Abstract
Acute aortic dissection (AAD) is a serious vascular disease. Currently the diagnosis relies on clinical and radiological means whereas serum biomarkers are lacking. The purpose of this study was to identify potential serum biomarkers for AAD using isobaric tags for relative and absolute quantitation (iTRAQ) approach. A total of 120 serum samples were collected from three groups: AAD patients (n = 60), patients with acute myocardial infarction (AMI, n = 30), and healthy volunteers (n = 30), whereas the first 10 samples from each group were used for iTRAQ analysis. Using iTRAQ approach, a total of 174 proteins were identified as significantly different between AAD patients and healthy subjects. Among them, forty-six proteins increased more than twofold, full-scale analysis using serum sample for the entire 120 subjects demonstrated that Lumican level was significantly increased relative to control and AMI samples. Further, Lumican level correlated with time from onset to admission in AAD but not AMI samples. Using iTRAQ approach, our study showed that Lumican may be a potential AAD-related serum marker that may assist the diagnosis of AAD.
1. Introduction
Acute aortic dissection (AAD) has become a treatable disease due to recent advances in new therapeutic approaches for the management of heart and arterial diseases; however, development of quick and economic diagnostic methods remains a challenge. Variability in disease presentation often obscures diagnoses, and imaging modalities such as computed tomography (CT), magnetic resonance imaging (MRI), and esophagus ultrasound remain prohibitive due to cost and availability. Aortic dissection remains a frequent target of medicolegal litigations with accusations of failure to diagnose against treating physicians and hospitals [1]. Some progress in the biochemical diagnosis of AAD has been made in the last decade [2, 3]; several acute phase proteins and coagulation parameters were identified to increase in AAD patients, but these are nonspecific biomarkers for AAD as they may be also aberrantly expressed in other disease conditions such as acute myocardial infarction (AMI).
Recently a quantitative proteomic assay, isobaric tags for relative and absolute quantitation (iTRAQ), has been developed and utilized to identify biomarkers for various disease conditions [4, 5]. This chemical labeling method involves the stable incorporation of isotopes into an amine tagging reagent, which can then be reliably detected by mass spectrometry, thereby permitting comparative quantitation of various proteins in a multiplex manner. It has been suggested to be suitable for the discovery of biomarkers in a wide range of body fluids and tissues, including serum and plasma [5, 6]. With this method, we expect to find the potential biomarkers which are released from the disruption of the aortic media and can provide sufficient specificity and longer time window for the diagnosis of AAD.
2. Materials and Methods
2.1. Samples
The study included a total of 30 healthy individuals and 90 patients (60 AAD, 30 AMI). All the patients were selected in a consecutive manner from the period of July 2009 to November 2011 from Fudan University affiliated Zhongshan Hospital (Shanghai, China). iTRAQ analysis was performed for the first twenty patients (10 AAD and 10 AMI) and ten healthy individuals. All patients presented within 72 hours after an episode of chest and/or back pain lasting 5 minutes or more. The diagnosis of AAD was confirmed by computed tomographic arteriography (CTA). The AMI patient was confirmed by electrocardiography (ECG) and cardiac troponin T (cTNT) tests. All patients gave their informed consent for the study. The protocol was approved by the Ethics Committee of Zhongshan Hospital.
For each study subject, whole blood samples were immediately collected in BD Vacutainer SST tubes (BD Diagnostics, Plymouth, UK) after admission and centrifuged at 4000 rpm for 10 min at room temperature. The serum was frozen and stored in aliquots at −80°C until analysis.
2.2. Serum C-Reactive Protein and Myoglobin Test
Vitros 5.1 FS automatic biochemistry analyzer (Johnson & Johnson; Calif, USA) was used for serum C-reactive protein (CRP) test, and Cobas e411 immunoassay analyzer (Roche; Mannheim, Germany) was used for the serum myoglobin (Myo) test. The results were then interpreted in accordance with that tested by the International Federation of Clinical Chemistry (IFCC) recommended method. Analyses were performed immediately after the centrifugation of whole blood samples.
2.3. iTRAQ Sample Preparation: Strong Cation Exchange (SCX) Chromatography
iTRAQ reagents were purchased from Applied Biosystems (Foster City, USA). Fourteen interfering highly abundant proteins from serum samples were removed using Agilent multiple affinity removal liquid chromatography (LC) column-Human 14 (MARS) (shimadzu, Kyoto, Japan). One hundred micrograms of each extract were precipitated using acetone at −20°C and suspended in 20 μL of dissolution buffer (Applied Biosystems, Foster City, USA). After reduction and alkylation, each sample was digested with trypsin (w(trypsin) : w(protein) = 1 : 20) at 37°C overnight. The tryptic peptides were labeled with the iTRAQ reagents as follows: normal controls group was labeled with iTRAQ 113, AMI group was labeled with iTRAQ 114, and AAD group was labeled with iTRAQ 115. The peptides were pooled and desalted with Sep-Pak Vac C18 (Waters, Milford, USA). The peptide mixture was diluted with buffer A containing 10 mM KH2PO4 in 25% acetonitrile (ACN) at pH 2.6. The peptides were fractionated by 20AD high-performance liquid chromatography (HPLC) system (Shimadzu; Kyoto, Japan) equipped with a polysulfoethyl A column (2.1 mm × 100 mm, 5u, 200 A; The Nest Group, Southborough, Mass). The composition of buffer B was 350 mM KCl, 10 mM KH2PO4, and 25% ACN at pH 2.6. Separation was performed using a linear binary gradient of 0–80% buffer B in buffer A at a flow rate of 200 μL/min for 60 min. The fractions were combined into 20 groups.
