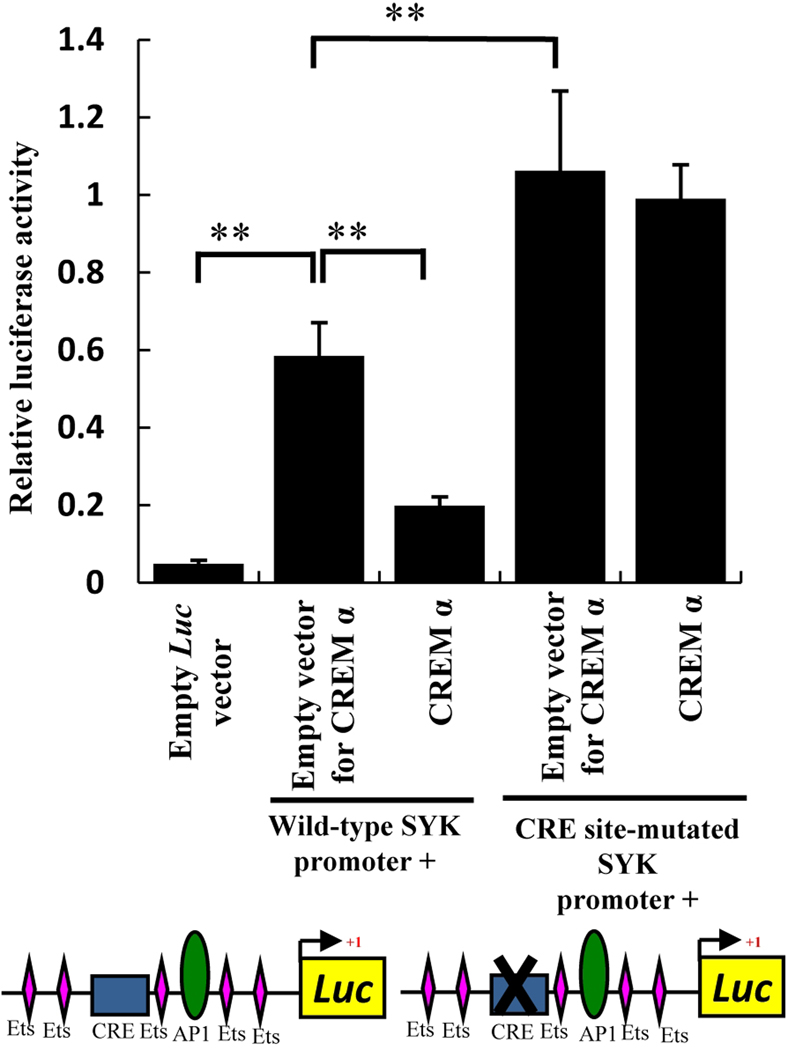
CREM α suppresses SYK promoter activity by binding to the CRE site
To verify that SYK inhibition by CREM α occurred indeed through binding to the CRE site of the SYK promoter we further extended our study by reporter assays using a SYK promoter-driven reporter construct. When the SYK promoter reporter construct was transfected into normal T cells, significant elevation in luciferase activity was detected compared to empty vector-transfected cells (Figure 4 – left side). When a CREM α expressing vector was co-transfected along with the SYK promoter-driven reporter construct, the promoter activity was inhibited significantly (Figure 4 – left side). Basal SYK promoter activity increased significantly when the CRE site was disrupted by site-directed mutagenesis (Figure 4– right side) suggesting that endogenous CREM α was not able to bind to this mutated CRE site and could not perform effective inhibition on the SYK promoter. Co-transfection of CREM α did not show any inhibitory effect on the SYK promoter activity where the CRE site was disrupted showing that even the overexpression of CREM α could not restore the inhibition of SYK when the CRE site was mutated.
Figure 4. CREM α overexpression reduces SYK promoter activity.
T cells were co-transfected with 2 µg wild-type SYK promoter-luciferase reporter construct, CRE site-mutated SYK promoter-luciferase construct or 500 ng CREM α expression plasmid and 50 ng Renilla plasmid vector as internal control. DNA concentrations were normalized by empty vector. Luciferase readings from cell extract was measured and normalized with Renilla luciferase and the data was plotted as mean ± SEM based on 6 different experiments. Construct cartoon shows upstream cis elements on SYK promoter. Rectangular box indicate CRE site and AP-1 site shown as oval shape. Out of several Ets binding sites on SYK promoter a few are shown as diamond shapes, those are close to CRE and AP-1 sites.
