Abstract
Background
GABRG1 and GABRA2, genes that encode the γ1 and α2 subunits, respectively, of the GABA-A receptor, are located in a cluster on chromosome 4p. Association of alcohol dependence (AD) with markers located at the 3′ region of GABRA2 has been replicated in several studies, but recent studies suggested the possibility that the signal may be attributable to the adjacent gene, GABRG1, located 90kb distant in the 3′ direction. Due to strong linkage disequilibrium in European-Americans, the origin, or origins, of the association signal are very difficult to discern, but our previous population-based study suggested that decreased LD across the GABRG1- GABRA2 region in African-Americans (AAs) may be useful for fine mapping and resolution of the association signal in that population.
Methods & Results
To examine these associations in greater detail, we genotyped 13 single nucleotide polymorphisms (SNPs) spanning GABRG1 and GABRA2 in 380 AAs with AD and in 253 AA controls. Although there was no association between any individual SNP and AD, a highly significant difference was shown between AD subjects and controls in the frequency of a 3-SNP GABRA2 haplotype (global p=0.00029). A similar level of significance was obtained in 6-SNP haplotypes that combined tagging SNPs from both genes (global p=0.00994). High statistical significance was also shown with a 6-SNP haplotype (T-G-C-G-T-A), p=0.0033. The T-G-C-G-T-A haplotype contains the most significant GABRA2 3-SNP haplotype (p= 0.00019), G-T-A.
Conclusions
These findings reflect the interrelationship of these two genes and the likelihood that risk loci exist in each of them. Study of an AA population allowed evaluation of these associations at higher genomic resolution than is possible in a European-American population, owing to the much lower linkage disequilibrium across these loci in AAs.
Introduction
Alcohol dependence (AD) is a genetically complex disorder in which both genetic and environmental factors play crucial roles. Heritability has been estimated to be at least 50% for both men and women (Reich et al., 1998). Adaptation of GABAA receptors to ethanol exposure may play a role in the pathophysiology of AD (Harris et al., 1995, 1997, 1998; Iwata et al., 2000).
Nineteen heterogeneous subunits of the GABAA receptor (6 α, 3 β, 3 γ, 1 δ, 1 ε, 1 θ, 1 π and 3 ρ) have been identified (Belelli and Lambert, 2005), with 70% sequence homology within classes and 30% homology between classes. Functional receptors are pentameric, with the receptor usually comprised of 2 α subunits, 2 β subunits, and one γ subunit. Genetic variation in genes coding receptor subunits, including, for example, variants affecting exon splicing mechanisms (Barnard et al., 1998; Sieghart and Sperk, 2002; Simon et al., 2004), may be responsible for differences in receptor composition and therefore differences in multimeric receptor properties, including location in the neuron, change in sensitivity to neurotransmitter, kinetics of the channel, other pharmacological and physiological properties, and expression patterns (Belelli and Lambert, 2005).
Several association studies have implicated genes encoding GABA-A subunits in the risk for AD. GABRA2, which encodes the α2 subunit, has been the most widely studied, with the majority of studies (Covalt et al., 2004; Drgon et al., 2006; Edenberg et al., 2004; Lappalainen et al., 2005, Enoch et al., 2006; Fehr et al., 2006; Matthews et al., 2007; Soyka et al., 2008), providing evidence that markers located at the 3′ region of the gene are associated to the risk for AD. GABRA2 maps to the GABA gene cluster on human chromosome 4 (4p12-p13), which also includes GABRG1, GABRB1 and GABRB4, genes that encode the γ1, β1 and β4 subunits of GABA-A, respectively (Buckle et al., 1989; Kirkness et al., 1991; McLean et al., 1995; Wilcox et al., 1992). No functional variant has yet been found in the GABRA2 gene that could account for the association. The previously reported associations between GABRA2 and AD could be attributable in part to the adjacent gene, GABRG1, which is located 90kb distant in the 3′ direction, and is in strong LD with GABRA2 in many populations (Ittiwut et al 2008). This explanation was first suggested by Covault et al., (2008) in a study of European Americans and was replicated and extended in a study of Plains Indians and Finns (Enoch et al., 2009). The aim of the present study was to investigate these two candidate genes in AAs, where linkage disequilibrium is lower than that in other populations (Ittiwut et al., 2008), possibly permitting the source of an association signal (or signals) to be resolved.
