Abstract
Xenopus laevis provides a unique animal model, alternative to mouse, to study immunology. Even though, several methodologies have been developed for the generation of transgenic Xenopus, to date none have been adapted for the Xenopus laevis/gilli (LG) isogenetic clones that are essential for immunological studies. Since LG clones are generated via gynogenesis, transgenic methods using transgene integration into the sperm nuclei are not suited. Therefore, we have tested three alternative methods for LG transgenesis: the phiC31 integrase, the Sleeping Beauty transposase and the I-SceI meganuclease. All three techniques produced transgenic LG clones; however, the I-SceI meganuclease was most effective. It resulted in high transgenesis efficiency (35–50%), bright non-mosaic GFP expression as well as stable germline transmission with 100% of the progeny carrying the transgene. Production of transgenic LG clones will allow us to modulate immune gene expression and further strengthen Xenopus laevis as a biomedical model.
Keywords: Amphibian, Xenopus laevis, immunology, transgenesis, phiC31 integrase, Sleeping Beauty transposase
Introduction
The frog Xenopus laevis is a unique comparative model system used to study fundamental aspects of immunology. The immune system of Xenopus is similar to that of mammals and it is one of the best defined non-mammalian immune systems (Du Pasquier et al., 1989; Robert and Ohta, 2009). One of the advantages of the Xenopus model is the availability of MHC-defined isogenic cloned frogs such as the LG-6 and LG-15 (Kobel and Du Pasquier, 1975, 1977). These clones are hybrids between X. laevis and X. gilli, whose progenies are generated via gynogenesis. During this process the diploid LG eggs are activated by UV irradiated spermatozoids which do not contribute any genetic material to the progeny. These clones share the same MHC haplotype (a/c) but differ at multiple minor histocompatability loci which makes them very useful in immunological assays such as adoptive cell transfers since cells can be pooled from several animals (Maniero and Robert, 2004). Also this system has proven invaluable in studying T cell responses and the role of heat shock proteins in skin grafting and anti-tumor immunity (Robert and Cohen, 1998; Robert et al., 2004; Robert et al., 2002; Robert et al., 1994, 1995). However, the ability to modulate the expression of immunologically relevant genes (e.g., inducible transgenes or RNAi knockdown) in LG clones would considerably strengthen Xenopus as a biomedical model.
Transgenesis is well established in Xenopus and has been used successfully by developmental biologist for several decades. The most common technique used is the Restriction Enzyme Mediated Integration (REMI) which requires integration of the transgene into sperm nuclei which are then transplanted into unfertilized eggs (Amaya and Kroll, 1999; Kroll and Amaya, 1996). Even though this procedure has proven very effective in generating transgenic Xenopus, its use for producing transgenic LG clones is problematic since the clones are generated by gynogenesis. Recently, several new techniques, which do not require DNA integration into the sperm nuclei, were shown effective in producing high-throughput transgenesis in Xenopus. These methods include the phiC31 integrase (Allen and Weeks, 2005), the Sleeping Beauty (SB) transposase (Sinzelle et al., 2006; Yergeau et al., 2009) and the I-SceI meganuclease (Ogino et al., 2006; Pan et al., 2006).
In the present work we set out to generate the first transgenic LG clones. Using a GFP reporter we tested all three transgenesis techniques; the phiC31 integrase, the SB transposase and the I-SceI meganuclease. We found that although all three techniques were able to generate GFP transgenic LG clones, the I-SceI meganuclease was the most efficient and reliable for our objectives. Notably, the I-SceI technique resulted in 100% germline transmission of the transgene, which makes segregation analysis unnecessary.
Results
To determine the feasibility of transgenesis in Xenopus LG isogenetic clones, we tested three different transgenic techniques; the phiC31 integrase, the SB transposase and the I-SceI meganuclease.