Our data clearly show that the newly identified CRE site is important in SYK gene regulation and CREM α suppresses the expression of SYK by binding to this CRE site of the SYK promoter.
CREM α fails to bind to the SYK promoter in SLE T cells
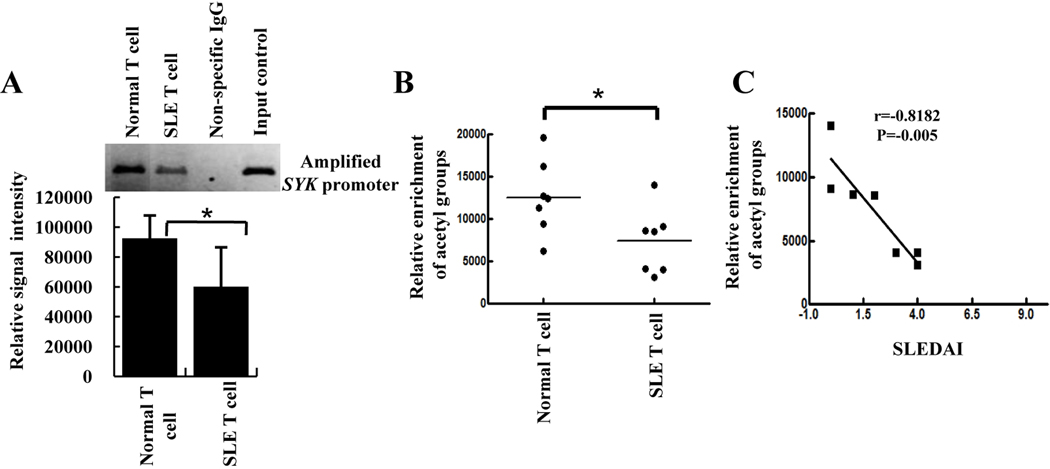
Based on the above findings we suggest that the elevated level of SYK in SLE T cells indeed triggers CREM α expression and may function as a negative feedback mechanism to mitigate SYK levels. However it remained unclear why elevated level of CREM α in SLE T cells is unable to suppress SYK expression in these cells. To gain insight into this question we considered the ability of CREM α to bind to the SYK promoter. As shown in Figure 5A, chromatin immunoprecipitation assays demonstrated that CREM α does not bind to the SYK promoter in SLE T cells as robustly as in normal T cells.
Figure 5. CREM α does not bind to the SYK promoter in SLE T cells.
A, Chromatin was prepared from T cells of 5 SLE patients and 5 age- and sex-matched controls and immunoprecipitated with CREM α antibody. One characteristic example of chromatin-immunoprecipitated SYK promoter with CREM α antibody and densitometry scan of 5 separate samples of control and SLE patients are shown as mean ± SEM of relative intensity of immunoprecipitated bands. B, Chromatin was prepared from 7 SLE patients and 7 age- and sex-matched controls and immunoprecipitated with anti-acetyl-histone H3 antibody. Individual immunoprecipitated bands were measured by densitometry and plotted as random scatter. C, Relative enrichment of acetyl group (as depicted on the X axis) was plotted against the respective SLE disease activity index (SLEDAI; Y axis) using Prism graph pad software. Statistical correlation is given as r value.
Histone acetylation and deacetylation regulates chromatin accessibility by adding or removing acetyl groups to lysine residue in the N-terminal histone domain. Acetylation neutralizes the positive charge of histone lysines, thus allowing transcription factors to bind on DNA. To determine whether altered histone acetylation could explain the limited binding of CREM α to the SYK promoter, we performed chromatin immunoprecipitation experiments using anti-acetyl-H3 antibody and observed less acetylation in SLE T cells compared to control T cells. Low level of acetylation on proximal SYK promoter explain that repression domain of SYK promoter might not be accessible to CREM α to suppress SYK expression in SLE T cells and resulting as weaker binding of CREM α on SYK promoter (Figure 5B). In addition, we observed that the levels of histone hypo-acetylation in SLE T cells correlated with disease severity (SLEDAI index) of individual patients (Figure 5C).