Materials and Methods
Populations
Three hundred eighty subjects diagnosed with AD (cases) and 253 healthy control subjects (screened to exclude major psychiatric illness) were recruited at three centers, including the University of Connecticut Health Center (Farmington) [224 cases (93 females (mean age 38.1 ± 11.3 yr) / 131 males (mean age 40.3 ± 11.29 yr) and 110 controls (77 females (mean age 39.9 ± 9.98 yr) / 33 males (mean age 38.9 ± 9.52 yr)], Yale University School of Medicine (New Haven, CT) [133 cases (52 females (mean age 38.6 ± 10.8 yr) / 81 males (mean age 38.0 ± 11.4 yr) and 52 controls (35 females (mean age 34.2 ± 10.2 yr) / 17 males (mean age 33.7 ± 11.4 yr)], and the Medical University of South Carolina (Charleston) [23 cases (6 females (mean age 39.2 ± 14.1 yr) / 17 males (mean age 35.0 ± 11.0 yr) and 91 controls (77 females (mean age 35.1 ± 11.4 yr) / 14 males (mean age 42.5 ± 12.3 yr)]. All subjects were of substantial African descent and all provided written informed consent as approved by the institutional review board at each institution. The race of all subjects was defined according to self-identification and confirmed by Bayesian model-based clustering using genetic marker information (Gelernter et al., 2005). Psychiatric diagnosis was based on responses obtained using the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) (Pierucci-Lagha et al., 2005) or the Structured Clinical Interview for DSM-IV disorders (SCID) (Spitzer et al., 1992; Williams et al., 1992).
DNA samples were obtained from blood, saliva, or immortalized cell lines. There is no overlap in subjects between the present study and the report by Covault et al., (2008).
Single nucleotide polymorphism (SNP) selection and genotyping
Our earlier work (Ittiwut et al., 2008) identified four regions of strong LD defined by D'>0.8 based on data from a set of 13 SNP markers in 48 AA subjects (Figure 1). We used the htSNP approach in HAPLOVIEW software version 3.32 (available at http://www.broad.mit.edu/mpg/haploview) (Barrett et al., 2005) to identify 6 haplotype tagging (ht) SNPs spanning a 312.6 kb region including GABRG1 and GABRA2. These SNPs, all of which were intronic, had a minor allele frequency (MAF) ≥15%, and included rs1497571, rs10938426, rs10033451, rs567926, rs279869, and rs279837. We will refer to these SNPs as A, D, F, G, J and L after the nomenclature used in our prior report (Ittiwut et al., 2008). SNPs A, D, and F map to GABRG1 and G, J, and L map to GABRA2. Of these, rs279837 (in intron 3 of GABRA2) tagged the 2-SNP LD block composed of that SNP and another synonymous SNP, rs279858, in exon 5 of the same gene. Genotyping was performed using a Taqman genotyping method as described previously (Ittiwut et al., 2008). All genotyping was performed in duplicate; genotyped plates with fewer than 3 mismatched samples (of 384; i.e., an error rate of <0.78%) were employed in the analysis; discordant genotypes were discarded. PCR amplification was carried out under the following conditions: 95° C for 10 min, followed by 15 s at 92° C, and then 60 s at 60° C for 40 cycles. Signals were analyzed with an ABI Prism 7900HT sequence detector using software version 2.1 from Applied Biosystems (Foster City, CA).
Figure 1.