The vector we used for the phiC31 integrase mediated integration consists of a GFP reporter gene driven by the CMV promoter that allows for constitutive transgene expression, and an attB site that allows integration of the plasmid into “pseudo” attP sites in the Xenopus genome (Allen and Weeks, 2005; Groth et al., 2000; Thyagarajan et al., 2001). In addition, this transgene was flanked by insulator sequences, which minimize chromatin-induced position effects. We co-injected integrase mRNA together with the vector into one-cell stage embryos. Tadpoles were screened for GFP expression at stage 56 (useful for immunological purposes) using a fluorescence stereomicroscope (Figure 1A). As shown in Table 1 we were able to produce transgenic LG clones with high transgene integration (30–40%, Figure 1Bb). Additionally, the survival rates of injected embryos (17±4%) were not significantly different control embryos (35±19%) although the variability of this later group was higher as indicated by the standard deviation (Table 1).
Figure 1. Generation of transgenic LG clones.
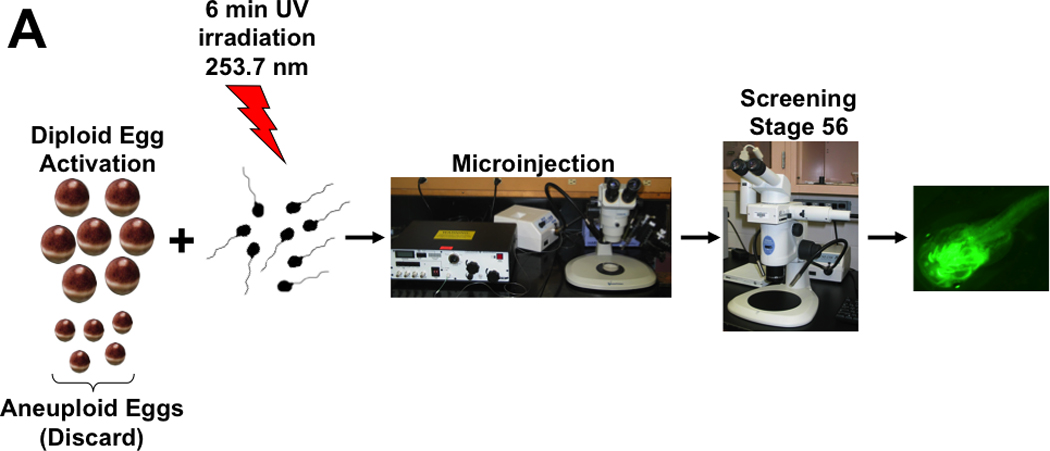

(A) General schema of the production of transgenic LG clones. LG female clones produce both diploid (large) and aneuploid (small) eggs. The small eggs were discarded while the diploid eggs were activated using UV irradiated sperm. After activation eggs were microinjected with the different constructs for transgenesis. Embryos were allowed to develop and were screened for GFP expression at stage 56 using a fluorescence stereomicroscope. Transgenic tadpoles were raised to adulthood in order to obtain F1 progeny. (B) Representative images of live non-transgenic (a) and transgenic larvae. Dejellied fertilized LG eggs were co-injected with (b) 1 ng of phiC31 integrase mRNA and 25 pg of CMV-GFP-DI-attB vector, (c) 0.1 ng of SB mRNA and 15 pg of HSV-GFP, (d) 1×10−3 I-SceI units and 80 pg of I-SceI-GFP in a total volume of 10 nl per egg.
Table 1.
