Abstract
Cartilage loss is a feature of chronic arthritis. It results from degradation of the extracellular matrix which is composed predominantly of aggrecan and type II collagen. Extracellular matrix degradation is mediated by aggrecanases and matrix metalloproteinases (MMPs). Recently, a number of endogenous matrix molecules, including fibronectin (FN), have been implicated in mediating cartilage degradation. We were interested in studying the C-terminal heparin-binding region of FN since it mediates aggrecan and type II collagen breakdown in cartilage, but the specific FN domains responsible for proteolytic enzyme activity and their receptors in cartilage are unknown. In this study, the ability of recombinant FN domains to induce cartilage breakdown was tested. We found that the FN III 13-14 domains in the C-terminal heparin-binding region of FN are potent inducers of aggrecanase activity in articular cartilage. In murine studies, the FN III 13-14-induced aggrecanase activity was inhibited in Toll-like receptor 4 (TLR4) knockout mice but not wild-type mice. FN III 13-14 domains also synergized with the known catabolic cytokines interleukin-1α and tumour necrosis factor and induced secretion of MMP-1, MMP-3, gp38 and serum amyloid-like protein A in chondrocytes. Our studies provide a mechanistic link between the innate immune receptor TLR4 and sterile arthritis induced by the FN III 13-14 domains of the endogenous matrix molecule FN.
Key Words: Fibronectin, Osteoarthritis, Toll-like receptor 4, Damage-associated molecular patterns, Aggrecanases, Matrix metalloproteinases
Introduction
A number of factors are implicated in joint damage during arthritis, including the cytokines tumour necrosis factor (TNF)-α and interleukin-1 (IL-1), superoxide release and the production of proteolytic enzymes including the aggrecanases and matrix metalloproteinases (MMPs) that degrade the main components of joint tissue [1]. Recent work has shown that ADAMTS-5 (a disintegrin and metalloproteinase with thrombospondin repeats 5) is the major aggrecanase regulating cartilage destruction in murine animal models [2,3]. Reports have also demonstrated how catabolic factors including IL-1 and retinoic acid upregulate aggrecanase production by chondrocytes, the only cell type present in cartilage [4]. However, the early steps which lead to tissue breakdown in osteoarthritis are not well understood.
It has recently been suggested that endogenous signals which arise in joint tissue upon injury induce further joint damage and promote the persistence of inflammation. These include the activation of catabolic pathways by endogenously expressed extracellular matrix proteins including fibronectin (FN) [5,6,7,8], fibromodulin [9], hyaluronan [10] and type II collagen [11]. Upon injury, there is upregulated expression of specific proteins including FN, fibromodulin and type II collagen [12,13]. The activation of pro-inflammatory pathways mediated by these proteins and their fragments has led to their description as damage-associated molecular patterns [14,15].
Of all the endogenous cartilage proteins implicated in joint degradation, a large amount of evidence has accumulated for specific regions of FN. FN is a glycoprotein comprising a domain structure of type I, II and III repeats that confer specific biological properties to this molecule [16]. FN fragments (FNfs) upregulate the release of MMPs and pro-inflammatory mediators in joint tissue, including cartilage [5,17] and synovial cells [18]. Recently, the Toll-like receptor 4 (TLR4) has been implicated in promoting the activation of pro-inflammatory pathways by interaction with specific damage-associated molecular patterns in chronic inflammation [14]. Increased TLR expression has been reported in arthritic tissue and proposed as an important mechanism for maintaining chronic inflammation [19,20]. The extra type III domain A (EDA) splice variant of FN, expressed in cellular but not plasma forms of FN, has recently been shown to mediate its pro-inflammatory effects via TLR4 in mast cells [21] and neutrophils [22]. However, the potential interaction of other FN domains with TLRs is unknown. Hashimoto et al. [23] reported that the C-terminal heparin-binding region of FN can regulate the activity of recombinant forms of ADAMTS-4. We were therefore interested in investigating which specific domain(s) in the C-terminal heparin-binding region of FN induce aggrecanase activity in cartilage models. In view of the EDA inducing joint damage via TLR4 [21], we also hypothesized that the C-terminal heparin-binding region may mediate its cartilage-degrading effects via TLR4. Here, we show for the first time that FN domains mapping to repeats FN III 13-14 in the C-terminal heparin-binding region upregulate aggrecanase activity via the activation of TLR4 in articular cartilage. We thereby describe a novel mechanism of how the innate immune receptor TLR4 activates catabolic pathways by TLR4 activation, thereby promoting ongoing joint destruction in arthritis.