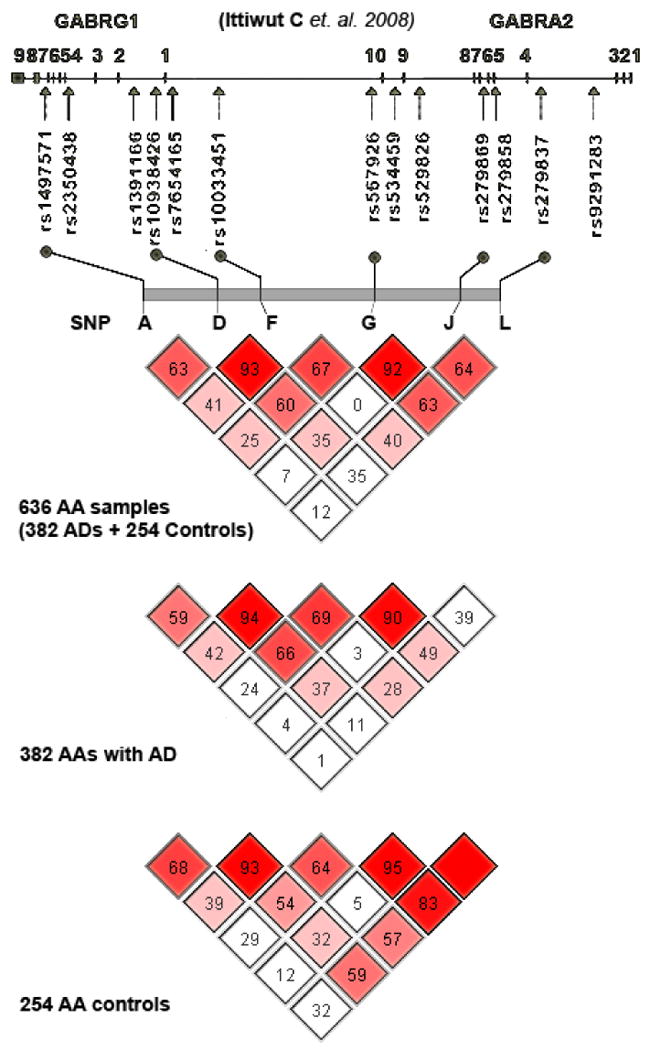
LD structure and location of 6 htSNPs over the GABRG1/GABRA2 region on chromosome 4p13-12 in African Americans (controls, AD subjects, and combined sample). Map shows placement of the present SNPs with respect to the larger set in our earlier report (Ittiwut et al 2008)
Inferred ancestry proportions
We ran the program STRUCTURE (Falush et al., 2003) to infer the individual ancestry proportions based on the genotype data for 37 ancestry informative markers (AIMs) (Yang et al., 2005a, 2005b; Stein et al., 2004). Four hundred seventeen of the 636 AA subjects had AIMs; an additional 105 European American (EA) subjects were included in the STRUCTURE run to set EA ancestral allele frequencies (to increase the accuracy when inferring admixture). The Monte Carlo simulation parameters were set to 50,000 burn-ins followed by 50,000 iterations, and K was set to 2. The inferred ancestry proportions were used as a covariate in the subsequent logistic regression.
Statistical analysis
Genotype and allele frequencies for each SNP were determined in the case and control groups. The chi-square test, with significance defined as p<0.05 for any single SNP comparison, was employed to determine goodness of fit between observed and expected genotype or allele frequencies comparing AD subjects with controls. Single marker association analysis was also carried out in a logistic regression with additive genetic model, and ancestry proportion and subject recruitment site as covariates. Analysis of haplotypic association was performed using the chi-square test to compare frequencies of haplotypes composed of all 6 tagging SNPs, spanning both loci. Bonferroni correction was employed to control for multiple testing. Frequencies for the commonly observed haplotypes were estimated using the computer program PHASE (Stephens and Donnelly, 2003) (version 2.1.1 available at fttp://www.stat.washington.edu/stephens/phase.html). We also computed global tests of significance for single SNP and the 6-SNP haplotypic associations between AD and controls subjects.
Results
The chi-square tests show that genotype and allele frequencies of all selected SNPs did not differ between the AD and control groups (Table 1). In the single marker association analysis, in the subset of the sample with AIM information, when the covariates of ancestry proportion and subject recruitment site were included as covariates, there was no significant single SNP association signal. A site effect for MUSC (p-value <0.0001) is likely due to the fact that the smallest proportion of AD subjects (6.1%) and the largest proportion of control subjects (36%) were recruited at that site. To gauge the contribution of each gene to AD risk, we tested differences in estimated haplotype frequency based on 3-SNP haplotypes spanning either GABRG1 or GABRA2. We designated GABRG1 haplotypes “G---” and GABRA2 haplotypes, “A---” (Tables 2 and 3). Notable significant differences were observed in 3-SNP haplotypes mapped to the GABRA2 gene, where significant differences were shown in 2 of 5 haplotypes, including the G-T-A and A-G-G (A2-4, and A2-5) haplotypes, which accounted for 10.6% and 3.2% in the case and control groups, respectively (Table 3). This corresponds to a global p-value of 0.00029 (df=4, χ2=18.87). Both haplotypes remained significant (p= 0.00019 and p= 0.00042) using a Bonferroni-corrected threshold of 0.01 (0.05/5).