2.4. LC-MS Analysis
Each SCX fraction was dried down by the rotary vacuum concentrator, dissolved in buffer C (0.1% formic acid, 5% ACN, 95% water), and analyzed on Qstar XL (Applied Biosystems; Foster City, USA). The HPLC gradient was 5–35% buffer D (95% ACN, 0.1% formic acid) in buffer C at a flow rate of 300 nL/min for 70 min. Analysis survey scans were acquired MS from m/z 400–1800 with up to 4 precursors selected for MS/MS from m/z 100–2000.
2.5. The Confirmative ELISA Analysis for Lumican
The confirmative ELISA analysis for Lumican was performed using the kits from CUSABIO BIOTECH CO, following manufacture's recommendation (CUSABIO BIOTECH CO., LTD., Wuhan, China).
2.6. Data Analysis
All statistical analyses were performed in SPSS 12.0 (SPSS Inc. Chicago, USA). Results were presented as Mean ± SD. A comparative analysis of multiple groups was performed with a one-way-ANOVA or Mann-Whitney/Kruskal-Wallis Test. Statistical significance was defined as P < 0.05. Peptide and protein identification was performed by searching the MS/MS spectra against the SwissProt database using the local Protein Pilot 2.0.1 software. Only peptides identified with confidence interval values of no less than 95% (Unused ProtScore >1.3) were used for protein identification compilation and subsequent quantitation calculation. Fold changes of >2 or <0.5 were set as cut-off values to designate significant differences in protein expression among the AAD group and the normal control group.
2.7. PANTHER Analysis
The PANTHER database was used to elucidate cellular components, biological processes, and the molecular functions associated with each individual protein (http://www.pantherdb.org/).
3. Results
3.1. Clinical Features of Study Subjects
The clinical features of the AAD patients, AMI patients, and normal controls are summarized in Table 1. There were no differences in age distribution and sex composition among the three groups involved either for ELISA analysis (N = 120) (P = 0.351 and 0.378, resp.,) or iTRAQ analysis (P = 0.241 and 0.873, resp.). There was no differences in the time from onset to admission between AAD and AMI group either (P = 0.776).
Table 1.
Clinical characteristics in three groups.
| AAD | AMI | Normal controls | P value | ||
|---|---|---|---|---|---|
| n | 60 | 30 | 30 | ||
| ELISA test (N = 120) | Age (Mean ± SD) | 55.63 ± 16.39 | 59.70 ± 13.98 | 59.50 ± 12.65 | 0.351a |
| Gender, n (%), male | 30 (50) | 17 (56.67) | 16 (53.33) | 0.378b | |
| Admission after onset hours (Mean ± SD) | 20.19 ± 18.09 | 19.33 ± 15.31 | / | 0.776c | |
| Type A n (%) | 31 (51.67) | / | / | ||
| Type B n (%) | 29 (48.33) | / | / | ||
| Marfan n (%) | 7 (11.67) | / | / | ||
| n | 10 | 10 | 10 | ||
| iTRAQ test (N = 30) | Age (Mean ± SD) | 51.60 ± 13.22 | 61.00 ± 5.25 | 49.9 ± 15.02 | 0.241a |
| Gender, n (%), male | 6 (60) | 6 (60) | 5 (50) | 0.873b | |
| Admission after onset hours (Mean ± SD) | 27.20 ± 24.56 | 18.40 ± 23.12 | / | 0.363c | |
| Type A n (%) | 5 (50) | / | / | ||
| Type B n (%) | 5 (50) | / | / | ||
| Marfan n (%) | 2 (20) | / | / | ||
a One-way-ANOVA; bChi-square Test; cMann-Whitney Test.
3.2. Functional Classification of Identified Proteins by iTRAQ
A total of 174 proteins with confidence interval values of no less than 95% were identified (Unused ProtScore > 1.3). However, after manually rechecking the MS/MS data thoroughly peak by peak, 155 proteins (89.08%) had a relative quantitation of one or more peptides. Fifteen proteins had no quantifiable peptides that could be ascertained, and four proteins had peptides with confidence interval values that were less than 95%.
In total, 174 proteins were sorted using the PANTHER classification system, which sorts the proteins into respective categories based on their molecular functions. The major groups include: signaling molecules (13%), enzyme modulators (12%), transfer/carrier proteins (11%), and proteases (10%). Other groups include: structural proteins (1%), cell adhesion molecules (1%), cytoskeletal proteins (3%), extracellular matrix proteins (2%), and cell junction proteins (1%).
As a way to cross-check the reliability of quantitation of iTRAQ reagent, serum CRP and Myo levels were assessed using both conventional biochemical and immunoassay tests and iTRAQ analysis on the same specimens. With biochemical and immunoassay analysis, CRP was 41.31 ± 32.76 mg/mL and Myo was 66.42 ± 81.23 mg/mL in AAD group, while in normal controls, the former was 5.88 ± 1.42 and the latter was 32.07 ± 14.14 mg/mL. CRP and Myo levels of AAD patients were 7.03-fold and 2.07-fold higher, respectively, than normal controls. Using iTRAQ, the AAD/normal controls ratios of CRP and Myo were similar at 9.12-fold (Table 2) and 1.47-fold. The ratios of CRP and Myo among three groups were similar with either biochemical and immunoassay or iTRAQ analysis, confirming the reliability of iTRAQ analysis.