Transgenesis efficiency of LG clones for different techniques
| Technique | Treatment | Experiments (N) |
Injected Eggs (N) |
Average Survival Rate at Hatching (% ± SD1) |
Transgenesis Efficiency (%) |
|---|---|---|---|---|---|
| phiC31 Integrase |
1ng Integrase mRNA + 20-50pg DNA |
10 | 601 | 17 ± 4 | 30 – 40 |
| non-injected | 10 | 660 | 35 ± 19 | --- | |
| SB Transposase |
0.1ng SB mRNA + 15-20pg DNA |
3 | 350 | 20 ± 13 | 12 – 35 |
| non-injected | 3 | 355 | 37 ± 12 | --- | |
| I-Sce I Meganuclease |
1x10-3 U I-Sce I + 80pg DNA |
8 | 881 | 36 ± 18 | 35 – 50 |
| non-injected | 8 | 722 | 52 ± 29 | --- | |
SD: standard deviation
For the SB transposase method we engineered two different vectors. The first contained the SB transposase (used for mRNA synthesis) and the second had the GFP reporter under the control of the human EF1α promoter flanked by Inverted Repeat/Direct Repeat (IR/DR) sequences which aid in incorporation of the transgene by the typical cut and paste mechanism of the transposase. However it has been shown that both in X. laevis and X. tropicalis transgene integration is actually mediated by a noncanonical process (Doherty et al., 2007; Sinzelle et al., 2006; Yergeau et al., 2009). Using this method we were able to generate transgenic LG clones albeit with lower transgenesis efficiency ranging from 12–30% (Table 1). Also as previously published for outbred frogs (Doherty et al., 2007; Sinzelle et al., 2006; Yergeau et al., 2009), the transgene integration into LG eggs was mosaic in appearance although we used a strong ubiquitous promoter (Figure 1Bc).
Finally, we tested the I-SceI meganuclease ability to stably integrate transgenes into the genome of the LG clones. It is currently unclear how this meganuclease mediates transgene integration since there are presumably no endogenous I-SceI sites in the X. laevis and X. tropicalis genomes. However, it is speculated that I-SceI binds to the cut ends of the transgene preventing concatemer formation and DNA degradation thereby increasing the transgenesis efficiency (Thermes et al., 2002). We used the same GFP reporter as with the SB technique; however, the reporter cassette was flanked by the 18bp I-SceI recognition sequences. The plasmid carrying the reporter was digested with I-SceI and the entire digest was injected into single-cell stage embryos. This method resulted in the highest transgenesis (35–50%) and survival rates (Table 1). Importantly, compared to the phiC31 integrase and the SB transposase the use of I-SceI resulted in strong and uniform non-mosaic expression of the GFP reporter in both F0 tadpoles and adults (Figure 1B).
Collectively, these data demonstrate that we can successfully generate transgenic LG clones using all three methods tested. However, because of the ease of use, the highest transgenesis and survival rates and the consistent non-mosaic transgene expression, we conclude that the best method for generating transgenic LG clones is the I-SceI meganuclease.
To further assess the reliability of the I-SceI technique for generating transgenic clones, we determined the occurrence of germline transmission. It has already been shown that transgenes can be transmitted and expressed in progeny from outbred Xenopus (Ogino et al., 2006; Pan et al., 2006). However, this requires outcrossing transgenic founders with non-transgenic animals and selecting offspring carrying the transgene. Therefore, a large number of progeny has to be screened for transgene integration. Since the LG clones are generated by gynogenesis we hypothesized that all of their progeny should carry the transgene, which would eliminate the need for outcrossing and screening the offspring. We first determined if the eggs from our transgenic LG founders carry the GFP transgene. We took individual unfertilized eggs and extracted DNA. Using GFP specific primers we showed that all eggs tested (57/57 from 2 different founders, 1 is shown in Figure 2) had the GFP transgene integrated into their genome.
Figure 2. All eggs from GFP transgenic founder LG clones carry the transgene.

Representative PCR of 28 unfertilized eggs from a GFP transgenic founder LG-6 frog (A) and 14 unfertilized eggs from a non-transgenic frog (B). GFP specific primers (35 cycles) and EF1-α primers (control, 35 cycles) were used. Each lane represents a single egg. (NT) no template.