Table 1. GABRG1-GABRA2 SNP information and genotype and allele frequencies for African American subjects*.
| SNP information | Genotype or Allele | Frequencies |
p-value (χ2) (ADs vs Controls) |
||||
|---|---|---|---|---|---|---|---|
| Gene | Location | SNP (nt change) | ADs (N=380) |
Controls (N=253) |
Alleles | Genotypes | |
| GABRG1 | intron 7 (46059250) |
SNP A: rs1497571 (C/T) |
CC | 0.143 | 0.147 | 0.99 | |
| CT | 0.477 | 0.478 | |||||
| TT | 0.379 | 0.375 | |||||
| C* | 0.382 | 0.386 | 0.87 | ||||
| T | 0.618 | 0.614 | |||||
| intron 1 (46117985) |
SNP D: rs10938426 (A/G) |
AA | 0.047 | 0.075 | 0.29 | ||
| AG | 0.375 | 0.390 | |||||
| GG | 0.578 | 0.535 | |||||
| A* | 0.235 | 0.272 | 0.14 | ||||
| G | 0.765 | 0.728 | |||||
| 26kb at 5′ (46152082) |
SNP F: rs10033451 (T/C) |
TT | 0.401 | 0.410 | 0.85 | ||
| TC | 0.467 | 0.446 | |||||
| CC | 0.133 | 0.145 | |||||
| T | 0.634 | 0.633 | 0.96 | ||||
| C* | 0.366 | 0.367 | |||||
| GABRA2 | 9kb at 3′ (46241519) |
SNP G: rs567926 (G/A) |
GG | 0.078 | 0.061 | 0.56 | |
| GA | 0.377 | 0.414 | |||||
| AA | 0.545 | 0.525 | |||||
| G* | 0.269 | 0.268 | 0.99 | ||||
| A | 0.731 | 0.732 | |||||
| intron 6 (46307745) |
SNP J: rs279869 (G/T) |
GG | 0.111 | 0.120 | 0.92 | ||
| GT | 0.380 | 0.369 | |||||
| TT | 0.509 | 0.510 | |||||
| G* | 0.300 | 0.305 | 0.84 | ||||
| T | 0.700 | 0.695 | |||||
| intron 3 (46339073) |
SNP L: rs279837 (A/G) |
AA | 0.554 | 0.542 | 0.90 | ||
| AG | 0.382 | 0.386 | |||||
| GG | 0.064 | 0.072 | |||||
| A | 0.745 | 0.735 | 0.68 | ||||
| G* | 0.255 | 0.265 | |||||
Allele nucleotide designations refer to NCBI sequence NT_006238. The RefSNP (rs) IDs of SNP A, D, F, G, J, and L as shown in this table represent the following Celera IDs; (A) hCV3030378, (D) hCV1445604, (F) hCV11763588, (G) hCV7537087, (J) hCV8262927, (L) hCV8263070 respectively. Asterisks indicate minor allele frequency of each SNP. Locations of each SNP on chromosome 4 (in parentheses) are located following the UCSC hg19/GRCh37 human genome assembly.
Table 2.
Haplotypic association between 3 GABRG1 SNPs (A, D, and F) and AD in African Americans.
| Haplotype | ADs | Controls | Odds Ratio | 95% CI | ||||
|---|---|---|---|---|---|---|---|---|
| Name | ADF | haplotype (N) |
E(freq) | haplotype (N) |
E(freq) | p-value | ||
| G1-1 | TGC | 234 | 0.308 | 157 | 0.310 | 1 | 1 | (0.74-1.34) |
| G1-2 | TGT | 200 | 0.263 | 134 | 0.265 | 1 | 1 | (0.74-1.36) |
| G1-3 | CAT | 144 | 0.189 | 116 | 0.229 | 0.292 | 0.83 | (0.60-1.16) |
| G1-4 | CGT | 106 | 0.139 | 50 | 0.099 | 0.080 | 1.42 | (0.95-2.15) |
| G1-5 | CGC | 42 | 0.055 | 28 | 0.055 | 1 | 1.00 | (0.58-1.76) |
|
| ||||||||
| 0.954 | 0.958 | |||||||
The p-value for each haplotype was generated by the Fisher exact test. The odds ratio was calculated with haplotype G1-1 as reference.