Table 2.
List of the increased (>2-fold) protein targets identified and their corresponding class, associated biological process, and cellular component.
| N | Unuseda | Peptidesb | Accession # | Name | Biological process | Cellular component | Protein class | AAD/CON ratio | AAD/AMI ratio |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2.7 | 1 | Q13509 | Tubulin beta-3 chain | Cellular component morphogenesis | Cytoskelelton | Cytoskeletal protein/tubulin | 39.8406 | 0.0711 |
| 2 | 2 | 1 | P01600 | Ig kappa chain V-I region Hau | Unclassified | 31.3480 | 3.0760 | ||
| 3 | 6.67 | 3 | P02743 | Serum amyloid P-component | Response to stress | Defense/immunity protein/antibacterial response | 24.4499 | 9.1241 | |
| 4 | 13.53 | 10 | P05546 | Heparin cofactor 2 | Protein metabolic process | Enzyme modulator | 19.7628 | 11.2740 | |
| 5 | 17.67 | 9 | P36955 | Pigment epithelium-derived factor | Protein metabolic process | Enzyme modulator | 19.7628 | 9.8135 | |
| 6 | 6.24 | 4 | P05543 | Thyroxine-binding globulin | Protein metabolic process | Enzyme modulator | 12.7065 | 3.7665 | |
| 7 | 12.63 | 18 | P01834 | Ig kappa chain C region | Response to stimulus | Immunoglobulin complex | Defense/immunity protein | 11.6959 | 1.8031 |
| 8 | 55.14 | 43 | P02751 | Fibronectin | Blood coagulation | Extracellular matrix | Transfer/carrier protein | 11.5875 | 3.7327 |
| 9 | 35.94 | 29 | P01011 | Alpha-1-antichymotrypsin | Protein metabolic process | Enzyme modulator | 10.5708 | 8.0906 | |
| 10 | 2.22 | 1 | P02741 | C-reactive protein | Response to stress | Defense/immunity protein | 9.1241 | 5.9701 | |
| 11 | 2 | 1 | Q9UK55 | Protein Z-dependent protease inhibitor | Protein metabolic process | Enzyme modulator | 8.7873 | 2.4888 | |
| 12 | 100.78 | 51 | P04114 | Apolipoprotein B-100 | Lipid metabolic process | Transfer/carrier protein | 8.6281 | 2.8050 | |
| 13 | 9.19 | 4 | P35858 | Insulin-like growth factor-binding protein complex acid labile chain | Cell-cell adhesion | Extracellular matrix | Receptor | 8.4746 | 4.0177 |
| 14 | 47.88 | 42 | P19827 | Inter-alpha-trypsin inhibitor heavy chain H1 | Protein metabolic process | Enzyme modulator | 5.8072 | 5.6497 | |
| 15 | 106.72 | 89 | P00450 | Ceruloplasmin | Blood coagulation | Extracellular matrix | Transporter | 5.1046 | 9.6339 |
| 16 | 37.56 | 23 | P04196 | Histidine-rich glycoprotein | Blood coagulation | Unclassified | 4.6125 | 3.2206 | |
| 17 | 4.15 | 2 | P08571 | Monocyte differentiation antigen CD14 | Immune system process | Receptor | 4.0933 | 2.4888 | |
| 18 | 2.72 | 1 | Q96KN2 | Beta-Ala-His dipeptidase | Protein metabolic process | Protease | 4.0933 | 1.9231 | |
| 19 | 31.66 | 20 | P01871 | Ig mu chain C region | Response to stimulus | Defense/immunity protein | 3.8023 | 2.3552 | |
| 20 | 12.12 | 6 | P51884 | Lumican | Cell-cell adhesion | Extracellular matrix | Receptor | 3.6311 | 1.2942 |
| 21 | 2.71 | 1 | P00740 | Coagulation factor IX | Blood coagulation | Protease | 3.4037 | 2.0137 | |
| 22 | 6.27 | 3 | P08185 | Corticosteroid-binding globulin | Protein metabolic process | Enzyme modulator | 3.2808 | 5.4945 | |
| 23 | 2.58 | 21 | P00739 | Haptoglobin-related protein | Unclassified | 3.2206 | 1.7538 | ||
| 24 | 4.98 | 4 | P04003 | C4b-binding protein alpha chain | Blood coagulation | Transfer/carrier protein | 3.1626 | 5.0582 | |
| 25 | 6.02 | 6 | P02745 | Complement C1q subcomponent subunit A | Response to stimulus | Transfer/carrier protein | 3.1046 | 3.6982 | |
| 26 | 5.83 | 3 | P22792 | Carboxypeptidase N subunit 2 | Cell adhesion | Extracellular matrix | Receptor | 3.0202 | 1.3805 |
| 27 | 15.99 | 11 | P05155 | Plasma protease C1 inhibitor | Protein metabolic process | Enzyme modulator | 2.9647 | 1.2824 | |
| 28 | 2.02 | 2 | P01742 | Ig heavy chain V-I region EU | Unclassified | 2.8580 | 1.1695 | ||
| 29 | 2 | 1 | P09486 | SPARC | Cell-cell signaling | Transfer/carrier protein | 2.7042 | 0.7312 | |
| 30 | 18.06 | 9 | P02760 | Protein AMBP | Blood coagulation | Enzyme modulator | 2.6062 | 3.5651 | |
| 31 | 8.99 | 5 | P12259 | Coagulation factor V | Blood coagulation | Extracellular matrix | Transporter | 2.6062 | 3.2206 |
