Abstract
Objectives
Shiga toxin–producing Escherichia coli (STEC) are pathogenic strains, whose virulence depends on induction of Shiga toxin–converting prophages and their subsequent lytic development. We explored which factors or conditions could inhibit development of these phages, potentially decreasing virulence of STEC.
Materials and Methods
Lytic development of Shiga toxin–converting bacteriophages was monitored after mitomycin C-provoked prophage induction under various conditions. Phage DNA replication efficiency was assessed by measurement of DNA amount in cells using quantitative polymerase chain reaction.
Results
We demonstrated that the use of citrate delayed Shiga toxin–converting phage development after prophage induction. This effect was independent on efficiency of prophage induction and phage DNA replication. However, an excess of glucose reversed the effect of citrate. Amino acid starvation prevented the phage development in bacteria both able and unable to induce the stringent response.
Conclusions
Lytic development of Shiga toxin–converting bacteriophages can be inhibited by either the presence of citrate or amino acid starvation. We suggest that the inhibition caused by the latter condition may be due to a block in prophage induction or phage DNA replication or both.
Applications
Our findings may facilitate development of procedures for treatment of STEC-infected patients.
Introduction
Escherichia coli is a bacterium naturally occurring in the mammalian intestine, occupying this habitat as a commensal (Hartl and Dykhuizen, 1984). However, some strains of this bacterium carry virulence genes and are thus capable of causing disease in humans. Shiga toxin–producing E. coli (STEC) strains are examples of such pathogenic bacteria (Hunt et al., 2010). Most naturally occurring E. coli strains are lysogenic, which usually does not change their relations with the mammalian host. On the other hand, the main virulence factors of STEC are Shiga toxins (Schmidt, 2001), encoded by genes located on genomes of lambdoid prophages, called Shiga toxin–converting prophages (Law, 2000). Production and release of these toxins depend on the induction of these prophages and subsequent phage lytic development (Law, 2000; Schmidt, 2001).
Although STEC are not pathogenic for cattle, the major reservoir of these bacteria, they cause serious symptoms after infecting human intestine, which are especially severe in the case of a subset of STEC, called enterohemorrhagic E. coli. These symptoms include bloody diarrhea, which in some patients progress to hemorrhagic colitis and/or hemolytic uremic syndrome. The latter complication is a severe disease, especially dangerous for children and elderly patients (Besser et al., 1999; Gyles et al., 2007; Serna and Boedeker, 2008). Treatment of patients infected with STEC is problematic. This is mainly due to the fact that many antibiotics used to cure patients with bacterial infections stimulate induction of Shiga toxin–converting prophages, enhancing severity of the disease symptoms (Serna and Boedeker, 2008). Hence, treatment with antibiotics is restricted if infection with STEC is confirmed or even suspected. In this light, understanding the mechanisms of prophage induction in human intestine is important to facilitate development of procedures preventing or alleviating Shiga toxin–caused diseases.
Contrary to laboratory conditions used for years in studies on lambdoid phages, our knowledge on factors causing prophage induction in natural habitats of E. coli, such as human intestine, is very limited. Only recently, it was reported that hydrogen peroxide may be one of such factors. Namely, oxidative stress, caused by hydrogen peroxide, efficiently induced lambdoid prophages carrying Shiga toxin genes (Łoś et al., 2009, 2010). Studies on OxyR, one of the major factors of the oxidative stress response, confirmed that this protein may play an important role in the regulation of maintenance of lambdoid prophages (Glinkowska et al., 2010). In addition, induction of Shiga toxin–converting prophages can be triggered also by factors used for food preservation, such as high hydrostatic pressure (Aertsen et al., 2005) and 60Co irradiation (Yamamoto et al., 2003), which may contribute to the propagation of these phages.