Global p-value = 0.1701 (df=4, χ2=6.417)
Significance at threshold 0.01 based on Bonferroni correction correspond to 0.05/5
Table 3. GABRA2 3-SNP haplotype association (SNPs G, J, and L).
| Haplotype | ADs | Controls | Odds Ratio | 95% CI | ||||
|---|---|---|---|---|---|---|---|---|
| Name | GJL | haplotype (N) |
E(freq) | haplotype (N) |
E(freq) | p-value | ||
| A2-1 | ATA | 289 | 0.380 | 202 | 0.399 | 1 | 1 | (0.77-1.30 |
| A2-2 | AGA | 208 | 0.274 | 154 | 0.304 | 0.73 | 0.94 | (0.71-1.26) |
| A2-3 | GTG | 133 | 0.175 | 117 | 0.231 | 0.158 | 0.79 | (0.58-1.09) |
| A2-4 | GTA | 65 | 0.086 | 16 | 0.032 | *0.00019 | 2.83 | (1.57-5.41) |
| A2-5 | AGG | 15 | 0.020 | 0 | 0 | *0.00042 | --- | --- |
|
| ||||||||
| 0.935 | 0.966 | |||||||
The p-value for each haplotype was generated by the Fisher exact test. The odds ratio was calculated with haplotype A2-1 as reference. The odds ratio for haplotype A2-5 could not be calculated as it is completely absent in the controls.
p-value < 0.05 is in bolded text.
The odds ratio for haplotype A2-5 could not be calculated as it is completely absent in controls.
Global p-value = 0.00029 (df= 4, χ2 = 18.87)
Significance at threshold 0.0125 based on Bonferroni correction corresponding to 0.05/4.
Haplotypes were inferred from all six tagging SNPs for both GABRG1 and GABRA2 (Table 4) to evaluate an interactive effect of these genes on risk of AD. The combination of all tagging SNPs yielded 8 haplotypes; each represented ≥2% of observations and together they accounted for 72.2% of the chromosomes among AD subjects and 80.8% among controls. Global analysis showed a nominally significant difference in haplotype frequency between AD and control groups, p=0.00994 (df=7, χ2=18.49). This compares with a Bonferroni-corrected p=0.00625. Specific associations of each common haplotype compared to others were also analyzed. Among these common haplotypes, which spanned 280 kb, the most significant difference was observed in the T-G-C-G-T-A (H8) haplotype (p=0.0033, OR=2.63, 95%CI=1.32-5.54) and accounts for 6.4% of haplotypes in the AD group but only 2.6% in controls. No other haplotype was even nominally significant different between AD subjects vs. controls. Considering the rarity of observations in affecteds and lack of this haplotype in controls, this finding based on estimated haplotype frequencies must be regarded as tentative.
Table 4. GABRG1-GABRA2 6-SNP haplotype association (SNP A to L).
| Haplotype | AD Subjects | Controls | p-value | Odd Ratio | 95% CI | |||
|---|---|---|---|---|---|---|---|---|
| Name | ADFGJL | haplotype (N) |
E(freq) | haplotype (N) |
E(freq) | |||
| H1 | TGTATA | 156 | 0.205 | 109 | 0.215 | 1 | 1 | (0.70-1.43) |
| H2 | TGCGTG | 71 | 0.093 | 70 | 0.138 | 0.115 | 0.71 | (0.46-1.09) |
| H3 | CATAGA | 68 | 0.089 | 63 | 0.125 | 0.198 | 0.75 | (0.48-1.18) |
| H4 | TGCAGA | 68 | 0.089 | 50 | 0.099 | 0.823 | 0.95 | (0.60-1.51) |
| H5 | CATATA | 53 | 0.070 | 46 | 0.091 | 0.405 | 0.81 | (0.49-1.32) |
| H6 | CGTATA | 50 | 0.066 | 29 | 0.057 | 0.515 | 1.20 | (0.70-2.11) |
| H7 | CGCGTG | 35 | 0.046 | 29 | 0.057 | 0.574 | 0.84 | (0.47-1.52) |
| H8 | TGCGTA | 49 | 0.064 | 13 | 0.026 | 0.0033 | 2.63 | (1.32-5.54) |
|
| ||||||||
| 0.722 | 0.808 | |||||||
The p-value for each haplotype was generated by the Fisher exact test. The odds ratio was calculated with haplotype H1 as reference.
p-value < 0.05 is in bolded text.