| 32 | 8.31 | 6 | P27169 | Serum paraoxonase/arylesterase 1 | Immune system process | Oxidoreductase | 2.5349 | 5.6497 | |
| 33 | 2.32 | 1 | P19320 | Vascular cell adhesion protein 1 | Cell-cell adhesion | Defense/immunity protein | 2.3121 | 2.1478 | |
| 34 | 2.01 | 1 | O00187 | Mannan-binding lectin serine protease 2 | Response to stimulus | Protease | 2.3121 | 1.5277 | |
| 35 | 43.19 | 29 | P02749 | Beta-2-glycoprotein 1 | Blood coagulation | Transfer/carrier protein | 2.2287 | 7.1124 | |
| 36 | 3.52 | 2 | P11226 | Mannose-binding protein C | Response to stimulus | Defense/immunity protein | 2.2080 | 2.6062 | |
| 37 | 87.93 | 67 | P00738 | Haptoglobin | Blood coagulation | Protease | 2.1678 | 3.5651 | |
| 38 | 5.7 | 4 | Q13790 | Apolipoprotein F | Lipid metabolic process | Transporter | 2.1478 | 3.9448 | |
| 39 | 65.42 | 41 | P02790 | Hemopexin | Vitamin transport | Transfer/carrier protein | 2.1478 | 3.9078 | |
| 40 | 2.96 | 1 | P20851 | C4b-binding protein beta chain | Blood coagulation | Transfer/carrier protein | 2.1281 | 4.8309 | |
| 41 | 2 | 1 | P61769 | Beta-2-microglobulin | Response to stimulus | Defense/immunity protein | 2.1088 | 2.1281 | |
| 42 | 20.86 | 10 | P02748 | Complement component C9 | Response to stimulus | Receptor | 2.1088 | 0.6252 | |
| 43 | 6.96 | 3 | P07477 | Trypsin-1 | Protein metabolic process | Protease | 2.1088 | 0.3436 | |
| 44 | 13.64 | 7 | P05156 | Complement factor I | Response to stimulus | Protease | 2.0325 | 0.9120 | |
| 45 | 15.17 | 10 | P01842 | Ig lambda chain C regions | Response to stimulus | Immunoglobulin complex | Defense/immunity protein | 2.0137 | 2.1678 |
| 46 | 4.29 | 1 | P35030 | Trypsin-3 | Protein metabolic process | Protease | 2.0137 | 1.3931 |
a Unused > 1.3 means at least 95% confidence; bnumber of peptides with 95% confidence; AAD: acute aortic dissection; AMI: acute myocardial infarction; CON: normal controls.
3.3. Proteins with Over Twofold Differential Expression
A total of 155 proteins had a relative quantitation difference for AAD patients compared with the normal control group of which 46 proteins increased more than twofold (Table 2), while 36 proteins decreased more than twofold among the AAD patients (Table 3). Among the identified proteins with increased levels in AAD, there were a number of acute phase reactants (CRP, Beta-2-microglobulin, Complement factor I), blood coagulation marker (Haptoglobin, Coagulation factor V, Coagulation factor IX), and cellular components (Lumican, Tubulin beta-3 chain, Fibronectin). However when compared to AMI patients, 14 of the 46 protein showed less than 2-fold increase, including complement component 9, complement factor1, Plasma protein C1 inhibitor, and Ig Kappa chain C-region (Table 2). Interestingly, some acute phase proteins such as CRP remains on the list as it showed the differential expression between the two conditions.
Table 3.
List of the decreased (<0.5 folds) protein targets identified and their corresponding class, associated biological process, and cellular component.
| N | Unuseda | Peptidesb | Accession # | Name | Biological process | Cellular component | Protein classification | AAD/CON ratio | AAD/AMI ratio |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 19.31 | 17 | P02775 | Platelet basic protein | Blood coagulation | Transfer/carrier protein | 0.0209 | 0.0398 | |
| 2 | 44.6 | 31 | P02671 | Fibrinogen alpha chain | Blood coagulation | Extracellular matrix | Transfer/carrier protein | 0.0370 | 0.0203 |
| 3 | 19.82 | 30 | P02656 | Apolipoprotein C-III | Lipid metabolic process | Transporter | 0.0570 | 0.0240 | |
| 4 | 5.7 | 5 | P01717 | Ig lambda chain V-IV region Hil | Unclassified | 0.0655 | 0.0679 | ||
| 5 | 8.45 | 4 | P02768 | Serum albumin | Transport | Transfer/carrier protein | 0.0724 | 0.8472 | |
| 6 | 3.22 | 1 | P04264 | Keratin, type II cytoskeletal 1 | Cellular component morphogenesis | Cytoskelelton | Structural protein | 0.0780 | 0.9639 |
| 7 | 2.05 | 1 | P04070 | Vitamin K-dependent protein C | Blood coagulation | Protease | 0.0794 | 0.0441 | |