Methods
Reagents
Materials were purchased from the following sources: fetal calf serum from Labtech International (UK), Dulbecco's modified Eagle's medium (DMEM), penicillin and streptomycin from Biowhittaker (UK), dimethylmethylene blue (DMMB) and shark chondroitin sulphate from Sigma-Aldrich Ltd. (UK), unstained and pre-stained precision protein standards for SDS-PAGE from Biorad (UK), Superdex-200, Superdex-75 pre-packed columns, a GFX DNA extraction kit and an mRNA purification kit from GE-Healthcare (UK), nickel-nitrilotriacetic acid (NTA) agarose resin from Qiagen (Hilden, Germany), amphotericin B, PVDF membrane, superscript II reverse transcriptase (RT) and the polymerase chain reaction (PCR) Blunt-TOP10 vector system from Invitrogen (Paisley, UK), chondroitinase ABC and keratanase from Seikagaku (Tokyo, Japan), anti-rabbit alkaline phosphatase (AP)-linked antibody, anti-mouse AP-linked antibody and AP substrate (5-bromo-4-chloro-3-indolyl-1-phosphate and nitroblue tetrazolium) from Promega (Southampton, UK), HepG2 cells from ECACC (Salisbury, UK), XL1-blue bacteria, pET3a vector, BL21-(DE3) cells, carbenicillin and kanamycin from Novagen (Nottingham, UK), IPTG (isopropyl β-D-thiogalactopyranoside) from Biogene (Cambridge, UK), T4 DNA ligase, restriction endonucleases from New England Biolabs (UK), Pfu Turbo from Stratagene Europe (Amsterdam, The Netherlands) and expired human plasma for FN purification from the London Blood Bank Services. An RNA extraction kit was from Qiagen (Crawley, UK). The Limulus amoebocyte assay kit was purchased from Cambrex (USA). Monoclonal antibody BC-3 that recognizes the aggrecanase-generated N-terminal neoepitope 374ARGSV of aggrecan and BC-14 that recognizes the MMP-generated N-terminal neoepitope FFGVS were gifts from Prof. B. Caterson and Dr. C. Hughes from the University of Cardiff, UK. A polyclonal antibody to anti-ALGS peptide was a gift from Dr. A. Fosang from the University of Melbourne, Australia. IL-1α was a gift from Prof. J. Saklatvala from the Imperial College, London, UK. Phenol-chloroform-purified Escherichia coli lipopolysaccharides (LPSs; rough and smooth) were from Alexis (Birmingham, UK). Porcine cartilage from pig trotters was obtained 4–8 h after slaughter and provided by Fresh Tissue Supplies (London, UK).
Murine Experiments
Homozygous TLR4-deficient mice on a C57BL/6 background were obtained from B & K Universal (Hull, UK) [24,25]. Homozygous MyD88-deficient mice on a C57BL/6 background were provided by the Sanger Institute (Cambridge, UK). Age-matched congenic inbred wild-type C57BL/6 mice were obtained from Charles River (Margate, UK). All animals were fed standard rodent chow and water ad libitum, and were housed (<6 mice/cage) in sawdust-lined cages in an air-conditioned environment with 12-hour light/dark cycles. All animal procedures were approved by the Institutional Ethics Committee.
Cartilage Culture with Catabolic Factors
Porcine articular cartilage from the metacarpophalangeal joints of 3- to 9-month-old pigs was dissected into small pieces (3 × 2–3 × 0.5 mm; wet weight approx. 10 mg). Following dissection, the cartilage rested for 48 h at 37°C under 5% CO2 in DMEM containing 5% fetal calf serum, penicillin, streptomycin and amphotericin B (100 units/ml each). After resting, cartilage was washed 3 times in serum-free DMEM. Each cartilage piece was placed in the well of a round-bottom 96-well plate with 200 μl of serum-free medium with FNfs and IL-1α. After 2 days, the conditioned media and cartilage were harvested separately and stored at −20°C until use.
Murine cartilage was obtained from 6-week-old mice and their femoral heads were dissected. After resting for 48 h at 37°C under 5% CO2 in DMEM containing 5% fetal calf serum, penicillin and streptomycin (100 units/ml) plus amphotericin B (100 units/ml), cartilage was washed 3 times in serum-free DMEM and then stimulated with FNfs or other catabolic factors including IL-1 and LPS. After 2 days, conditioned media were harvested and stored at −20°C until use.
Analysis of Glycosaminoglycan Release
Glycosaminoglycan (GAG) released into the conditioned medium was measured using the DMMB assay as described by Farndale et al. [26]. A volume of 250 μl of DMMB reagent was mixed with 5 μl of sample. Each sample was assayed in duplicate. A standard curve using shark chondroitin sulphate (0–2.6 μg) was included in each plate. The treatments were tested on cartilage in triplicate, for which the absorbance at 540 nm was used for GAG release. Analyses were performed using the Graphpad prism software (version 4; San Diego, Calif., USA) (see statistics section).
SDS-PAGE and Western Blot Analyses of Aggrecan Fragments Released Using Neoepitope Antibodies for Detecting Aggrecan Neoepitopes
Proteins were resolved by SDS/PAGE using ammediol gels [27], and stained with either Coomassie brilliant blue R-250 or silver according to Schevchenko et al. [28]. To detect responsible metalloproteinases that degrade cartilage aggrecan, the media containing 0–100 μg GAG was digested with chondroitinase ABC and keratanase, following which, samples were subjected to Western blotting analyses using BC-3 monoclonal antibody or anti-ALGS antibody for aggrecanase-generated fragments and BC-14 monoclonal antibody for MMP-generated fragments, as described by Gendron et al. [29].
Purification of FN, Expression and Purification of Recombinant FNfs
FN was purified from plasma using gelatin-Sepharose affinity chromatography as described by Weiss and Reddi [30]. The concentration of the purified FN was calculated by molar extinction coefficients.
Human FN cDNA was obtained by RT-PCR of RNA extracted from human HepG2 cells. A total of 5 μg total RNA was obtained from 300 × 106 cells. mRNA was purified from total RNA using an mRNA purification kit and reverse transcribed using superscript II RT into cDNA. cDNAs encoding various recombinant FNfs were generated by PCR using primers described in table 1. The PCR products were ligated into the PCR-Blunt vector and transformed into TOP10 cells. After confirming DNA sequences, respective fragments were ligated into pET-3a for recombinant FN III domains spanning the III 12, 13, 14 and V region. All recombinant proteins were histidine (6×)-tagged at the N-terminus and the pET vector harbouring FNf cDNA was introduced into BL21-(DE3) cells. Transformed cells were grown until the optical density reached 0.5 at 600 nm and then incubated for 3.5 h, after 0.1 mM IPTG had been added. Bacterial pellets were harvested by centrifugation, resuspended in TBS and passed through a French press (ThermoSpectraphor) 3 times, then spun at 10,000 g for 15 min.
Table 1.