Even less is known about conditions that may delay or inhibit lytic development of Shiga toxin–converting phages, a process occurring subsequent to prophage induction and being necessary for expression of the stx genes encoding Shiga toxin subunits. It was previously found that lytic development of bacteriophage λ (a paradigm of the family of lambdoid phages) can be slowed down significantly in bacteria cultured under conditions of nutrient limitation (Gabig et al., 1998; Słomińska et al., 1999) or even stopped in starved hosts (Łoś et al., 2007). One of the main agents causing inhibition of phage λ development in starved bacteria appears to be guanosine tetraphosphate (ppGpp), the stringent control alarmone, which is intensively produced upon starvation for various nutrients, but particularly for amino acids. This specific nucleotide interacts with RNA polymerase, causing global changes in genes' expression (Potrykus and Cashel, 2008), and with primase (Maciąg et al., 2010), the enzyme necessary for DNA replication. In fact, recent studies indicated that plasmids derived from Shiga toxin–converting phages could not replicate in amino acid–starved E. coli cells (Nejman et al., 2009, 2011). Therefore, the aim of this work was to test whether development of a model phage from this group, a derivative of 933W, can be efficiently inhibited under conditions of nutrient limitation or amino acid starvation.
Materials and Methods
Bacterial strains and bacteriophage
Otherwise isogenic E. coli strains CF1648 (synonymous to MG1655; wild type, relA+) and CF1652 (ΔrelA251::kan), described previously (Xiao et al., 1991), were used in all experiments. These strains were lysogenized with bacteriophage 933WΔtox (Gamage et al., 2004), a derivative of the Shiga toxin–converting phage 933W, which was devoid of the toxin genes for safety reasons, but retained all other features of this phage.
Media and growth conditions
Either 1/15-Luria-Bertani (LB) medium (Sambrook and Russell, 2001) with 15-fold less concentrations of nutrients, that is, 0.33 g yeast extract and 0.67 bacto-peptone/L, and standard amount (10 g/L) of NaCl or a minimal medium MMGlu (Jasiecki and Węgrzyn, 2003) were used. In some experiments, Orsalit (IBSS Biomed Co., Cracow, Poland), an oral rehydration mixture with decreased osmolarity, was added to culture medium at the amount of 2.2 g per 100 mL (equal to the recommended dose for patients). The concentrations of particular Orsalit's compounds in such a medium were as follows: 13.5 g/L (75 mM) glucose, 1.7 g/L (75 mM) sodium ions, 2.3 g/L (65 mM) chloride, 0.8 g/L (20 mM) potassium ions, 1.9 g/L (10 mM) citrate; the osmolarity of the solution was 245 mOsm/L. Bacteria were cultured at 37°C in shake flasks with agitation. Isoleucine starvation of bacteria growing in the MMGlu medium was induced by addition of l-valine to a final concentration of 1 mg/mL. The pH value of all variants of LB and MMGlu was 7.2.
Monitoring of phage development after prophage induction
Lysogenic bacteria were cultured in indicated medium to an A600 of 0.1. Prophage induction was provoked by addition of mitomycin C to a final concentration of 1 μg/mL. Samples of the culture were withdrawn at indicated times. To each sample (volume of 0.5 mL), 30 μL of chloroform was added, and following vortexing for 20 sec, the mixture was centrifuged in a microfuge (2000 g for 5 min). The lysate was titrated on E. coli C600 strain (Sambrook and Russell, 2001) by the plating method, which allows to obtain well-visible plaques of phages, which otherwise form very small ones, including 993W and its derivatives, as previously described (Łoś et al., 2008).
Estimation of phage and bacterial DNA content in cells
Bacteria were cultured as described in the preceding paragraph. At indicated times after prophage induction, samples (1 mL each) were withdrawn and boiled for 10 min. Following centrifugation (4000 g for 10 min), supernatants (1.5 μL from each sample) were used as the source of templates for quantitative polymerase chain reaction (PCR). The following primers were used (concentration of each primer stock was 600 nM, and the volume of each primer solution added to the reaction mixture was 1.5 μL): for determination of the amount of phage 933WΔtox DNA, phageF (5′-CAG GGG GAC ACA AAA GAC AC) and phageR (5′-GCC CAT GAC AGG AAG TTG TT)—complementary to the fragment comprising parts of the O and P genes; for determination of bacterial genomic DNA, bacterialF (5′-CAA GAG TTT GAT CCT GGC TCA G) and bacterialR (5′-CAT ACG GCT ACC TTG TTA CGA CTT)—complementary to the fragment of the gene coding for 16S RNA. After initial denaturation (94°C, 5 min), 22 cycles of amplification (94°C for 45 sec, 60°C for 45 sec, and 72°C for 75 sec) were performed. PCR products were separated electrophoretically in agarose gels (Sambrook and Russell, 2001), and amounts of DNA were estimated densitometrically on the basis of comparison of the intensities of investigated bands with those of markers of known DNA amounts (GeneRuler™ 100 bp Plus DNA Ladder; Fermentas International, Inc., Vilnius, Lithuania), run on the same gel. In preliminary experiments, we have proven the quantitative determination of DNA amount by using samples with various dilutions of known amounts of tested DNA (data not shown).