Global p-value = 0.00994 (df=7, χ2=18.49).
Significance at threshold 0.00625 based on Bonferroni correction corresponding to 0.05/8.
Discussion
We genotyped 6 tagging SNPs spanning GABRA2 and GABRG1 from among 13 previously identified SNPs in a sample of 380 AA subjects with AD and 253 AA control subjects. Although we did not find significant evidence for single SNP association with AD, haplotype-based analysis of GABRG1-GABRA2 showed promising results in both global and specific comparisons. There was evidence for haplotypic association with both genes, but the evidence for association with GABRA2 was much stronger than that for GABRG1. To investigate the interactions of multiple SNPs both within each gene and spanning both genes, haplotypes were reconstructed combining three SNPs for each gene separately and all six tagging SNPs together. Three-SNP haplotypes were reconstructed from SNPs A, D, and F to investigate the role of GABRG1. Three-SNP haplotypes containing SNPs G, J, and L were reconstructed for the GABRA2 analysis. A nominally significant difference was observed in the G1-4 haplotype (C-G-T in Table 2), which reflects the potential for a small role of GABRG1 in AD risk in AAs, but the difference in GABRA2 haplotype frequencies was much more statistically significant (Table 3). The most significantly-associated risk haplotype (p=0.00019, OR=2.83, 95%CI=1.57-5.41) was A2-4 (G-T-A), which was estimated to be present on 65 chromosomes (8.6%) in AD subjects compared to only 16 chromosomes (3.2%) in the control group. Another haplotype that was less significant in the AD group than the control group, A2-5 (A-G-G with p=0.00042), was completely absent in the control group (such that the OR could not be calculated).
The p-value for the GABRA2 A2-4 (G-T-A) haplotype (p=0.00019) can be compared to that for the 6-SNP H8 haplotype: T-G-C-G-T-A (Table 4), which includes rs1497571, rs10938426, rs10033451, rs567926, rs279869, and rs279837, and yielded a p-value of 0.0033) (OR=2.63; 95%CI=1.32-5.54). It should be noted that the 6-SNP H8 haplotype was the only 6-SNP haplotype carrying the A2-4 (GTA) GABRA2 haplotype associated to AD. The GABRG1 component (G1-1, TGC) of this 6-SNP haplotype did not show any association to AD alone (Table 2). In conclusion, the findings reported here in an AA sample are globally consistent with the findings obtained previously by Covault et al., (2008) in EAs. Specifically, it appears that GABRG1 and GABRA2 genes are interrelated or that independent risk loci for AD exist in each of them. Previous studies have suggested an association of AD with either GABRA2 (Covault et al., 2004; Edenberg et al., 2004; Lappalainen et al., 2005) or GABRG1 (Covault et al., 2008; Enoch et al., 2009) markers. In our study, GABRA2 haplotypes accounted for most of the association signal and joint effects of the two loci. Inspection of the LD patterns in EA and AA populations (Ittiwut et al., 2008) suggests that the effects of these two loci are very difficult to disentangle, by study of EA samples alone. Because there is much less LD in AAs, we sought to distinguish effects of the two loci by studying association with AD in this population group. We found evidence for association with markers in both genes, but the evidence for GABRA2 was much stronger. This demonstrates the utility of using a low-LD population to resolve an issue of fine-scale LD mapping.
Acknowledgments
This work was supported by NIH grants R01 AA11330, R01 AA017535, D43 TW06166, R01 DA12690, R01 DA12849, K01DA24758, K24 DA15105, K24 DA022288, K24 AA13736 and M01 RR06192; and the U.S. Department of Veterans Affairs (the VA Connecticut–Massachusetts Mental Illness Research, Education and Clinical Center [MIRECC]). Dr. Kathleen Brady participated in the subject recruitment and assessment part of the study. Ann Marie Lacobelle and Greg Kay provided excellent technical assistance. John Farrell provided excellent database support.