Primer sequences for recombinant FN domains
| Primer | Sequence |
|---|---|
| Recombinant FNf III 12–14 | |
| Sense | 5′ ATA CAT ATG CAT CAT CAT CAT CAT CAT CCT GCA CCA ACT GAC CTG AAG 3′ |
| Antisense | 5′ GCC GGA TCC TTA TGT CTT TTT CCT TCC AAT CAG GGG 3′ |
| Recombinant FNf III 13–14 | |
| Sense | 5′ ATA CAT ATG CAT CAT CAT CAT CAT CAT AAT GTC AGC CCA CCA AGA AGG 3′ |
| Antisense | 5' GCC GGA TCC TTA TGT CTT TTT CCT TCC AAT CAG GGG 3′ |
| Recombinant FNf III 12 | |
| Sense | 5′ ATA CAT ATG CAT CAT CAT CAT CAT CAT CCT GCA CCA ACT GAC CTG AAG 3′ |
| Antisense | 5′ GCC GGA TCC TTA CTC CAG AGT GGT GAC AAC ACC 3′ |
| Recombinant FNf III 13 | |
| Sense | 5′ ATA CAT ATG CAT CAT CAT CAT CAT CAT AAT GTC AGC CCA CCA AGA AGG 3′ |
| Antisense | 5' GCC GGA TCC TTA AGT GGA GGC GTC GAT GAC 3′ |
| Recombinant FNf III 14 | |
| Sense | 5′ ATA CAT ATG CAT CAT CAT CAT CAT CAT GCC ATT GAT GCA CCA TCC ACC C 3′ |
| Antisense | 5′ GCC GGA TCC TTA TGT CTT TTT CCT TCC AAT CAG GGG 3′ |
| Recombinant FNf V region | |
| Sense | 5′ ATA CAT ATG CAT CAT CAT CAT CAT CAT GAC GAG CTT CCC CAA CTG GTA AC 3′ |
| Antisense | 5′ GCC GGA TCC TTA TCC TAC ATT CGG CGG GTA TGG TC 3′ |
Primers for the desired region in FN were made. Constructs contained NdeI (CAT ATG) and BamHI (GGA TCC) restriction enzyme sites for vector insertion. The CAT repeating sequence denotes a histidine tag at the N-terminus.
Recombinant FNfs were purified from the soluble fraction by applying the soluble proteins to a nickel-NTA affinity column. The column was washed with 10 column volumes of 50 mM Tris/HCl pH 7.5, 150 mM NaCI and 60% isopropanol to remove LPS, as described previously [31]. It was then eluted by 100 mM imidazole in 50 mM Tris/HCl pH 7.5 and 150 mM NaCl. After the nickel-NTA column purification step all three proteins were homogeneous by SDS-PAGE. Levels of LPS in the recombinant FNfs and control media were measured using the Limulus amoebocyte assay (Cambrex) and were detectable in the range 0–100 pg/ml. For all purified recombinant proteins and conditioned media, levels were undetectable using this assay. LPS levels required for cartilage stimulation are in the μg/ml range [20].
RT-PCR Experiments on Isolated Porcine Chondrocytes
Cartilage was incubated with Pronase E (1 mg/ml per 1 g cartilage) for 30 min at 37°C, followed by collagenase (1 mg/ml per 1 g cartilage) for 5 h at 37°C. The digest was passed through a 70-μM cell strainer, then centrifuged for 5 min at 1,400 rev/min. Pellets were washed, resuspended in DMEM containing 10% fetal calf serum. Cells were counted and plated on 24-well plates (1.5-cm diameter) at a density of 1 million cells per well (100% confluent). Isolated chondrocytes were cultured in DMEM (2 ml) that had been supplemented with 10% fetal calf serum, HEPES (25 mM), penicillin (1.25 units/ml), streptomycin (100 μg/ml) and amphotericin B (2 μg/ml) in a humidified atmosphere of 95% air, 5% CO2 at 37°C.
For RT-PCR, porcine chondrocytes (1 × 106 cells/well) were stimulated with IL-1α (10 ng/ml), FNf domains (1 μM) or controls for 24 h. After culture, RNA was extracted from cells using an mRNA purification kit as directed by the manufacturer. For each treatment, 10 μg RNA was subjected to RT-PCR to obtain cDNA. The cDNA produced was then subjected to PCR for several gene products shown in table 2 as previously described [31].
Table 2.
Primers for RT-PCR performed on DNA extracted from porcine chondrocytes
| Primer | Sequence |
|---|---|
| ADAMTS-4 | |
| Sense | 5′ ACC ACT TTG ACA CAG CCA TTC TG 3′ |
| Antisense | 5′ ACC CCC ACA GGT CCG AGA GCA G 3′ |
| ADAMTS-5 | |
| Sense | 5′ TGT GCT GTG ATT GAA GAC GAT 3′ |
| Antisense | 5′ GAC TGC AGG AGC GGT AGA TGG 3′ |
| GAPDH | |
| Sense | 5′ CAT GGA GAA GGC TGG GGC TC 3′ |
| Antisense | 5′ ATG AGG TCC ACC ACC CTG TT 3′ |
Metabolic Labelling of Chondrocytes with 35S
Confluent porcine chondrocytes (1 × 106 cells/well) were stimulated overnight with IL-1, recombinant FN domains, FN or medium alone. The following morning, the stimulus was removed, the cells were washed with 2 ml Met/Cys-free medium for 1 h, then 15 μCi 35S was added to each well in a volume of 150 μl. Cells were incubated for a further 6 h at 37°C. After this, the conditioned medium was spun at 13,000 rpm for 5 min. The supernatant was removed and added to 40 μl 4× sample buffer and boiled for 10 min. Samples were then run on SDS-PAGE using 10% Tris-glycine gels. The gels were removed and silver-stained. Gels were dried on a gel dryer for 2 h and then exposed to autoradiography overnight. Bands of interest were subjected to proteomic analysis as described below.