Statistical analysis
Each experiment was performed three times, and mean and SD values from three independent experiments were calculated. The statistical significance was assessed by using the independent two-sample t-test. The values were considered significantly different when p<0.05.
Results
An oral rehydration solution delays development of Shiga toxin–converting bacteriophage
In our studies we aimed to mimic, to some extent, conditions occurring in human intestine infected by STEC. As concentrations of nutrients in human intestine are usually significantly lower than those employed in standard laboratory experiments with E. coli (Salonen et al., 2009), we decided to use a medium, called 1/15-LB, containing 15-fold lower concentrations of nutrients relative to standard LB medium. STEC infections usually cause diarrhea in patients. As long as a specific diarrhea-causing agent is not determined, one of the treatment strategies is to administer products that help maintain appropriate levels of minerals. Orsalit (produced by IBSS Biomed Co.), an oral rehydration mixture, is one of such products. To test whether this product has any effect on development of Shiga toxin–converting prophage, we have monitored the titer of the 933WΔtox phage after prophage induction in lysogenic E. coli cells.
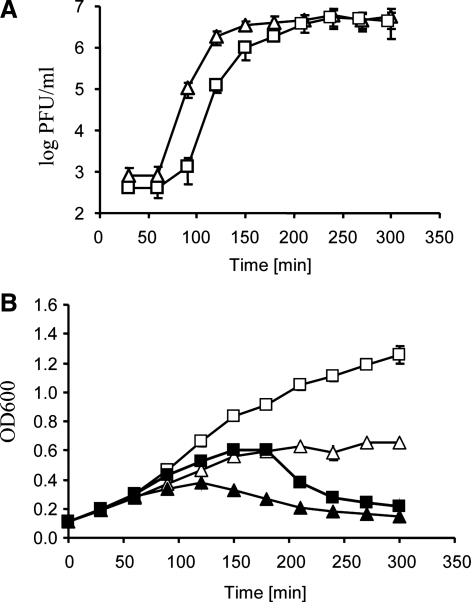
We found that in the presence of Orsalit (concentrations of particular compounds of this product in the medium were equal to those recommended by the manufacturer for preparation of the liquid to be orally administered to patients) development of the 933WΔtox phage was delayed relative to conditions of bacterial cultivation without this product (Fig. 1A). These results were corroborated by measurement of optical density of bacterial cultures. Prophage induction and subsequent phage lytic development eventually lead to bacterial cell lysis, which can be observed as a decrease in turbidity of the culture. We observed a delay in the drop of A600 of the Orsalit-containing culture of E. coli CF1648 (933WΔtox) after prophage induction relative to the analogous culture without Orsalit (Fig. 1B). Nevertheless, both phage titer and turbidity of bacterial culture achieved similar final values in both cultures containing Orsalit or devoid of this product (Fig. 1).
FIG. 1.
Development of bacteriophage 933WΔtox in Escherichia coli CF1648 (wild-type) host in the presence (squares) or absence (triangles) of Orsalit after prophage induction with mitomycin C (1 μg/mL) at time 0. Panel A shows normalized numbers of pfu/mL of the culture, calculated by subtraction of the pfu/mL values in cultures untreated with mitomycin C from the pfu/mL values in mitomycin C–treated cultures. Average values from three independent experiments are presented, with error bars indicating SD. The values were significantly different (p<0.05) at times 90, 120, 150, and 180 min. Panel B shows A600 (OD600) values of bacterial cultures either treated (closed symbols) or untreated (open symbols) with mitomycin C. Average values from three independent experiments are presented, with error bars indicating SD. The differences between bacteria cultured in the presence and absence of Orsalit (in both variants: with and without mitomycin C) were significantly different (p<0.05) at times between 90 and 300 min. pfu, plaque-forming units; SD, standard deviation.