Although we showed that indeed all diploid eggs from the transgenic females carried the transgene, we wanted to test if the F1 progeny will also express GFP. Consequently, we generated progeny from transgenic LG founders and allowed the offspring to develop into young adults. These animals were screened for GFP expression using a fluorescence stereomicroscope. As hypothesized, all F1 frogs tested had constitutive GFP expression (Table 2, Figure 3A). Furthermore, red blood cells and lymphocytes from randomly selected F1 frogs were analyzed for GFP expression by flow cytometry (Figure 3B). This analysis supported our microscopy data and showed that all of the F1 progeny had significant GFP expression when compared to control LG frogs. Hence, we have demonstrated that the GFP transgene is transmitted to the F1 progeny in LG clones with 100% transmission efficiency.
Table 2.
Germline transmission in LG clones generated by I-SceI meganuclease
| F0 LG founder | Total Embryos | GFP-Positive Embryos | Transmission |
|---|---|---|---|
| # 1 | 43 | 43 | 100% |
| # 2 | 40 | 40 | 100% |
Figure 3. Germline transmission of the GFP transgene to F1 LG progeny.
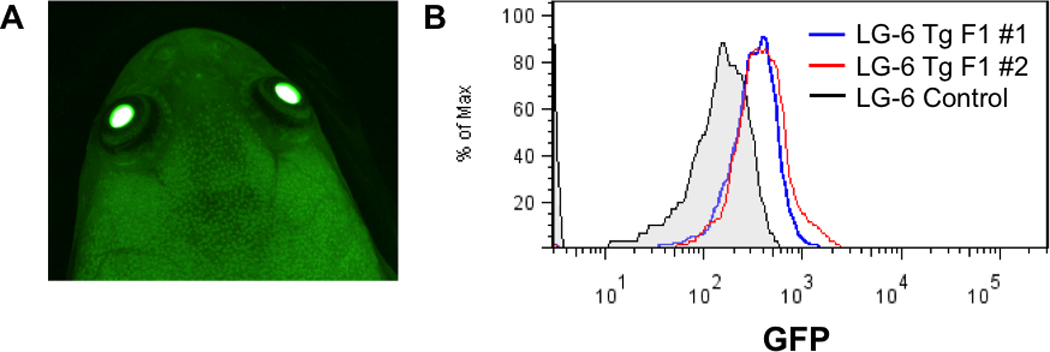
F1 transgenic progeny were reared to adulthood and screened for GFP fluorescence. (A) Representative image of an adult GFP positive F1 LG transgenic clone. (B) Representative flow cytometry analysis of blood (including both red blood cells and lymphocytes) from two F1 transgenic LG clones (blue and red histograms) as well as a non-transgenic control frog (gray shaded histogram). 50,000 events were collected, gated on live cells and analyzed for GFP expression using the FlowJo software.
Discussion
We have adapted and optimized a transgenic method using the I-SceI meganuclease in our Xenopus isogenetic LG clonal model system that will be valuable for future comparative immunological studies. We have shown that while we can generate transgenic LG clones using both the phiC31 integrase and the SB transposase, the I-SceI meganuclease is a superior method for LG high-throughput transgenesis. Attractive features of this procedure include its high transgenesis efficiency rate (35–50%) coupled with a high survival rate. Additionally, I-SceI mediated transgenesis is more straightforward since it only requires the commercially available meganuclease. In contrast, the other two techniques require mRNA synthesis and are therefore critically dependent on the mRNA quality, which makes them technically more challenging.
Most importantly for our objectives, transgenic frogs generated using the I-SceI meganuclease display a non-mosaic ubiquitous transgene expression. Indeed, our results show that transgenic LGs both in the F0 as well as the F1 generation had uniform constitutive GFP expression driven by the EF1α promoter. It has been shown that there are only 1–2 different integration sites in the Xenopus genome (Pan et al., 2006).Therefore, the absence of mosaic transgene expression in transgenic LGs suggests that there is no chromatin-induced position effects. The non-mosaic transgene expression will be advantageous to accelerate certain experiments by using F0 animals directly. F0 transgenics can easily be screened at early larval stage for GFP expression before assessing the effect of the immune transgene of interest on immune function (e.g., overexpression of MHC class I genes).