| 8 | 34.98 | 27 | P02652 | Apolipoprotein A-II | Lipid metabolic process | Transporter | 0.0991 | 0.1067 | |
| 9 | 20.3 | 10 | P02654 | Apolipoprotein C-I | Lipid metabolic process | Transporter | 0.1159 | 0.0847 | |
| 10 | 94.94 | 108 | P02765 | Alpha-2-HS-glycoprotein | Protein metabolic process | Extracellular matrix | Extracellular matrix protein | 0.1180 | 0.1472 |
| 11 | 39.07 | 25 | P01008 | Antithrombin-III | Protein metabolic process | Enzyme modulator | 0.1202 | 0.0973 | |
| 12 | 72.69 | 57 | P00734 | Prothrombin | Blood coagulation | Enzyme modulator | 0.1225 | 0.2051 | |
| 13 | 6.88 | 3 | P27918 | Properdin | Response to stimulus | Unclassified | 0.1419 | 0.1905 | |
| 14 | 6.31 | 4 | P69905 | Hemoglobin subunit alpha | Blood circulation | Transfer/carrier protein | 0.1500 | 0.2312 | |
| 15 | 8.97 | 15 | Q03591 | Complement factor H-related protein 1 | Blood coagulation | Transfer/carrier protein | 0.1706 | 0.0991 | |
| 16 | 27.03 | 22 | P10909 | Clusterin | Apoptosis | Unclassified | 0.1905 | 0.1076 | |
| 17 | 2 | 1 | Q15942 | Zyxin | Cellular component morphogenesis | Enzyme modulator | 0.1923 | 0.2109 | |
| 18 | 20.4 | 26 | P01024 | Complement C3 | Protein metabolic process | Transfer/carrier protein | 0.2070 | 0.1406 | |
| 19 | 7.43 | 3 | P17936 | Insulin-like growth factor-binding protein 3 | Cell-matrix adhesion | Unclassified | 0.2089 | 0.3162 | |
| 20 | 2.23 | 2 | P55290 | Cadherin-13 | Cell-cell adhesion | Cell junction | Receptor | 0.2188 | 0.1660 |
| 21 | 1.41 | 1 | P13598 | Intercellular adhesion molecule 2 | Cell-cell adhesion | Transfer/carrier protein | 0.2355 | 0.1600 | |
| 22 | 96.24 | 64 | P00751 | Complement factor B | Blood coagulation | Transfer/carrier protein | 0.2466 | 0.2729 | |
| 23 | 28.82 | 16 | P09871 | Complement C1s subcomponent | Blood coagulation | Protease | 0.2606 | 0.4169 | |
| 24 | 13.74 | 9 | P01019 | Angiotensinogen | Protein metabolic process | Enzyme modulator | 0.2630 | 0.5058 | |
| 25 | 95.34 | 74 | P19823 | Inter-alpha-trypsin inhibitor heavy chain H2 | Protein metabolic process | Enzyme modulator | 0.2805 | 0.5649 | |
| 26 | 5.15 | 2 | P18065 | Insulin-like growth factor-binding protein 2 | Cell-matrix adhesion | Unclassified | 0.2884 | 0.3281 | |
| 27 | 16.84 | 10 | P07996 | Thrombospondin-1 | Blood coagulation | Extracellular matrix | Transfer/carrier protein | 0.3404 | 0.8317 |
| 28 | 5.31 | 3 | P26927 | Hepatocyte growth factor-like protein | Blood coagulation | Transfer/carrier protein | 0.3436 | 0.3436 | |
| 29 | 7.85 | 4 | O14791 | Apolipoprotein L1 | Lipid metabolic process | Transporter | 0.3908 | 0.2831 | |
| 30 | 94.76 | 57 | P06727 | Apolipoprotein A-IV | Lipid metabolic process | Transporter | 0.4018 | 0.1837 | |
| 31 | 14.1 | 9 | P35527 | Keratin, type I cytoskeletal 9 | Cellular component morphogenesis | Structural protein | 0.4055 | 0.7244 | |
| 32 | 9.48 | 6 | P00746 | Complement factor D | Blood coagulation | Protease | 0.4571 | 0.6668 | |
| 33 | 48.09 | 24 | P02649 | Apolipoprotein E | Lipid metabolic process | Transporter | 0.4656 | 0.2051 | |
| 34 | 38.8 | 32 | P02735 | Serum amyloid A protein | Immune system process | Transporter | 0.4742 | 0.0319 | |
| 35 | 9.46 | 5 | P10720 | Platelet factor 4 variant | Blood coagulation | Transfer/carrier protein | 0.4742 | 0.5105 | |
| 36 | 53.31 | 29 | P02774 | Vitamin D-binding protein | Transport | Transfer/carrier protein | 0.4831 | 2.2287 |
a Unused > 1.3 means at least 95% confidence; bnumber of peptides with 95% confidence; AAD: acute aortic dissection; AMI: acute myocardial infarction; CON: normal control.
Among proteins with decreased expression in AAD patients compared with normal controls, there were a number of molecules involved in protein metabolism (Inter-alpha-trypsin inhibitor heavy chain H2, Alpha-2-HS-glycoprotein), lipid metabolic process (Apolipoprotein A-IV, Apolipoprotein E, Apolipoprotein C-I), blood coagulation marker (Fibrinogen alpha chain, Prothrombin), and cellular components (Alpha-2-HS-glycoprotein, thrombospondin-1 (TSP-1)). When compared to AMI patients, 8 of 36 proteins did not reach the 2-fold differential expression (Table 3).