Mass-Spectrometric Analysis
Silver- or Coomassie-stained protein bands of SDS/PAGE were excised with a scalpel and digested in gel with trypsin using an Investigator ProGest (Huntingdon, UK) robotic digestion system [32]. Tandem electrospray mass spectra were recorded using a Q-TOF mass spectrometer (Waters, Manchester, UK) interfaced to a Waters CapLC capillary chromatograph. Samples were dissolved in 0.1% aqueous formic acid, injected onto a 300-μm × 15-mm Pepmap C18 column (LC Packings, Amsterdam, The Netherlands), and eluted with an acetonitrile/0.1% formic acid gradient. A survey scan over the m/z range of 400–1,300 was used to identify protonated peptides with charge states of 2, 3 or 4, which were automatically selected for data-dependent MS/MS analysis, and fragmented by collision with argon. The resulting product ion spectra were transformed onto a singly charged m/z axis using a maximum entropy method (MaxEnt 3, Waters) and proteins were identified by correlation of uninterpreted spectra to entries on Swiss-Prot and TrEMBL, using the ProteinLynx Global Server (version 1.1, Waters). Hits which rested on a single matching peptide were confirmed by manual interpretation of sequence-specific fragment ions using the MassLynx program PepSeq (Waters).
Statistical Analysis
The GAG release assay in cartilage explants was evaluated using Graphpad Prism software (version 4). Mean values were calculated for each treatment in triplicate and expressed as mean ± SEM (n = 3). Significance was analyzed with an unpaired 2-tailed Student t test and defined as p < 0.05. For band densitometry of Western blots and RT-PCR, results were scanned using a Biorad densitometer and pixel intensity quantified using Phoretix software (Totallab, Newcastle, UK). Data was plotted for mean band intensity ± SEM (n = 3).
Results
Activity of Recombinant FNfs
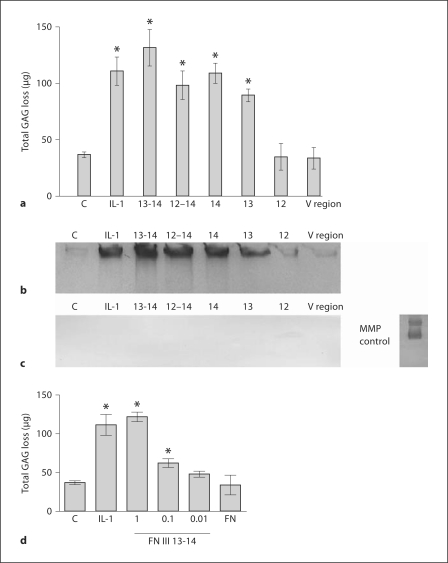
Specific domains of the C-terminal heparin-binding region of FN were generated to test how they regulate aggrecanase activity. Fragments comprising the III 12, III 13, III 14, III 12–14, III 13-14 and the V region were expressed as recombinant proteins using an E. coli expression system (fig. 1). All proteins were histidine tagged and purified using nickel-NTA affinity chromatography (fig. 1). The recombinant fragments were tested using normal porcine articular cartilage explants. Levels of GAG release were measured after a 2-day culture period using the DMMB assay (fig. 2). The FN III 12–14 (1 μM) domains maintained high levels of cartilage-degrading activity comparable to the positive control IL-1α (10 ng/ml). When domains were tested singly, the III 12 domain had no activity and was comparable to background levels. Furthermore, fragments consisting of the III 13 and III 14 domains had high levels of GAG-releasing and aggrecanase activity. Since the FN III 13-14 domains have previously been shown to have synergistic binding [33], they were expressed as a recombinant fragment and were found to stimulate the highest levels of GAG release (fig. 2). In order to investigate specific aggrecanase activity induced by the distinct FN domains, the conditioned medium from the experiment in figure 2a was deglycosylated with chondroitinase and keratanase and then subjected to Western blotting using neoepitope antibodies specific for aggrecanase- and MMP-cleaved neoepitopes. Results showed that the highest activity was observed for FN III 13-14, suggesting that these domains are critical for aggrecanase induction by the C-terminal heparin-binding region in cartilage (fig. 2). Of note, FN III 12 and the V region were unable to stimulate aggrecanase activity in cartilage (fig. 2). Results using the MMP cleavage-dependent neoepitope antibody showed that cleavage of aggrecan was predominantly dependent on aggrecanases and not MMPs in this 2-day culture experiment, since conditioned medium from the same experiment did not show any MMP-cleaved neoepitopes (fig. 2b, c). In order to test the dose-response effect of FN III 13-14, the levels of aggrecan loss by this recombinant fragment were compared with full-length FN and IL-1α served as a positive control (fig. 2d). FN III 13-14 induced a dose-dependent release of aggrecan loss in the micromolar range which was comparable to the positive control IL-1α (10 ng/ml) at the highest concentration. In comparison, full-length FN induced no significant levels of GAG release.
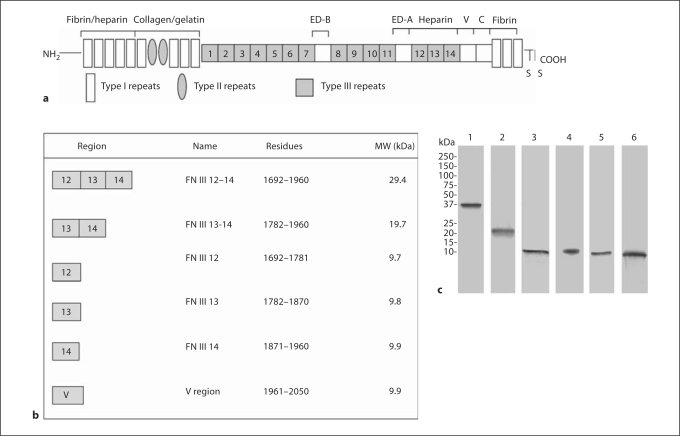
Fig. 1.