Finally, for immunological applications requiring large number of transgenic adults, our LG clonal system permits 100% germline transmission via gynogenesis (i.e., no contribution of male DNA). Notably, once we have generated transgenic founders, there will be no need for further outcrossing and screening of the offspring, which is quite laborious and time consuming.
In conclusion, we have established a reliable and highly efficient technique for production of transgenic LG clones, which will allow us to generate animals with modulated expression of immunologically-relevant genes such as MHC class I molecules, and investigate their role in anti-tumor and anti-viral immunity as well as skin graft rejection.
Methods
Animals
LG-6 and LG-15 animals were obtained from our Xenopus laevis Research Resource for Immunology at the University of Rochester (http://www.urmc.rochester.edu/smd/mbi/xenopus/index.htm). All animals were handled under strict laboratory and UCAR regulations (Approval number 100577/2003-151), minimizing discomfort at all times.
Plasmid Construction
The plasmids used for phiC31 mediated transgenesis, pET11-phiC31-poly(A) (used to generate integrase mRNA) and the CMV-GFP-DI-attB, were provided by Dr. Weeks (University of Iowa, Allen and Weeks, 2005). For SB mediated transgenesis we generated two different vectors. To synthesize SB mRNA we generated pCMV-SPORT6-SB by inserting the SB sequence into the NotI and EcoRI sites of the pCMV-SPORT6 vector. We also made a reporter cassette by removing the human EF-1α promoter from pBudCE4.1 and cloning it into the pEGFP-C2 vector from which we previously removed the pCMV promoter and the multiple cloning site. The resulting 2.2 Kb reporter cassette, consisting of the human EF-1α promoter followed by EGFP and the SV40 polyA, was cloned into the ClaI site of the HSV T-MCS (provided by Dr. Bowers, University of Rochester) vector that’s flanked by the IR/DR sites (Bowers et al., 2006). Finally we generated the I-SceI-GFP vector by cloning our 2.2 Kb GFP reporter cassette into the SacI and PstI sites of the I-SceIpBSIISk+ (provided by Dr. Grainger, University of Virginia) vector flanked by the I-SceI 18 bp recognition sites.
mRNA synthesis
We linearized the pET11-phiC31-poly(A) plasmid with EcoRI and the pCMV-SPORT6-SB plasmid with KpnI in order to generate either integrase or SB transposase mRNA respectively. We used 5 µg of digested DNA as a template. mRNA was synthesized in vitro using the T7 mMessage Machine Kit (Ambion) using manufacturer’s instruction.
Microinjection of LG Eggs
LG-6 and LG-15 females were primed with 10–20 IU and boosted with 20–40 IU of human chorionic gonadotrophin (hCG, Sigma) one day before egg collection. Additional injection of 100 IU of hCG may be necessary the morning of the days of transgenic injection. Testis from outbred Xenopus laevis males were homogenized in 2 ml Deboer’s solution (100 mM NaCL, 1.3 mM KCl, 0.4 mM CaCl2) and UV irradiated (UV light source at 253.7 nm, Gelman Instrument) for 6 minutes before use. Eggs were squeezed into a Petri dish and fertilized in vitro with 500 µl of the irradiated sperm. 10 minutes later the eggs were dejellied with 2% cysteine pH 8.0 (made in 0.1 × MBS), washed with 0.1 × MBS (13°C) several times, and placed into injection media (4% Ficoll 400 in 0.3 × MBS). For all three techniques we employed a standard microinjection technique where activated one-cell stage LG embryos were injected (PLI-100, Harvard Apparatus) with 10 nl total volume into the animal pole close to the site of sperm entry. For phiC31 mediated transgenesis we co-injected 1 ng of integrase mRNA together with 20–50 pg of RNase free CMV-GFP-DI-attB vector per egg. 0.1 ng of SB mRNA was injected per egg together with 15–20 pg of RNase free HSV-GFP in order to generate transgenic LG clones with the SB method. Finally for the I-SceI method, the I-SceI-GFP vector was digested with the I-SceI meganuclease (New England Biolabs) for 40 mins at 37°C and 80 pg DNA along with 1×10−3 U I-SceI were injected per egg. The digests were used within an hour. After injection all embryos were incubated at 13°C for four hours in order to delay cell division providing extended time for transgene integration. The embryos were then transferred in 0.3 × MBS with 50 µg/ml gentamycin and reared at 18°C until hatching. After that larvae were raised in declorinated water at room temperature.