References
- Barnard EA, Skolnick P, Olsen RE, Mohler H, Sieghart W, Biggio G, Braestrup C, Bateson AN, Langer SZ. International Union of Pharmacology. XV. Subtypes of gamma-aminobutyric acidA receptors: classification on the basis of subunit structure and receptor function. Pharmacol Rev. 1998;50:291–313. [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Belelli D, Lambert JJ. Neurosteroids: endogenous regulators of the GABA(A) receptor. Nat Rev Neurosci. 2005;6:565–575. doi: 10.1038/nrn1703. [DOI] [PubMed] [Google Scholar]
- Buckle VJ, Fujita N, Ryder-Cook AS, Derry JM, Barnard PJ, Lebo RV, Schofield PR, Seeburg PH, Bateson AN, Darlison MJ, Barnard EA. Chromosomal localization of GABAA receptor subunit genes: relationship to human genetic disease. Neuron. 1989;3:647–654. doi: 10.1016/0896-6273(89)90275-4. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2004;129B:104–109. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Jensen K, Anton R, Kranzler HR. Markers in the 5′-region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology. 2008;33:837–848. doi: 10.1038/sj.npp.1301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drgon T, D'Addario C, Uhl GR. Linkage disequilibrium, haplotype and association studies of a chromosome 4 GABA receptor gene cluster: candidate gene variants for addictions. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:854–860. doi: 10.1002/ajmg.b.30349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, Jr, O'Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Hodgkinson CA, Yuan Q, Albaugh B, Virkkunen M, Goldman D. GABRG1 and GABRA2 as independent predictors for alcoholism in two populations. Neuropsychopharmacology. 2009;34:1245–1254. doi: 10.1038/npp.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Schwartz L, Albaugh B, Virkkunen M, Goldman D. Dimensional anxiety mediates linkage of GABRA2 haplotypes with alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:599–607. doi: 10.1002/ajmg.b.30336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C, Dahmen N, Schmidt LG, Szegedi A. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet. 2006;16:9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B, Poling J, Wilcox M, Farrer L, Kranzler HR. Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet B Neuropsychiatr Genet. 2005;136B(1):45–52. doi: 10.1002/ajmg.b.30189. 2005 Jul 5. [DOI] [PubMed] [Google Scholar]
- Harris RA, Mihic SJ, Brozowski S, Hadingham H, Whiting PJ. Ethanol, flunitrazepam, and pentobarbital modulation of GABAA receptors expressed in mammalian cells and Xenopus oocytes. Alcohol Clin Exp Res. 1997;21:444–451. doi: 10.1111/j.1530-0277.1997.tb03789.x. [DOI] [PubMed] [Google Scholar]
- Harris RA, Proctor WR, McQuilkin SJ, Klein RL, Mascia MP, Whatley V, Whiting PJ, Dunwiddie TV. Ethanol increases GABAA responses in cells stably transfected with receptor subunits. Alcohol Clin Exp Res. 1995;19:226–232. doi: 10.1111/j.1530-0277.1995.tb01496.x. [DOI] [PubMed] [Google Scholar]
- Harris RA, Valenzuela CF, Brozowski S, Chuang L, Hadingham K, Whiting PJ. Adaptation of gamma-aminobutyric acid type A receptors to alcohol exposure: studies with stably transfected cells. J Pharmacol Exp Ther. 1998;284:180–188. [PubMed] [Google Scholar]
- Ittiwut C, Listman J, Mutirangura A, Malison R, Covault J, Kranzler HR, Sughondhabirom A, Thavichachart N, Gelernter J. Interpopulation linkage disequilibrium patterns of GABRA2 and GABRG1 genes at the GABA cluster locus on human chromosome 4. Genomics. 2008;91:61–69. doi: 10.1016/j.ygeno.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata N, Virkkunen M, Goldman D. Identification of a naturally occurring Pro385-Ser385 substitution in the GABA(A) receptor alpha6 subunit gene in alcoholics and healthy volunteers. Mol Psychiatry. 2000;5:316–319. doi: 10.1038/sj.mp.4000706. [DOI] [PubMed] [Google Scholar]