Citrate inhibits Shiga toxin–converting phage development
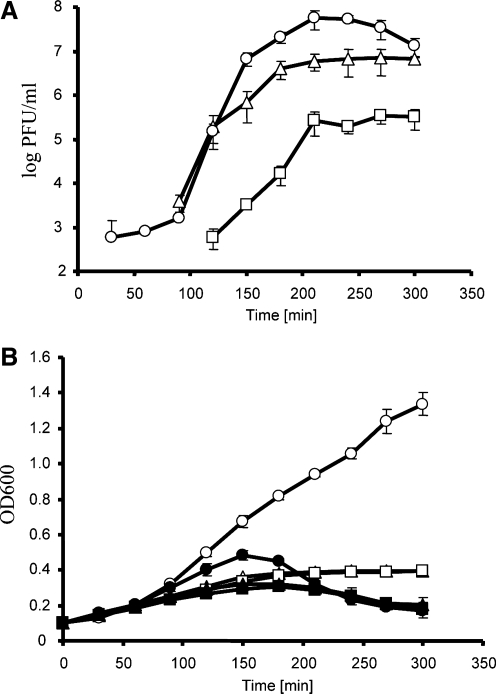
We then tested which compound present in Orsalit is responsible for the delay in 933WΔtox phage development. Therefore, we tested all compounds separately in experiments analogous to those described in the preceding paragraph. We found no significant effects of glucose, sodium ions, potassium ions, and chloride (results not shown). However, in the presence of 10 mM citrate, development of 933WΔtox after mitomycin C–provoked prophage induction was both significantly delayed and significantly inhibited (Fig. 2A). Although the delay in development was similar to that observed in experiments with Orsalit, over 10-fold less-efficient production of progeny phages relative to the medium without citrate was the new phenomenon relative to the presence of Orsalit (Fig. 2A). Again, these observations were compatible with results of determination of bacterial culture turbidity (Fig. 2B). Interestingly, the presence of glucose at a concentration of 1.6% reversed the effect of citrate, that is, the development of the 933WΔtox was no longer inhibited (Fig. 2).
FIG. 2.
Development of bacteriophage 933WΔtox in E. coli CF1648 (wild-type) host in the presence of 10 mM citrate and 1.6% glucose (circles), in the presence of 10 mM citrate and absence of glucose (squares), or in the absence of both citrate and glucose (triangles), after prophage induction with mitomycin C (1 μg/mL) at time 0. Panel A shows normalized numbers of pfu/mL of the culture, calculated by subtraction of the pfu/mL values in cultures untreated with mitomycin C from the pfu/mL values in mitomycin C–treated cultures. Average values from three independent experiments are presented, with error bars indicating SD. When comparing each pair of the experiments separately, significant differences (p<0.05) were found in all cases at times between 150 and 300 min. Panel B shows A600 (OD600) values of bacterial cultures either treated (closed symbols) or untreated (open symbols) with mitomycin C. Average values from three independent experiments are presented, with error bars indicating SD. When considering cultures treated with mitomycin C, significant differences (p<0.05) were found between experiments represented by closed circles and closed squares (the presence of citrate and glucose vs. the presence of citrate and absence of glucose), and those represented by closed circles and closed triangles (the presence of citrate and glucose vs. the absence of citrate and glucose) at times 90, 120, 150, and 180 min.