Fluorescence Microscopy
LG transgenic larvae (st. 58) were screened for GFP expression using SMZ1500 Nikon stereomicroscope equipped with a DS-Qi1 Monochrome Cooled Digital Camera (Nikon). Tadpoles that screened positive were raised to adults and bred to produce F1 progeny.
PCR
DNA was extracted from individual unfertilized eggs from LG GFP+ transgenic founders using the DNeasy Blood and Tissue Kit (Qiagen) and was quantified by spectrophotometry. PCR was performed for the GFP transgene with GFP specific primers (F 5’-ACGGCCACAAGTTCAGCGTG-3’, R 5’-GTCCATGCCGAGAGTGATCC-3’) and 100 ng of egg DNA as template at 64°C annealing temperature and 35 cycles. Primers for EF-1α (F 5’-CCTGAATCACCCAGGCCAGATTGGTG-3’, R 5’-GAGGGTAGTCTGAGAAGCTCTCCACG-3’) were used as a control.
Flow Cytometry
For this analysis we used six month adult LG transgenic F1 progeny as well as LG control frogs. Frogs were anesthetized by immersion in a 0.1% aqueous solution of tricaine methane sulfonate (TMS, MS-222) buffered with sodium bicarbonate and were bled from the dorsal tarsus vein using a pulled glass needle (Nedelkovska et al., 2010). The blood was collected in a 10 ml solution of ice cold Amphibian PBS (APBS) and 500 units of heparin in order to prevent clotting, washed once with 10 ml APBS and resuspended at 1×106 cells in 500 µl of staining buffer (APBS + 1% BSA + 0.01% NaN3). The cells were then analyzed by flow cytometry for GFP expression on a FACSCantoII (BD Biosciences) using the Blue 488 laser, channel Blue E and 50,000 events were collected. Analysis was performed using the FlowJo software (Tree Star Inc.).
Acknowledgments
We would like to thank David Albright and Tina Martin for the expert animal husbandry and Nikesha Haynes and Eva-Stina Edholm for critical reading of the manuscript. This research was supported by the NIH T32-AI 07285 (H.N.), 1R03-HD061671-01, R24-AI-059830-06.
References
- Allen BG, Weeks DL. Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat Methods. 2005;2:975–979. doi: 10.1038/nmeth814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaya E, Kroll KL. A method for generating transgenic frog embryos. Methods Mol Biol. 1999;97:393–414. doi: 10.1385/1-59259-270-8:393. [DOI] [PubMed] [Google Scholar]
- Bowers WJ, Mastrangelo MA, Howard DF, Southerland HA, Maguire-Zeiss KA, Federoff HJ. Neuronal precursor-restricted transduction via in utero CNS gene delivery of a novel bipartite HSV amplicon/transposase hybrid vector. Mol Ther. 2006;13:580–588. doi: 10.1016/j.ymthe.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Doherty JR, Johnson Hamlet MR, Kuliyev E, Mead PE. A flk-1 promoter/enhancer reporter transgenic Xenopus laevis generated using the Sleeping Beauty transposon system: an in vivo model for vascular studies. Dev Dyn. 2007;236:2808–2817. doi: 10.1002/dvdy.21321. [DOI] [PubMed] [Google Scholar]
- Du Pasquier L, Schwager J, Flajnik MF. The immune system of Xenopus. Annu Rev Immunol. 1989;7:251–275. doi: 10.1146/annurev.iy.07.040189.001343. [DOI] [PubMed] [Google Scholar]
- Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci U S A. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobel HR, Du Pasquier L. Production of Large Clones of Histocompatible, Fully Identical Clawed Toads (Xenopus) Immunogenetics. 1975:87–91. [Google Scholar]