DISCUSSION
Having observed that 1) SYK expression is increased in SLE T cells (5, 11, 12), 2) SYK inhibition in lupus prone mice results in significant disease improvement (23) and 3) CREM α is increased in SLE T cells and suppresses IL-2 production (16), we were interested in identifying possible links between the aberrant expression of these two molecules.
Our data clearly demonstrate the presence of a CRE motif in the promoter of SYK which binds CREM α and in turn suppresses its expression (Figures 1 and 2). Such suppression would provide negative feedback to the increased SYK expression which occurs in normal T cells cultured in vitro (5). The increased levels of both SYK and CREM α in SLE T cells suggest a failure of this control feedback mechanism. Such a defective mechanism would prevent downregulation of SYK in activated T cells which subsequently should display a hyper-responsive phenotype (9). Indeed, CREM α was found to bind to the SYK promoter of SLE T cells in less quantity compared to normal T cells (Figure 5). Notwithstanding the quantitative limitations of chromatin immunoprecipitation assays, it is tempting to propose that limited CREM α binding may prevent the expected CREM α-mediated suppression of the activity of the SYK promoter. Altered access of transcription factors to gene regulatory elements is well known in SLE T cells (24, 25) and may alter regulation of gene expression.
Epigenetic modifications are typically ascribed responsibility for altered access of transcription factors to regulatory elements of genes. Epigenetic abnormalities and particularly DNA methylation have been studied extensively and reported abnormal in SLE T cells (26). Histone acetylation abnormalities have been reported in human (27) and murine (28) lupus T cells. Chromatin immunoprecipitation studies (Figure 5) demonstrated limited presence of acetylated histone in the SYK promoter explaining the observed limited binding of CREM α. Limited binding of CREM α can readily explain the increased levels of SYK in SLE T cells despite the fact that they have increased levels of CREM α (Figure 6).
Figure 6.
Model demonstrating CREM α activation by cAMP pathway and subsequent binding on the CRE site of the SYK promoter to result in downregulating SYK expression in normal T cells. In SLE T cells hypoacetylated SYK promoter inhibits CREM α binding on the CRE site and CREM α-mediated suppression of SYK gene expression.
Gene expression may be regulated through a balance between histone acetylation and deacetylation. Recently it was shown that many transcriptional activators can physically interact with cofactors with histone acetyltransferase function and the ability to recruit these histone modifying enzymes is closely intertwined with the ability of the transcription factor to activate gene expression (29, 30, 31). So it is the amalgamation of more subtle changes in nucleosome structure in chromatin. Our findings provide additional support on the role of epigenetic abnormalities (histome acetylation and DNA methylation) in the development of SLE and other autoimmune diseases (25).
Previously, it was suggested that histone deacetylase inhibitors limit disease in lupus-prone mice (32). One mechanism for such an effect may involve enhanced presence of acetylated histones in the SYK promoter which would enable the binding of the transcriptional repressor CREM α resulting in suppression of the expression of SYK. Therefore, the stipulated introduction of histone deacetylase inhibitors in the treatment of SLE is supported by our findings. In addition, SYK inhibition has been shown to be of value in the treatment of rheumatoid arthritis (33) and may represent an appropriate modality in the treatment of patients with SLE. Understanding all these molecular bases of its increased expression will facilitate the development of future improved therapeutic approaches for SLE patients. Additional studies are needed to further understand the role of histone acetylation and deacetylation on the expression of other genes which are expressed abnormally in SLE T cells such as CD3 ζ, CREM α, CamKIV and PP2A (34).
ACKNOWLEDGEMENT
This work was supported by an NIH grant RO1 AI42269 to GCT.
ABBREVIATIONS
- SYK
spleen tyrosine kinase
- CRE
cAMP response element
- CREM
cAMP response-element modulator protein
Footnotes
Conflict statement: None of the authors has an apparent conflict of interest with any company or other entity.