Citrate-mediated inhibition of phage 933WΔtox development is independent on DNA replication
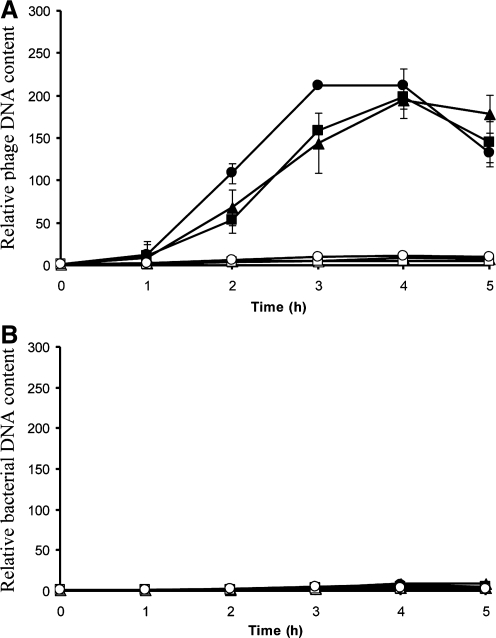
Theoretically, inhibition of lytic development of a lambdoid phage after prophage induction may occur either at the stage of phage genome replication or at the stage of production of progeny viruses. To test at which step this inhibition occurs, we have monitored levels of phage DNA after prophage induction in cells cultured in the presence or absence of citrate. We did not find any significant differences in the kinetics of the increase of phage DNA amount after prophage induction between cultures with and without citrate (Fig. 3). As expected, an increase in either uninduced prophage or bacterial DNA amount was negligible relative to replicating phage DNA (Fig. 3). Therefore, we conclude that citrate-mediated inhibition of phage 933WΔtox development does not result from problems with phage DNA replication.
FIG. 3.
Relative amount of phage (panel A) or bacterial (panel B) DNA in E. coli CF1648 (wild-type) cells either treated (closed symbols) or untreated (open symbols) with mitomycin C (1 μg/mL) at time 0, in the presence of 10 mM citrate and 1.6% glucose (circles), in the presence of 10 mM citrate and absence of glucose (squares), or in the absence of both citrate and glucose (triangles). Average values from three independent experiments are presented, with error bars indicating SD. When analyzing results of experiments depicted in panel A, the only significant differences (p<0.05) could be found between cultures treated and untreated with mitomycin C (closed and open symbols, respectively). No statistically significant differences occurred between values determined in experiments with mitomycin C–treated bacteria cultured under various conditions (combinations of the presence or absence of citrate and glucose). No statistically significant differences were also found between any values shown in panel B.
Amino acid starvation prevents phage 933WΔtox development
Since during patient's diarrhea the intestinal bacteria may experience conditions of transient starvation, and previous studies on bacteriophage λ indicated that such conditions may inhibit development of this phage (Łoś et al., 2007), we tested effects of starvation on lytic development of phage 933WΔtox. Amino acid starvation conditions were employed as a model for nutrient deprivation. This model was used as amino acid starvation can be easily and effectively induced in E. coli and mechanisms of physiological response to this kind of starvation are relatively well understood (Potrykus and Cashel, 2008).
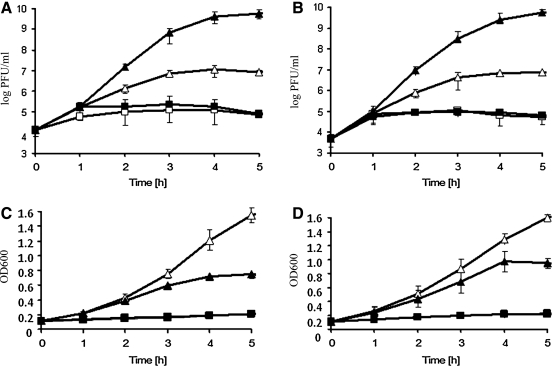
We found that upon treatment of the lysogenic bacteria with mitomycin C, bacteriophage 933WΔtox is not able to develop efficiently (i.e., to produce phage progeny) in amino acid-starved cells (Fig. 4). This was the case in both bacteria able to induce the stringent response to amino acid starvation (the relA+ strain) and bacteria revealing the relaxed response (the relA− mutant), characterized by a lack of production of ppGpp, an alarmone of the stringent response (Fig. 4).
FIG. 4.
Development of bacteriophage 933WΔtox in E. coli CF1648 (wild-type, panels A and C) and CF1652 (ΔrelA251::kan mutant, panels B and D) hosts in the presence (squares) or absence (triangles) of l-valine (1 mg/mL), which provoked isoleucine starvation, after prophage induction with mitomycin C (1 μg/mL) at time 0 (closed symbols) or without such an induction (open symbols). Panels A and B show pfu/mL values of bacteriophage 933WΔtox, and panels C and D shows A600 (OD600) values of bacterial cultures. Average values from three independent experiments are presented with error bars indicating SD. When considering experiments with mitomycin C–mediated induction of prophages, statistically significant differences (p<0.05) were found between values obtained for bacteria cultured in the presence and absence of l-valine (each panel). Analogous significant differences (p<0.05) were noted between l-valine–treated and untreated cultures when considering experiments without mitomycin C.