3.4. The ELISA Analysis of Serum Concentrations of Lumican
Based on the iTRAQ findings above we selected two targets, Fibronectin and Lumican, the protein markers potentially associated with vascular injury, for the validation using ELISA method. At the initial analysis using 10 AAD and 10 normal samples we found that statistical significant difference between AAD and normal individual was seen for Lumican but not Fibronectin (data not shown). Therefore, we carried a full validation study only for Lumican, using the entire 120 samples collected (see Table 1). A statistical significant difference between AAD, AMI, and normal individual was seen in serum concentrations of Lumican (2.66 ± 4.58 ng/mL in AAD group, 0.69 ± 0.34 ng/mL in AMI group, and 0.85 ± 0.53 ng/mL in normal control, P = 0.003). The difference for AAD and AMI also reached statistical significance (P < 0.05) suggesting the specificity of this marker for AAD (Figure 1). We further analyzed the correlation between Lumican levels with time from onset of symptoms to admission. As shown in Figure 2, a correlation was seen in AAD group (r = 0.256, P = 0.048) but not in AMI group (r = 0.077, P = 0.685), further confirming the specificity of Lumican as a marker for AAD.
Figure 1.
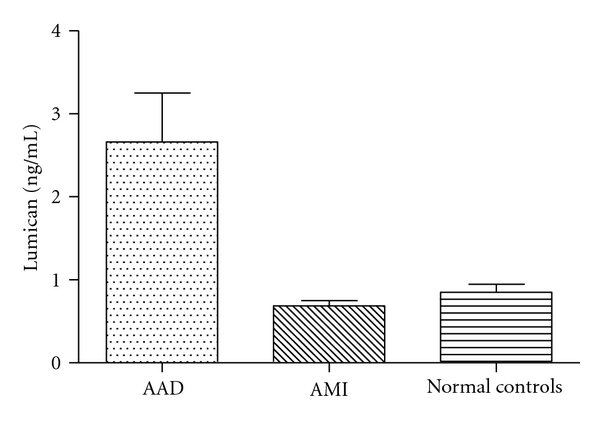
Lumican levels were significant difference between AAD, AMI, and normal individuals (Mean ± SEM; P = 0.003).
Figure 2.
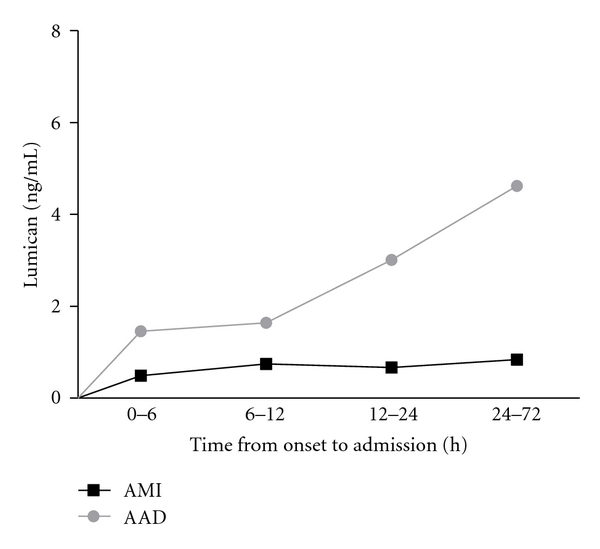
Lumican levels were correlation with time from onset to admission in AAD group (r = 0.256, P = 0.048), but not in AMI group (r = 0.077, P = 0.685).
4. Discussion
iTRAQ analysis is recently been used as a potentially more effective biomarker discovery method than traditional proteomic methods. The high reproducibility optimizes this technique for embarking on “fishing-expeditions” as an initial screening for potential useful biomarkers [6–8]. As a means of internal validation, the iTRAQ method was compared with CRP biochemical assay and Myo immunoassay. In our study there were no significant differences in the serum levels determined by the different methods. Thus, the iTRAQ method we employed appears in this preliminary analysis to be suitable for the detection of relevant proteins.
To identify differentially expressed proteins, in many studies, the cut-off points were set at 20% to 50% average variance [7, 9, 10]. However, such approaches may result in finding markers with low specificity [2, 3]. We therefore appropriated to increase the cut-off point at 100% variance in serum levels of candidate proteins between AAD patients and normal subjects. Thus, only twofold changes below or above normal controls were considered significant. In our study, total of 155 proteins had a relative difference between AAD patients and healthy volunteers. Therefore, with higher specificity, these candidate proteins are more likely to be potential biomarkers for AAD.
In the group of significantly increased proteins, there were numerous acute phase reactants, such as Beta-2-microglobulin (P61769), which could be indicative of an increased inflammatory response among AAD patients. CRP (P02741), a protein found to be elevated in patients who presented with symptoms or rupture of AAD and abdominal aortic aneurysm, was also identified using iTRAQ [11, 12]. CRP is a nonspecific biomarker associated with AAD and a predictor for long-term adverse events [13], and it can be used to monitor evolution of false lumen thrombosis [14]. Unfortunately, CRP is also produced in coronary plaques [15], acute myocardial infarction [16], and so forth. The elevations of these acute phase reactants represent a generalized reaction to vascular injury, and as such, they are nonspecific biomarkers. In addition, many proteins identified are associated with blood coagulation and fibrinolytic system. Among which, ten had increased serum levels (e.g., P00450-Ceruloplasmin, P02751-Fibronectin, P00738-Haptoglobin…), and twelve had decreased serum levels (e.g., P02671-Fibrinogen alpha chain, P00751-Complement factor B, P00734-Prothrombin…). The pathophysiological mechanism for the appearance of these proteins may be explained by the release of tissue factors from the dissected aortic wall then the activation of the extrinsic coagulation system [17–19]. In addition, platelets can be activated by injuries to the vessel wall, activation of the coagulation cascade, or by activating factors released from stimulated endothelial cells and platelets (e.g., ADP, thromboxane, von Willebrand Factor). It also has been found that platelet functions were affected secondary to acute massive consumption coagulopathy in the false lumen in AAD patient [20, 21].