Recombinant FN domains synthesized. a Domain structure of FN comprising the type I, II and III repeats. The affinity of certain domains for specific molecules e.g. fibrin, heparin and collagen and the location of splice variants ED-B, ED-A, V and C regions are shown. b Regions covered by the recombinant proteins that were synthesized with the corresponding amino acid residues and the molecular weight of each protein. c Coomassie-stained gel showing 10 μg of each recombinant protein analyzed by SDS-PAGE (12% gel) under reducing conditions. Lanes: 1 = FN III 12–14, 2 = FN III 13–14, 3 = FN III 12, 4 = FN III 13, 5 = FN III 14, 6 = V region.
Fig. 2.
Proteoglycan loss induced by FN domains in cartilage explant cultures. a Porcine cartilage was stimulated with IL-1α (10 ng/ml) as a positive control, recombinant FN domains at 1 μM concentration each or control (medium alone) for 2 days. The conditioned medium was then harvested and analyzed for GAG release using the DMMB assay (n = 3). Statistically significant values: ∗ p < 0.05. Equal volumes of conditioned media (50 μl) from the experiment (a) were deglycosylated and Western blots performed for aggrecanase-cleaved neoepitopes using the BC-3 antibody recognizing the N-terminal sequence 374ARGSV (b) and the BC-14 antibody recognizing the N-terminal sequence 342FFGVG (c). c A positive control of aggrecan cleaved with MMP-3 which was then immunoblotted using the 342FFGVG antibody is shown on the right. d The dose-response effect of the most potent FN III domains, namely FN III 13–14, was tested in the 0.01–1 μM range. IL-1α (10 ng/ml) served as a positive control and control medium and full-length FN (1 μM) were negative controls. Statistically significant values: ∗ p < 0.05.
FN III 13-14 Upregulate ADAMTS-4 and ADAMTS-5 in Chondrocytes
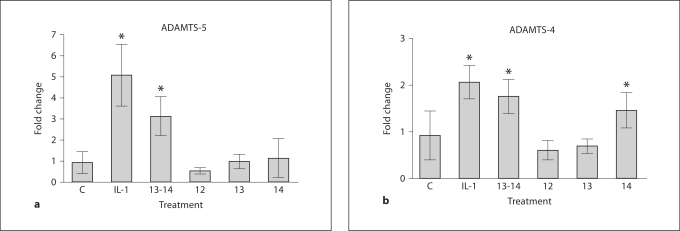
Since the highest activity was observed for FN III 13-14, these domains were tested for their ability to regulate ADAMTS-4 and ADAMTS-5. Porcine chondrocytes were treated for 24 h with recombinant III 13-14, full-length FN and IL-1α (10 ng/ml) as a positive control. Results showed that IL-1α upregulated ADAMTS-5 messenger RNA by RT-PCR (fig. 3). ADAMTS-4 mRNA was also upregulated by FN III 13-14, but to a lesser degree than ADAMTS-5 (fig. 3). When the specific domains were compared, FN III 12 induced virtually no ADAMTS-4 or ADAMTS-5 activity in chondrocytes. In comparison, the levels of induction of ADAMTS-4 and ADAMTS-5 for FN III 13 and FN III 14 alone were higher than FN III 12, but not as high as FN III 13-14, suggesting that FN III 13-14 have synergistic effects.
Fig. 3.
Analysis of aggrecanase gene products induced by FN domains. Primary porcine chondrocytes were isolated using collagenase digestion. After resting overnight, cells (1 < 106/well) were washed 3 times in serum-free medium and stimulated for 24 h with IL-1 (10 ng/ml), FN III 12, FN III 13, FN III 14 and FN III 13–14, all at 1 μM in serum-free medium. After the culture period, RNA was isolated from cells and subjected to RT-PCR for ADAMTS-4 and ADAMTS-5 with GAPDH as controls. Results show fold change for ADAMTS-4 or ADAMTS-5 compared with GAPDH controls (n = 3). Statistically significant values: ∗ p < 0.05.
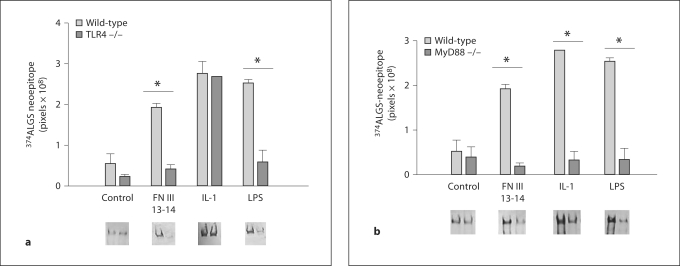
FN III 13-14 Mediate Aggrecanase Activity via TLR4 in Cartilage
Previous work by other groups has suggested that chronic inflammatory pathways stimulated by the EDA of FN are mediated through TLR4 [21,22]. Such work prompted us to investigate the catabolic effects of FN III 13-14 on aggrecanase induction using C57BL6 wild-type and TLR4–/– null mice. Conventional catabolic factors for cartilage degradation, namely IL-1 and also LPS, which is known to signal through TLR4 [34], were used as positive controls in our experiments. In these experiments, murine hip cartilage was cultured with treatments for a 48-hour period. Following this, conditioned medium was harvested and assayed for aggrecanase neoepitope activity using the anti-ALGS antibody (which is the corresponding sequence to ARGS in mice). Signal intensity from Western blots was measured and results showed that in the wild-type mice, the positive controls IL-1 (10 ng/ml) and LPS (10 ng/ml) were effective at inducing strong aggrecanase-generated neoepitopes (fig. 4a). FN III 13-14 at 1 μM induced strong aggrecanase activity in the murine system equivalent to the positive controls. When the same experiment was performed in the TLR4–/– null mice, signals for LPS were strongly diminished. It is known that LPS activity is mediated through TLR4; therefore, these results were expected. Furthermore, the FN III 13-14 signal was strongly diminished in the TLR4–/– mice, suggesting that the activity of FN III 13-14 is also mediated via TLR4. Studies were also conducted in TLR2–/– mice, but no change in aggrecanase neoepitopes compared to the wild-type mice was observed (data not shown).