Discussion
Infections of humans by STEC often result in severe symptoms, and treatment of patients suffering from them is problematic (Serna and Boedeker, 2008; Hunt et al., 2010). This is mainly because virulence of STEC depends on induction of Shiga toxin–converting prophages and their further lytic development, and many antibiotics stimulate prophage induction. Therefore, in the light of restrictions in effective treatment with antibiotics, it appears that finding conditions either preventing prophage induction or inhibiting phage lytic development may be important in developing medical procedures to treat STEC-infected patients.
As Shiga toxin–converting bacteriophages belong to the group of lambdoid phages, previous studies on bacteriophage λ development may have implications for understanding the physiology of Shiga toxin production by STEC. Therefore, reports indicating that starvation conditions may inhibit both λ prophage induction (Czyz et al., 2001) and λ phage lytic development (Łoś et al., 2007) encouraged us to study effects of starvation on Shiga toxin–converting phage development. This is an important consideration, because there are opposing recommendations for treatment of patients with acute diarrhea, a symptom that is characteristic for STEC infections. One approach favors either reducing oral intake or even fasting during illness, whereas another approach recommends continued feeding (Brown, 1994; Grimwood and Forbes, 2009; Koletzko and Osterrieder, 2009).
Results of our studies indicated that the medicinal product (called Orsalit) used for maintaining appropriate amounts of minerals during diarrhea can delay development of Shiga toxin–converting bacteriophage 933WΔtox (Fig. 1). Importantly, we have identified that citrate, present also in this product, is responsible for this delay as well as for inhibition of efficiency of formation of progeny phages (Fig. 2). Moreover, glucose can alleviate this inhibition. These results may suggest that the fasting strategy, combined with providing minerals and citrate, rather than continued feeding, may be more appropriate in management of STEC infections.
To learn about potential mechanism(s) of citrate-mediated inhibition of 933WΔtox development, we tested kinetics of the increase in phage DNA amount after prophage induction and found that this process is not downregulated in the presence of citrate (Fig. 3). Therefore, we speculate that either production of phage structural proteins or progeny virion assembly is affected by citrate. Other processes, such as phage adsorption on the host cell, may be excluded as we tested only intracellular development of the viruses. As citrate is known to be a chelator of divalent cations, one could suspect that this activity may be considered when looking for the precise mechanism of the inhibition of 933WΔtox development. On the other hand, reversion of the effect of citrate by an excess of glucose (Fig. 2) suggests that some specific changes in cellular metabolism, mediated by citrate and overcome by glucose, may be crucial in this phenomenon. Further studies with the use of other chelators may indicate whether this activity of citrate is crucial in inhibiting Shiga toxin phage development.
Finally, our results indicated that amino acid starvation resulted in complete inhibition of phage 933WΔtox development (Fig. 4). This may be due to inhibition of prophage induction or inhibition of lytic development or both, as these processes were found to be negatively regulated in starved E. coli cells in the case of bacteriophage λ (Czyz et al., 2001; Łoś et al., 2007). On the other hand, although phage λ development is influenced by the stringent response alarmone, ppGpp (Słomińska et al., 1999), the effects of amino acid starvation on 933WΔtox development appear to be independent on this specific nucleotide as similar results were obtained in experiments with the wild-type strain and a mutant (relA) unable to produce ppGpp in starved cells. These results are compatible with previous findings that DNA synthesis of replicons derived from Shiga toxin–converting phages can be inhibited in amino acid–starved cells irrespective of the induction of the stringent response (Nejman et al., 2011).
Conclusions
Results presented in this report indicate that both the presence of citrate and starvation conditions may delay or inhibit development of Shiga toxin–converting phage. Therefore, these findings may support the fasting strategy rather than the recommendation of the continued feeding when STEC infection is confirmed or suspected in patients suffering from acute diarrhea. The reported results may also facilitate development of novel procedures for treatment of STEC-infected patients. One should note that this report describes the work on a single phage. Thus, it should be considered as a model study, providing a rationale for further research on effects of citrate and starvation on development of various Shiga toxin–converting bacteriophages in STEC, including natural isolates.