REFERENCES
- 1.Rudd CE. Adaptors and molecular scaffolds in immune cell signaling. Cell. 1999;96(1):5–8. doi: 10.1016/s0092-8674(00)80953-8. [DOI] [PubMed] [Google Scholar]
- 2.Sada K, Takano T, Yanagi S, Yamamura H. Structure and function of SYK protein-tyrosine kinase. J Biochem. 2001;130(2):177–186. doi: 10.1093/oxfordjournals.jbchem.a002970. [DOI] [PubMed] [Google Scholar]
- 3.Palacios EH, Weiss A. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene. 2004;23(48):7990–8000. doi: 10.1038/sj.onc.1208074. [DOI] [PubMed] [Google Scholar]
- 4.Chu P, Pardo J, Zhao H, Li CC, Pali E, Shen MM, et al. Systematic identification of regulatory proteins critical for T-cell activation. J Biol. 2003;2(3):21. doi: 10.1186/1475-4924-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krishnan S, Warke VG, Nambiar MP, Tsokos GC, Farber DL. The FcR gamma subunit and SYK kinase replace the CD3 zeta-chain and ZAP-70 kinase in the TCR signaling complex of human effector CD4 T cells. J Immunol. 2003;170(8):4189–4195. doi: 10.4049/jimmunol.170.8.4189. [DOI] [PubMed] [Google Scholar]
- 6.Kadlecek TA, van Oers NS, Lefrancois L, Olson S, Finlay D, Chu DH, et al. Differential requirements for ZAP-70 in TCR signaling and T cell development. J Immunol. 1998;161(9):4688–4694. [PubMed] [Google Scholar]
- 7.Couture C, Baier G, Altman A, Mustelin T. p56lck-independent activation and tyrosine phosphorylation of p72SYK by T-cell antigen receptor/CD3 stimulation. Proc Natl Acad Sci U S A. 1994;91(12):5301–5305. doi: 10.1073/pnas.91.12.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan AC, van Oers NS, Tran A, Turka L, Law CL, Ryan JC, et al. Differential expression of ZAP-70 and SYK protein tyrosine kinases, and the role of this family of protein tyrosine kinases in TCR signaling. J Immunol. 1994;152(10):4758–4766. [PubMed] [Google Scholar]
- 9.Tsokos GC, Nambiar MP, Tenbrock K, Juang YT. Rewiring the T-cell: signaling defects and novel prospects for the treatment of SLE. Trends Immunol. 2003;24(5):259–263. doi: 10.1016/s1471-4906(03)00100-5. [DOI] [PubMed] [Google Scholar]
- 10.Juang YT, Wang Y, Jiang G, Peng HB, Ergin S, Finnell M, et al. PP2A dephosphorylates Elf-1 and determines the expression of CD3zeta and FcRgamma in human systemic lupus erythematosus T cells. J Immunol. 2008;181(5):3658–3664. doi: 10.4049/jimmunol.181.5.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnan S, Kiang JG, Fisher CU, Nambiar MP, Nguyen HT, Kyttaris VC, et al. Increased caspase-3 expression and activity contribute to reduced CD3zeta expression in systemic lupus erythematosus T cells. J Immunol. 2005;175(5):3417–3423. doi: 10.4049/jimmunol.175.5.3417. [DOI] [PubMed] [Google Scholar]
- 12.Krishnan S, Juang YT, Chowdhury B, Magilavy A, Fisher CU, Nguyen H, et al. Differential expression and molecular associations of SYK in systemic lupus erythematosus T cells. J Immunol. 2008;181(11):8145–8152. doi: 10.4049/jimmunol.181.11.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66 doi: 10.1146/annurev.biochem.66.1.807. 807-802. [DOI] [PubMed] [Google Scholar]
- 14.Matthews RP, Guthrie CR, Wailes LM, Zhao X, Means AR, McKnight GS. Calcium/calmodulin-dependent protein kinase types II and IV differentially regulate CREB-dependent gene expression. Mol Cell Biol. 1994;14(9):6107–6116. doi: 10.1128/mcb.14.9.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loriaux MM, Brennan RG, Goodman RH. Modulatory function of CREB-CREMα heterodimers depends upon CREMα phosphorylation. J Biol Chem. 1994;269:28839–28843. [PubMed] [Google Scholar]
- 16.Tenbrock K, Juang YT, Tolnay M, Tsokos GC. The cyclic adenosine 5'-monophosphate response element modulator suppresses IL-2 production in stimulated T cells by a chromatin-dependent mechanism. J Immunol. 2003;170(6):2971–2976. doi: 10.4049/jimmunol.170.6.2971. [DOI] [PubMed] [Google Scholar]