Fig. 4.
FN III 13–14 mediate aggrecanase activity via TLR4. a Hip explants from 6-week-old C57B6 wild-type and TLR 4-/- null mice were dissected and rested in serum containing medium for 2 days. After this, explants were washed 3 times in serum-free medium and subjected to treatment with control medium (no additives), FN III 13–14 (1 μM), IL-1α (10 ng/ml) and LPS (10 ng/ml). After 2 days’ culture, conditioned medium was harvested and equal volumes of conditioned medium (50 μl) were deglycosylated using chondroitinase and keratanase. The samples were subjected to Western blotting using the anti-ALGS neoepitope antibody. The signal intensity of the samples (from triplicate wells, n = 3) was measured using Phoretics software and expressed as a mean and SEM. b Hip explants from Myd88-/- mice were treated with the same factors as described in a. Following 2 days’ culture, conditioned medium was harvested, deglycosylated and subjected to Western blotting as described in a with the anti-ALGS antibody.
TLRs require co-receptors for activity and include MD2 for TLR4 or CD36 for TLR2. TLR signalling requires adaptor proteins, including MyD88, Mal, TRIF and TRAM, which are recruited to specific receptors [34]. In order to investigate further the pathways involved in the activity of FN III 13-14, cultures were performed using cartilage from MyD88-null mice since MyD88 has been shown to be a downstream adaptor molecule for TLR4 stimulation. Results showed inhibition of aggrecanase neoepitope signal for IL-1 and LPS, as would be expected, since both molecules act via TLR4 and signal through MyD88 (fig. 4b). The signal for FN III 13-14 was reduced with a similar reduction to IL-1 and LPS, suggesting that the activity of FN III 13-14 is also mediated via the TLR4/MyD88 pathway.
FN III 13-14 Synergize with IL-1 and TNF to Induce Aggrecanase Activity
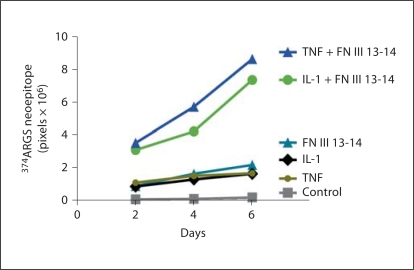
After establishing that FN III 13-14 induced aggrecanase activity in joint tissue, we were interested in investigating whether other ligands of the TLR superfamily synergize with FN domains to enhance cartilage degradation. We utilized TNF and IL-1α at 0.1 ng/ml and 0.1-μM FN III 13-14 domains (fig. 5). Results showed that over a 6-day culture period the aggrecanase neoepitope signal increased synergistically with IL-1α and TNF at 0.1 ng/ml combined with FN III 13-14 (0.1 μM), but not when each of the treatments was cultured alone with cartilage explants.
Fig. 5.
Synergy between FN III 13–14, IL-1 and TNF in the induction of proteoglycan loss. FN III 13–14 (0.1 μM), IL-1μ (10 ng/ml) or TNF-α (10 ng/ml) were cultured alone, or in combination with porcine articular cartilage over a 6-day culture period in serumfree medium. Conditioned medium was harvested every 2 days and replaced with fresh medium. At the end of the experiment, equal volumes of conditioned medium (50 μl) were deglycosylated using chondroitinase and keratanase. The samples were subjected to Western blotting and GAG release was measured using the anti-ARGSV neoepitope antibody. The signal intensity of the samples (from triplicate wells, n = 3) was measured using Phoretix software and expressed as mean and SEM.
FN III 13-14 Upregulate Secretion of Proteases and Pro-Inflammatory Molecules
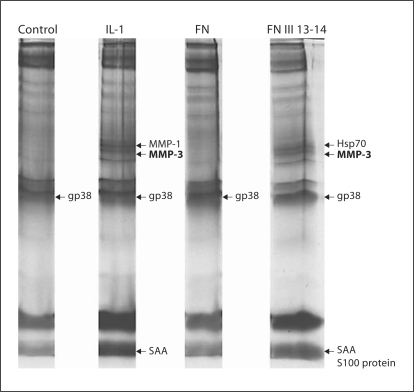
The C-terminal heparin-binding region has previously been shown to upregulate the production of collagen-binding proteins in cartilage including cartilage oligomeric matrix protein and chondroadherin [6]. To investigate whether the FN III 13-14 domains were regulating the production of other proteins in chondrocytes, a method previously described for stimulating isolated chondrocytes in monolayer culture was utilized [35]. The synthesis and secretion of proteins by metabolically labelling chondrocyte cultures with 35S-Met-Cys and separating the secreted proteins by SDS-PAGE was performed with full-length FN and FN III 13-14, and IL-1α served as a positive control. Conditioned medium from treated cells was collected and run on SDS-PAGE (fig. 6). The gels were silver-stained, dried and newly synthesized proteins were detected by autoradiography. Radiolabelled bands that corresponded with silver-stained bands were excised, digested with trypsin and analyzed by MALDI MS. For IL-1, the proteins identified included MMP-3, MMP-1 and gp38 (the porcine homologue of human gp39). As previously published, the IL-1α response stimulated 14-kDa serum amyloid-like protein A (SAA). In comparison, FN III 13-14 produced a distinct signature from IL-1 (fig. 6). In addition to some proteins whose secretion was induced by both FN III 13-14 and IL-1α, including MMP-3 and SAA, FN III 13-14 also upregulated hsp70. FN III 13-14 also induced the synthesis of S100 protein. This was in addition to MMP-3 and SAA whose synthesis was induced by both IL-1α and FN III 13-14. The only protein which was induced by both FN III 13-14 and full-length FN was gp38. These data suggest that FN III 13-14 regulate distinct pathways in comparison with full-length FN that only appears upon fragmentation.