- Kobel HR, Du Pasquier L. Strains and species of Xenopus for immunological research. In: Solomon JB, Horton JD, editors. Developmental Immunobiology. Amsterdam: Elsevier/North Holland Publishing Co.; 1977. pp. 299–306. [Google Scholar]
- Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- Maniero GD, Robert J. Phylogenetic conservation of gp96-mediated antigen-specific cellular immunity: new evidence from adoptive cell transfer in xenopus. Transplantation. 2004;78:1415–1421. doi: 10.1097/01.tp.0000140846.73210.91. [DOI] [PubMed] [Google Scholar]
- Nedelkovska H, Cruz-Luna T, McPherson P, Robert J. Comparative in vivo study of gp96 adjuvanticity in the frog Xenopus laevis. J Vis Exp. 2010 doi: 10.3791/2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino H, McConnell WB, Grainger RM. Highly efficient transgenesis in Xenopus tropicalis using I-SceI meganuclease. Mech Dev. 2006;123:103–113. doi: 10.1016/j.mod.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Pan FC, Chen Y, Loeber J, Henningfeld K, Pieler T. I-SceI meganuclease-mediated transgenesis in Xenopus. Dev Dyn. 2006;235:247–252. doi: 10.1002/dvdy.20608. [DOI] [PubMed] [Google Scholar]
- Robert J, Cohen N. Evolution of immune surveillance and tumor immunity: studies in Xenopus. Immunol Rev. 1998;166:231–243. doi: 10.1111/j.1600-065x.1998.tb01266.x. [DOI] [PubMed] [Google Scholar]
- Robert J, Gantress J, Cohen N, Maniero GD. Xenopus as an experimental model for studying evolution of hsp--immune system interactions. Methods. 2004;32:42–53. doi: 10.1016/s1046-2023(03)00186-5. [DOI] [PubMed] [Google Scholar]
- Robert J, Gantress J, Rau L, Bell A, Cohen N. Minor histocompatibility antigen-specific MHC-restricted CD8 T cell responses elicited by heat shock proteins. J Immunol. 2002;168:1697–1703. doi: 10.4049/jimmunol.168.4.1697. [DOI] [PubMed] [Google Scholar]
- Robert J, Guiet C, Du Pasquier L. Lymphoid tumors of Xenopus laevis with different capacities for growth in larvae and adults. Dev Immunol. 1994;3:297–307. doi: 10.1155/1994/37392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert J, Guiet C, Du Pasquier L. Ontogeny of the alloimmune response against a transplanted tumor in Xenopus laevis. Differentiation. 1995;59:135–144. doi: 10.1046/j.1432-0436.1995.5930135.x. [DOI] [PubMed] [Google Scholar]
- Robert J, Ohta Y. Comparative and developmental study of the immune system in Xenopus. Dev Dyn. 2009;238:1249–1270. doi: 10.1002/dvdy.21891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinzelle L, Vallin J, Coen L, Chesneau A, Du Pasquier D, Pollet N, Demeneix B, Mazabraud A. Generation of trangenic Xenopus laevis using the Sleeping Beauty transposon system. Transgenic Res. 2006;15:751–760. doi: 10.1007/s11248-006-9014-6. [DOI] [PubMed] [Google Scholar]
- Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev. 2002;118:91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol Cell Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau DA, Johnson Hamlet MR, Kuliyev E, Zhu H, Doherty JR, Archer TD, Subhawong AP, Valentine MB, Kelley CM, Mead PE. Transgenesis in Xenopus using the Sleeping Beauty transposon system. Dev Dyn. 2009;238:1727–1743. doi: 10.1002/dvdy.21994. [DOI] [PMC free article] [PubMed] [Google Scholar]