- Kirkness EF, Kusiak JW, Fleming JT, Menninger J, Gocayne JD, Ward DC, Venter JC. Isolation, characterization, and localization of human genomic DNA encoding the beta 1 subunit of the GABAA receptor (GABRB1) Genomics. 1991;10:985–995. doi: 10.1016/0888-7543(91)90189-l. [DOI] [PubMed] [Google Scholar]
- Lappalainen J, Krupitsky E, Remizov M, Pchelina S, Taraskina A, Zvartau E, Somberg LK, Covault J, Kranzler HR, Krystal JH, Gelernter J. Association between alcoholism and gamma-amino butyric acid alpha2 receptor subtype in a Russian population. Alcohol Clin Exp Res. 2005;29:493–498. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- Matthews AG, Hoffman EK, Zezza N, Stiffler S, Hill SY. The role of the GABRA2 polymorphism in multiplex alcohol dependence families with minimal comorbidity: within-family association and linkage analyses. J Stud Alcohol Drugs. 2007;68:625–633. doi: 10.15288/jsad.2007.68.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean PJ, Farb DH, Russek SJ. Mapping of the alpha 4 subunit gene (GABRA4) to human chromosome 4 defines an alpha 2-alpha 4-beta 1-gamma 1 gene cluster: further evidence that modern GABAA receptor gene clusters are derived from an ancestral cluster. Genomics. 1995;26:580–586. doi: 10.1016/0888-7543(95)80178-o. [DOI] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, Farrer L, Kranzler HR. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- Sieghart W, Sperk G. Subunit composition, distribution and function of GABA(A) receptor subtypes. Curr Top Med Chem. 2002;2:795–816. doi: 10.2174/1568026023393507. [DOI] [PubMed] [Google Scholar]
- Simon J, Wakimoto H, Fujita N, Lalande M, Barnard EA. Analysis of the set of GABA(A) receptor genes in the human genome. J Biol Chem. 2004;279:41422–41435. doi: 10.1074/jbc.M401354200. [DOI] [PubMed] [Google Scholar]
- Soyka M, Preuss UW, Hesselbrock V, Zill P, Koller G, Bondy B. GABA-A2 receptor subunit gene (GABRA2) polymorphisms and risk for alcohol dependence. J Psychiatr Res. 2008;42:184–191. doi: 10.1016/j.jpsychires.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Spitzer RL, Williams JB, Gibbon M, First MB. The Structured Clinical Interview for DSM-III-R (SCID). I: History, rationale, and description. Arch Gen Psychiatry. 1992;49:624–629. doi: 10.1001/archpsyc.1992.01820080032005. [DOI] [PubMed] [Google Scholar]
- Stein MB, Schork NJ, Gelernter J. A polymorphism of the beta1-adrenergic receptor is associated with low extraversion. Biol Psychiatry. 2004;56(4):217–224. doi: 10.1016/j.biopsych.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox AS, Warrington JA, Gardiner K, Berger R, Whiting P, Altherr MR, Wasmuth JJ, Patterson D, Sikela JM. Human chromosomal localization of genes encoding the gamma 1 and gamma 2 subunits of the gamma-aminobutyric acid receptor indicates that members of this gene family are often clustered in the genome. Proc Natl Acad Sci U S A. 1992;89:5857–5861. doi: 10.1073/pnas.89.13.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JB, Gibbon M, First MB, Spitzer RL, Davies M, Borus J, Howes MJ, Kane J, Pope HG, Jr, Rounsaville B, Wittchen WU. The Structured Clinical Interview for DSM-III-R (SCID). II. Multisite test-retest reliability. Arch Gen Psychiatry. 1992;49:630–636. doi: 10.1001/archpsyc.1992.01820080038006. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Zhao H, Kranzler HR, Gelernter J. Practical population group assignment with selected informative markers: characteristics and properties of Bayesian clustering via STRUCTURE. Genet Epidemiol. 2005 May;28(4):302–312. doi: 10.1002/gepi.20070. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Zhao H, Kranzler HR, Gelernter J. Characterization of a likelihood based method and effects of markers informativeness in evaluation of admixture and population group assignment. BMC Genet. 2005;6:50. doi: 10.1186/1471-2156-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]