Fig. 6.
Changes in synthesis of secreted proteins in isolated chondrocytes in response to IL-1α, FN III 13–14 domains and fulllength FN. Isolated chondrocytes cultured for 48 h were serumstarved (2 h) and left unstimulated (control C) or stimulated for 18 h with IL-1α (10 ng/ml), FN III 13–14 (1 μM) or full-length FN (1 μM). The following day, cells were washed for 30 min in Met-Cys-free medium and then incubated for 6 h with medium containing 35 S-Met-Cys. Medium was removed, clarified by centrifugation and subjected to SDS-PAGE (12% gel). The gel was silverstained, dried and autoradiographed to visualize newly synthesized proteins. For identification of radiolabelled bands that corresponded to silver-stained ones, the gel was rehydrated and the silver-stained bands were cut out and digested with trypsin. Tryptic digests were analyzed by mass spectrometry. The newly synthesized metabolically labelled proteins identified by mass spectrometry were: MMP-1, MMP-3, hsp70, S100 protein, gp38 and SAA.
Discussion
Extracellular matrix components form an essential scaffold in tissues, such as cartilage, that maintain cells within a structural framework. Degradation of the extracellular matrix by proteolytic enzymes is a key step in altering cellular behaviour during development, tissue remodelling and repair [36]. FNfs are produced during injury and their expression is increased in injured arthritic tissues [37,38]. Our study has shown that within the C-terminal heparin-binding domain of FN, the FN III 13-14 domains are endogenous molecules that induce catabolic pathways in joint tissue. Previously, a 40-kDa C-terminal heparin-binding fragment was shown to stimulate the production of MMPs in joint tissue. The C-terminal heparin-binding region induced MMP-3 and MMP-13 in cartilage [7]. This fragment also induced MMP-1, MMP-3 and MMP-13 production in RA synovial fibroblasts [18]. Our study has shown that the FN III 13-14 domains within the C-terminal heparin-binding region are critical for the induction of degradative pathways in joint tissue. Previous work by other groups has described how fragments of FN generated by proteolytic cleavage are detectable in arthritic tissue in the micromolar range [37,38]. Peters et al. [38] also described FNfs from the synovial fluid of subjects with osteoarthritis and rheumatoid arthritis that include the FN III 13 and 14 domains [38]. It is therefore likely that such fragments, once in their cleaved form, reveal cryptic epitopes that are then available to interact with receptors such as TLRs in mediating further tissue destruction and inflammation.
We have demonstrated that the FN III 13-14 domains regulate aggrecanase activity. In 2004, Hashimoto et al. [23] showed that the C-terminal heparin-binding FNf inhibits the activity of recombinant full-length ADAMTS-4 but not shorter ADAMTS-4 species lacking the spacer domain. Our study has found that FN III 13-14 regulate the aggrecanase activity in cartilage explant cultures. While we observed that FN III 13 and 14 alone caused significant GAG release (fig. 2), these fragments seemed to have only very limited effects on aggrecanase expression (fig. 3). It is possible that the FN III 13 and 14 domains upregulate the production of other proteases in cartilage to promote aggrecan cleavage via TLR4. For example, Zhang et al. [20] described how the activation of specific TLR1/2 and TLR6/2 ligands in chondrocytes upregulates collagenase gene production in these cells.
Our study is the first to show that aggrecanase activity induced by FN III 13-14 is mediated via TLR4. Several groups have recently described the presence of TLRs in joint tissue [19,20,21]. The only other FN domain reported to potentiate tissue destruction via TLRs in an arthritic model is the EDA [22]. Okamura et al. [39] also showed that the alternatively spliced EDA fragment of cellular FN acted via TLR4 in macrophages to stimulate MMP production. The FN III 13-14 domains share structural homology with EDA and all these domains comprise a β-pleated sheet structure confirmed by crystallography [33]. Furthermore, our work has shown that the FN III 13-14 domains utilize the MyD88-signalling pathway, since cartilage explants from MyD88 knockout mice treated with FN III 13-14 were protected from aggrecan loss.
Yasuda et al. [7] also suggested that the FN C-terminal heparin-binding region stimulates its effects by interaction with CD44. CD44 interacts with hyaluronan, and hyaluronan fragments have been implicated in the development of arthritic disease [10]. Taylor et al. [40] recently showed that TLR4, MD2 and CD44 form a unique complex that recognizes hyaluronan. They performed immunoprecipitation experiments that confirmed the physical association of TLR4 and CD44. Their work was a previously unknown mechanism for the initiation of sterile inflammation that involves recognition of released hyaluronan fragments as an endogenous signal of tissue injury. It is therefore possible that FN III 13-14 may also interact with both TLR4 and CD44 to maintain tissue injury. Future experiments will be required to establish this. The mechanism we described here may be an important pathway inducing the recruitment of degradative enzymes and pro-inflammatory mediators in the arthritic joint through the TLR4/MyD88 pathway.