- 17.Tenbrock K, Juang YT, Leukert N, Rith J, Tsokos GC. The transcriptional repressor cAMP response element modulator alpha interacts with histone deacetylase 1 to repress promoter activity. J Immunol. 2006;177(9):6159–6164. doi: 10.4049/jimmunol.177.9.6159. [DOI] [PubMed] [Google Scholar]
- 18.Tenbrock K, Kyttaris VC, Ahlmann M, Ehrchen JM, Tolnay M, Melkonyan H, et al. The cyclic AMP response element modulator regulates transcription of the TCR zeta-chain. J Immunol. 2005;175(9):5975–5980. doi: 10.4049/jimmunol.175.9.5975. [DOI] [PubMed] [Google Scholar]
- 19.Mishra N, Khan IU, Tsokos GC, Kammer GM. Association of deficient type II protein kinase A activity with aberrant nuclear translocation of RIIβ subunit in systemic lupus erythematosus T lymphocytes. J Immunol. 2000;165:2830–2840. doi: 10.4049/jimmunol.165.5.2830. [DOI] [PubMed] [Google Scholar]
- 20.Juang YT, Wang Y, Solomono EE, Li Y, Marwin C, Tenbrock K, Kyttaris VC, Tsokos GC. Systemic lupus erythematosus serum IgG increases CREM binding to the IL-2 promoter and suppress IL-2 production through CaMKIV. J Clinical Investigation. 2005;115:996–1004. doi: 10.1172/JCI200522854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foulkes NS, Borrelli E, Sassone-Corsi P. CREM gene: use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcription. Cell. 1991;64(4):739–749. doi: 10.1016/0092-8674(91)90503-q. [DOI] [PubMed] [Google Scholar]
- 22.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng GM, Liu L, Bahjat FR, Pine PR, Tsokos GC. Suppression of skin and kidney disease by inhibition of spleen tyrosine kinase in lupus-prone mice. Arthritis Rheum. 2010;62(7):2086–2092. doi: 10.1002/art.27452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel DR, Richardson BC. Epigenetic mechanisms in lupus. Current opinion in immunology. 2010;22:478–482. doi: 10.1097/BOR.0b013e32833ae915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richardson BC. Primer: epigenetics of autoimmunity. Nat Clin Pract Rheumatol. 2007;3(9):521–527. doi: 10.1038/ncprheum0573. [DOI] [PubMed] [Google Scholar]
- 26.Zhao S, Long H, Lu Q. Epigenetic perspectives in systemic lupus erythematosus: pathogenesis, biomarkers, and therapeutic potentials. Clinc Rev Allerg Immunol. 2010;39:3–9. doi: 10.1007/s12016-009-8165-7. [DOI] [PubMed] [Google Scholar]
- 27.Zhao M, Sun Y, Gao F, Wu X, Tang J, Yin HLY, Richardson B, Lu Q. Epigenetics and SLE: RFX1 downregulation causes CD11a and CD70 overexpression by altering modifications in lupus CD4+ T cells. J Autoimmunity. 2010;35:58–69. doi: 10.1016/j.jaut.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Garcia BA, Busy SA, Shabanowitz J, Hunt DF, Mishra N. Resetting the epigenetic histone code in the MRL-lpr/lpr mouse model of lupus by histone deacetylase inhibition. J Proteome Research. 2005;4:2032–2042. doi: 10.1021/pr050188r. [DOI] [PubMed] [Google Scholar]
- 29.Privalsky ML. Depudecin makes a debut. Commentry. Proc Natl Acad Sci. 1998;95:3335–3337. doi: 10.1073/pnas.95.7.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 31.Bird A, Jaenisch R. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 32.Mishra N, Reilly CM, Brown DR, Ruiz P, Gilkeson GS. Histone deacetylase inhibitors modulate renal disease in MRL-lpr/lpr mouse. J Clin invest. 2003;111:539–552. doi: 10.1172/JCI16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinblatt ME, Kavanaugh A, Genovese MC, Musser TK, Grossbard EB, Magilavy DB. An oral spleen tyrosine kinase (SYK) inhibitor for rheumatoid arthritis. N Engl J Med. 2010;363(14):1303–1312. doi: 10.1056/NEJMoa1000500. [DOI] [PubMed] [Google Scholar]
- 34.Tsokos GC. Systemic lupus erythematosus from pathogenesis to treatment. N Eng J Med. 2011 (In Press) [Google Scholar]