In the past few years, extracellular matrix (ECM) components of vessel walls such as elastin have been shown to be elevated in aortic dissections; however, such increases were less than twofold [3]. Our study found 9 extracellular matrix component proteins with greater than twofold differences, among these are Carboxypeptidase (P22792), Lumican (P51884), Fibronectin (P02751), Ceruloplasmin (P00450), and Thrombospondin-1 (TSP-1, P07996). Fibronectin is a polymorphic and multifunctional glycoprotein that plays wide-ranging roles in tissue injury [22–25]. TSP-1, which is an extracellular protein that participates in cell-to-cell and cell-to-matrix communication, can stimulate or inhibit the migration of vascular smooth muscle cells or endothelial cells. It has been known as a plasma marker of peripheral arterial disease [26].
Lumican is distributed in interstitial collagenous matrices throughout the body. In coronary arteries ischemic lesion, it is overexpressed by vascular smooth muscle cells (VSMCs) [27] and also synthesized in aortic smooth muscle cells [28]. In iTRAQ analysis, serum Lumican levels in patients with AAD were 1.29-fold and 3.63-fold higher than in patients with AMI and normal controls, respectively. It is interesting to note that with iTRAQ analysis, the level of difference between AAD and AMI for Lumican is less than that of Fibronectin (Table 2), yet the initial validation using ELISA method showed that only Lumican was significantly increased in AAD and AMI samples. While there may be variety reasons to explain the variations of the findings between the two methods, it highlights the importance of validation in biomarker studies. The finding that Lumican expression correlated with the time from onset to admission only in AAD but not in AMI sample further confirmed the specificity of this protein in association with AAD.
Proteomic approach provides an exciting platform to identify clinically useful protein biomarkers. As an initial step our study identified potential candidate protein biomarkers in the serum of AAD patients with the iTRAQ technique. However, the ultimate development of biomarkers which provide sufficient sensitivity or specificity for the diagnosis of AAD will require multiple validations and clinical testing, which may include nonprotein markers. Nevertheless our findings provide preliminary list of candidate biomarkers that should be further validated, either alone or in combination.
5. Conclusion
In this paper, we found that iTRAQ technique is a suitable approach for the detection of the new potential protein markers in the serum of AAD patients. Using iTRAQ approach, our study identified that Lumican may be a potentially interesting new serum marker of AAD, and upon further validation this marker may assist the clinical diagnosis of AAD.
Acknowledgments
This study was supported by Shanghai Committee of Science and Technology (114119a9000). The authors also thank the Department of Chemistry and the Institute of Biomedical Science, Fudan University, Peoples Republic of China, for providing support for the project.
References
- 1.Elefteriades JA, Barrett PW, Kopf GS. Litigation in nontraumatic aortic diseases: a tempest in the malpractice maelstrom. Cardiology. 2008;109(4):263–272. doi: 10.1159/000107790. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki T, Distante A, Eagle K. Biomarker-assisted diagnosis of acute aortic dissection: how far we have come and what to expect. Current Opinion in Cardiology. 2010;25(6):541–545. doi: 10.1097/HCO.0b013e32833e6e13. [DOI] [PubMed] [Google Scholar]
- 3.Shinohara T, Suzuki K, Okada M, et al. Soluble elastin fragments in serum are elevated in acute aortic dissection. Arteriosclerosis, Thrombosis, and Vascular Biology. 2003;23(10):1839–1844. doi: 10.1161/01.ATV.0000085016.02363.80. [DOI] [PubMed] [Google Scholar]
- 4.Boylan KLM, Andersen JD, Anderson LB, Higgins L, Skubitz APN. Quantitative proteomic analysis by iTRAQ for the identification of candidate biomarkers in ovarian cancer serum. Proteome Science. 2010;8, article 31 doi: 10.1186/1477-5956-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Latterich M, Abramovitz M, Leyland-Jones B. Proteomics: new technologies and clinical applications. European Journal of Cancer. 2008;44(18):2737–2741. doi: 10.1016/j.ejca.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Song X, Bandow J, Sherman J, et al. iTRAQ experimental design for plasma biomarker discovery. Journal of Proteome Research. 2008;7(7):2952–2958. doi: 10.1021/pr800072x. [DOI] [PubMed] [Google Scholar]
- 7.Kolla V, Jenö P, Moes S, et al. Quantitative proteomics analysis of maternal plasma in Down syndrome pregnancies using isobaric tagging reagent (iTRAQ) Journal of Biomedicine & Biotechnology. 2010;2010:10 pages. doi: 10.1155/2010/952047. Article ID 952047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee SG, Poh KC, Trong KP, Wright PC. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ) Journal of Proteome Research. 2007;6(2):821–827. doi: 10.1021/pr060474i. [DOI] [PubMed] [Google Scholar]