Acknowledgments
This work was operated within the Foundation for Polish Science Ventures Program co-financed by the EU European Regional Development Fund (Grant No. Ventures/2009-3/6 to B.N.-F.) and was supported in part by the Ministry of Science and Higher Education, Poland (Grant No. N N301 192439 to A.W.). The research by B.N.-F. was also supported by the Program of Introduction of Modern Education Elements at University of Gdańsk as well as by Foundation for Development of University of Gdańsk.
Disclosure Statement
No competing financial interests exist.
References
- Aertsen A. Faster D. Michiels CW. Induction of Shiga toxin–converting prophage in Escherichia coli by high hydrostatic pressure. Appl Environ Microbiol. 2005;71:1155–1162. doi: 10.1128/AEM.71.3.1155-1162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser RE. Griffin PM. Slutsker L. Escherichia coli O157:H7 gastroenteritis and the hemolytic uremic syndrome: an emerging infectious disease. Annu Rev Med. 1999;50:355–367. doi: 10.1146/annurev.med.50.1.355. [DOI] [PubMed] [Google Scholar]
- Brown KH. Dietary management of acute diarrheal disease: contemporary scientific issues. J Nutr. 1994;124:1455S–1460S. doi: 10.1093/jn/124.suppl_8.1455S. [DOI] [PubMed] [Google Scholar]
- Czyz A. Los M. Wrobel B. Węgrzyn G. Inhibition of spontaneous induction of lambdoid prophages in Escherichia coli cultures: simple procedures with possible biotechnological applications. BMC Biotechnol. 2001;1:1. doi: 10.1186/1472-6750-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Gabig M. Obuchowski M. Węgrzyn A. Szalewska-Pałasz A. Thomas MS. Węgrzyn G. Excess production of phage
 delayed early proteins under conditions supporting high Escherichia coli growth rates. Microbiology. 1998;144:2217–2224. doi: 10.1099/00221287-144-8-2217. [DOI] [PubMed] [Google Scholar]
delayed early proteins under conditions supporting high Escherichia coli growth rates. Microbiology. 1998;144:2217–2224. doi: 10.1099/00221287-144-8-2217. [DOI] [PubMed] [Google Scholar] - Gamage SD. Patton AK. Hanson JF. Weiss AA. Diversity and host range of Shiga toxin–encoding phage. Infect Immun. 2004;72:7131–7139. doi: 10.1128/IAI.72.12.7131-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinkowska M. Łoś JM. Szambowska A. Czyż A. Całkiewicz J. Herman-Antosiewicz A. Wróbel B. Węgrzyn G. Węgrzyn A. Łoś M. Influence of the Escherichia coli oxyR gene function on λ prophage maintenance. Arch Microbiol. 2010;192:673–683. doi: 10.1007/s00203-010-0596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwood K. Forbes DA. Acute and persistent diarrhea. Pediatr Clin North Am. 2009;56:1343–1361. doi: 10.1016/j.pcl.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Gyles CL. Shiga toxin–producing Escherichia coli: an overview. J Anim Sci. 2007;85:e45–e62. doi: 10.2527/jas.2006-508. [DOI] [PubMed] [Google Scholar]
- Hartl DL. Dykhuizen DE. The population genetics of Escherichia coli. Annu Rev Genet. 1984;18:31–68. doi: 10.1146/annurev.ge.18.120184.000335. [DOI] [PubMed] [Google Scholar]
- Hunt JM. Shiga toxin–producing Escherichia coli (STEC) Clin Lab Med. 2010;30:21–45. doi: 10.1016/j.cll.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasiecki J. Węgrzyn G. Growth-rate dependent RNA polyadenylation in Escherichia coli. EMBO Rep. 2003;4:172–177. doi: 10.1038/sj.embor.embor733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koletzko S. Osterrieder S. Acute infectious diarrhea in children. Dtsch Arztebl Int. 2009;106:539–547. doi: 10.3238/arztebl.2009.0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law D. Virulence factors of Escherichia coli O157 and other Shiga toxin–producing E. coli. J App Microbiol. 2000;88:729–745. doi: 10.1046/j.1365-2672.2000.01031.x. [DOI] [PubMed] [Google Scholar]