- 9.Magharious M, D'Onofrio PM, Hollander A, Zhu P, Chen J, Koeberle PD. Quantitative iTRAQ analysis of retinal ganglion cell degeneration after optic nerve crush. Journal of Proteome Research. 2011;10(8):3344–3362. doi: 10.1021/pr2004055. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, Zhang L, Hua Y, et al. Comparative proteomic analysis of plasma membrane proteins between human osteosarcoma and normal osteoblastic cell lines. BMC Cancer. 2010;10, article 206 doi: 10.1186/1471-2407-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trimarchi S, Sangiorgi G, Sang X, et al. In search of blood tests for thoracic aortic diseases. Annals of Thoracic Surgery. 2010;90(5):1735–1742. doi: 10.1016/j.athoracsur.2010.04.111. [DOI] [PubMed] [Google Scholar]
- 12.Domanovits H, Schillinger M, Müllner M, et al. Acute phase reactants in patients with abdominal aortic aneurysm. Atherosclerosis. 2002;163(2):297–302. doi: 10.1016/s0021-9150(02)00006-0. [DOI] [PubMed] [Google Scholar]
- 13.Sakakura K, Kubo N, Ako J, et al. Peak C-reactive protein level predicts long-term outcomes in type B acute aortic dissection. Hypertension. 2010;55(2):422–429. doi: 10.1161/HYPERTENSIONAHA.109.143131. [DOI] [PubMed] [Google Scholar]
- 14.Makita S, Ohira A, Tachieda R, et al. Behavior of C-reactive protein levels in medically treated aortic dissection and intramural hematoma. American Journal of Cardiology. 2000;86(2):242–244. doi: 10.1016/s0002-9149(00)00869-9. [DOI] [PubMed] [Google Scholar]
- 15.Yasojima K, Schwab C, McGeer EG, McGeer PL. Generation of C-reactive protein and complement components in atherosclerotic plaques. American Journal of Pathology. 2001;158(3):1039–1051. doi: 10.1016/S0002-9440(10)64051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Celik T, Iyisoy A, Kursaklioglu H, Turhan H, Isik E. Does the pre-existing inflammation play a role in the increased C-reactive protein levels in acute myocardial infarction? International Journal of Cardiology. 2006;112(1):134–135. doi: 10.1016/j.ijcard.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Vandercappellen J, Van Damme J, Struyf S. The role of the CXC chemokines platelet factor-4 (CXCL4/PF-4) and its variant (CXCL4L1/PF-4var) in inflammation, angiogenesis and cancer. Cytokine and Growth Factor. 2011;22(1):1–18. doi: 10.1016/j.cytogfr.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Blair P, Flaumenhaft R. Platelet α-granules: basic biology and clinical correlates. Blood Reviews. 2009;23(4):177–189. doi: 10.1016/j.blre.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Struyf S, Burdick MD, Proost P, Van Damme J, Stricter RM. Platelets release CXCL4L1, a nonallelic variant of the chemokine platelet factor-4/CXCL4 and potent inhibitor of angiogenesis. Circulation Research. 2004;95(9):855–857. doi: 10.1161/01.RES.0000146674.38319.07. [DOI] [PubMed] [Google Scholar]
- 20.Shi G, Morrell CN. Platelets as initiators and mediators of inflammation at the vessel wall. Thrombosis Research. 2011;127(5):387–390. doi: 10.1016/j.thromres.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka M, Kawahito K, Adachi H, Ino T. Platelet dysfunction in acute type A aortic dissection evaluated by the laser light-scattering method. Journal of Thoracic and Cardiovascular Surgery. 2003;126(3):837–841. doi: 10.1016/s0022-5223(03)00734-7. [DOI] [PubMed] [Google Scholar]
- 22.Romberger DJ. Fibronectin. The International Journal of Biochemistry & Cell Biology. 1997;29(7):939–934. doi: 10.1016/s1357-2725(96)00172-0. [DOI] [PubMed] [Google Scholar]
- 23.Peters JH, Grote MN, Lane NE, Maunder RJ. Changes in plasma fibronectin isoform levels predict distinct clinical outcomes in critically III patients. Biomarker Insights. 2011;6:59–68. doi: 10.4137/BMI.S7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucena S, Arocha Piñango CL, Guerrero B. Fibronectin. Structure and functions associated to hemostasis. Review. Investigacion Clinica. 2007;48(2):249–262. [PubMed] [Google Scholar]
- 25.Örem C, Çelik SO, Örem A, Calapolu M, Erdöl C. Increased plasma fibronectin levels in patients with acute myocardial infarction complicated with left ventricular thrombus. Thrombosis Research. 2002;105(1):37–41. doi: 10.1016/s0049-3848(01)00414-5. [DOI] [PubMed] [Google Scholar]
- 26.Smadja DM, D'Audigier C, Bièche I, et al. Thrombospondin-1 is a plasmatic marker of peripheral arterial disease that modulates endothelial progenitor cell angiogenic properties. Arteriosclerosis, Thrombosis, and Vascular Biology. 2011;31(3):551–559. doi: 10.1161/ATVBAHA.110.220624. [DOI] [PubMed] [Google Scholar]
- 27.Qin H, Ishiwata T, Asano G. Effects of the extracellular matrix on lumican expression in rat aortic smooth muscle cells in vitro. Journal of Pathology. 2001;195(5):604–608. doi: 10.1002/path.994. [DOI] [PubMed] [Google Scholar]
- 28.Naito Z. The role of small leucine-rich proteoglycan (SLRP) family in pathological lesions and cancer cell growth. Journal of Nippon Medical School. 2005;72(3):137–145. doi: 10.1272/jnms.72.137. [DOI] [PubMed] [Google Scholar]