- Łoś JM. Golec P. Węgrzyn G. Węgrzyn A. Łoś M. Simple method for plating Escherichia coli bacteriophages forming very small plaques or no plaques under standard conditions. Appl Environ Microbiol. 2008;74:5113–5120. doi: 10.1128/AEM.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łoś JM. Łoś M. Węgrzyn G. Węgrzyn A. Differential efficiency of induction of various lambdoid prophages responsible for production of Shiga toxin in response to different induction agents. Microb Pathog. 2009;47:289–298. doi: 10.1016/j.micpath.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Łoś JM. Łoś M. Węgrzyn A. Węgrzyn G. Hydrogen peroxide-mediated induction of the Shiga toxin–converting lambdoid prophages ST2-8624 in Escherichia coli O157:H7. FEMS Immunol Med Microbiol. 2010;58:322–329. doi: 10.1111/j.1574-695X.2009.00644.x. [DOI] [PubMed] [Google Scholar]
- Łoś M. Golec P. Łoś JM. Węglewska-Jurkiewicz A. Czyż A. Węgrzyn A. Węgrzyn G. Neubauer P. Effective inhibition of lytic development of bacteriophages λ, P1 and T4 by starvation of their host, Escherichia coli. BMC Biotechnol. 2007;7:13. doi: 10.1186/1472-6750-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciąg M. Kochanowska M. Łyżeń R. Węgrzyn G. Szalewska-Pałasz A. ppGpp inhibits the activity of Escherichia coli DnaG primase. Plasmid. 2010;63:61–67. doi: 10.1016/j.plasmid.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Nejman B. Los JM. Los M. Wegrzyn G. Wegrzyn A. Plasmids derived from lambdoid bacteriophages as models for studying replication of mobile genetic elements responsible for production of Shiga toxins by pathogenic Escherichia coli strains. J Mol Microbiol Biotechnol. 2009;17:211–220. doi: 10.1159/000242447. [DOI] [PubMed] [Google Scholar]
- Nejman B. Nadratowska-Wesołowska B. Szalewska-Pałasz A. Węgrzyn A. Węgrzyn G. Replication of plasmids derived from Shiga toxin–converting bacteriophages in starved Escherichia coli. Microbiology. 2011;157:220–233. doi: 10.1099/mic.0.042820-0. [DOI] [PubMed] [Google Scholar]
- Potrykus K. Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Salonen A. Palva A. de Vos WM. Microbial functionality in the human intestinal tract. Front Biosci. 2009;14:3074–3084. doi: 10.2741/3436. [DOI] [PubMed] [Google Scholar]
- Sambrook J. Russell DW. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor; 2001. [Google Scholar]
- Schmidt H. Shiga-toxin–converting bacteriophages. Res Microbiol. 2001;152:687–695. doi: 10.1016/s0923-2508(01)01249-9. [DOI] [PubMed] [Google Scholar]
- Serna A. Boedeker EC. Pathogenesis and treatment of Shiga toxin–producing Escherichia coli infections. Curr Opin Gastroenterol. 2008;24:38–47. doi: 10.1097/MOG.0b013e3282f2dfb8. [DOI] [PubMed] [Google Scholar]
- Słomińska M. Neubauer P. Węgrzyn G. Regulation of bacteriophage λ development by guanosine 5′-diphosphate-3′-diphosphate. Virology. 1999;262:431–441. doi: 10.1006/viro.1999.9907. [DOI] [PubMed] [Google Scholar]
- Xiao H. Kalman M. Ikehara K. Zemel S. Glaser G. Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- Yamamoto T. Kojio S. Taneike I. Nakagawa S. Iwakura N. Wakisaka-Saito N. 60Co irradiation of Shiga toxin (Stx)-producing Escherichia coli induces Stx phage. FEMS Microbiol Lett. 2003;222:115–121. doi: 10.1016/S0378-1097(03)00259-3. [DOI] [PubMed] [Google Scholar]