Data from our metabolic labelling experiments showed that some of the proteins we showed to be upregulated by FN III 13-14, such as hsp70 and S100 protein, are endogenous TLR ligands themselves [14]. Taken together, increased levels of FNfs such as FN III 13-14 in arthritic joints may not only directly interact with receptors such as TLR4 to induce aggrecanase activity in the arthritic joint, but also increase the production of other endogenous TLR ligands hsp70 and S100 that are also TLR4 ligands [41], thereby accentuating the activation of catabolic pathways in the arthritic joint by endogenous FNfs. Targeting the activation of TLR4-mediated pathways which promote the persistence of catabolic signals during arthritic joint disease may be an attractive target for the treatment of arthritis in the future.
Acknowledgements
We would like to thank Prof. Hideaki Nagase, Dr. Linda Troeberg (for technical expertise) and Dr. Claudia Monaco (for advice on TLR4–/– mice) from the Kennedy Institute of Rheumatology, Imperial College London. Dr. Clare Hughes and Prof. Bruce Caterson are acknowledged for their gift of BC-3 and BC-14 antibodies. This work was supported in part by the Arthritis Research Campaign, a clinical research training fellowship from the Wellcome Trust (Grant No. 070848) and the Hammersmith Hospital Trustees’ Research Committee grant awarded to Dr. Nidhi Sofat.
References
- 1.Nagase H, Kashiwagi M. Aggrecanases and cartilage matrix degradation. Arthritis Res Ther. 2003;5:94–103. doi: 10.1186/ar630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanton H, Rogerson FM, East CJ, Golub SB, Lawlor KE, Meeker CT, Little CB, Last K, Farmer PJ, Campbell IK, Fourie AM, Fosang AJ. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434:648–652. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- 3.Glasson SS, Askew R, Sheppard B, Carito B, Blanchet T, Ma HL, Flannery CR, Peluso D, Kanki K, Yang Z, Majumdar MK, Morris EA. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434:644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 4.Rogerson FM, Stanton H, East CJ, Golub SB, Tutolo L, Farmer PJ, Fosang AJ. Evidence of a novel aggrecan-degrading activity in cartilage: studies of mice deficient in both ADAMST-4 and ADAMTS-5. Arthritis Rheum. 2008;58:1664–1673. doi: 10.1002/art.23458. [DOI] [PubMed] [Google Scholar]
- 5.Homandberg GA, Meyers R, Xie DL. Fibronectin fragments cause chondrolysis of bovine articular cartilage slices in culture. J Biol Chem. 1992;267:3597–3604. [PubMed] [Google Scholar]
- 6.Johnson A, Smith R, Saxne T, Hickery M, Heinegard D. Fibronectin fragments cause release and degradation of collagen-binding molecules from equine explant cultures. Osteoarthritis Cartilage. 2004;12:149–159. doi: 10.1016/j.joca.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Yasuda T, Poole AR, Shimizu M, Nakagawa T, Julovi SM, Tamamura H, Fujii N, Nakamura T. Involvement of CD44 in induction of matrix metalloproteinases by a COOH-terminal heparin-binding fragment of fibronectin in human articular cartilage in culture. Arthritis Rheum. 2003;48:1271–1280. doi: 10.1002/art.10951. [DOI] [PubMed] [Google Scholar]
- 8.Forsyth CB, Pulai J, Loeser RF. Fibronectin fragments and blocking antibodies to alpha2beta1 and alpha5beta1 integrins stimulate mitogen-activated protein kinase signaling and increase collagenase 3 (matrix metalloproteinase 13) production by human articular chondrocytes. Arthritis Rheum. 2002;46:2368–2376. doi: 10.1002/art.10502. [DOI] [PubMed] [Google Scholar]
- 9.Heathfield TF, Onnerfjord P, Dahlberg L, Heinegard D. Cleavage of fibromodulin in cartilage explants involves removal of the N-terminal tyrosine sulfate-rich region by proteolysis at a site that is sensitive to matrix metalloproteinase-13. J Biol Chem. 2004;279:6286–6295. doi: 10.1074/jbc.M307765200. [DOI] [PubMed] [Google Scholar]
- 10.Knudson W, Casey B, Nishida Y, Eger W, Kuettner KE, Knudson CB. Hyaluronan oligosaccharides perturb cartilage matrix homeostasis and induce chondrocytic chondrolysis. Arthritis Rheum. 2000;43:1165–1174. doi: 10.1002/1529-0131(200005)43:5<1165::AID-ANR27>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 11.Klatt AR, Paul-Klausch B, Klinger G, Kuhn G, Renno JH, Banerjee M, Malchau G, Wielckens K. A critical role for collagen II in cartilage matrix degradation: collagen II induces pro-inflammatory cytokines and MMPs in primary human chondrocytes. J Orthop Res. 2009;27:65–70. doi: 10.1002/jor.20716. [DOI] [PubMed] [Google Scholar]
- 12.Aigner T, Zien A, Gehrsitz A, Gebhard PM, Mckenna L. Anabolic and catabolic gene expression pattern analysis in normal versus osteoarthritic cartilage using complementary DNA-array technology. Arthritis Rheum. 2001;44:2777–2789. doi: 10.1002/1529-0131(200112)44:12<2777::aid-art465>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 13.Aigner T, McKenna L, Zien A, Fan Z, Gebhard PM, Zimmer R. Gene expression profiling of serum- and interleukin-1 beta-stimulated primary human adult articular chondrocytes – a molecular analysis based on chondrocytes isolated from one donor. Cytokine. 2005;31:227–240. doi: 10.1016/j.cyto.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Foell D, Wittkowski H, Roth J. Mechanisms of disease: a ‘DAMP’ view of inflammatory arthritis. Nat Clin Pract Rheumatol. 2007;3:382–390. doi: 10.1038/ncprheum0531. [DOI] [PubMed] [Google Scholar]
- 15.Sofat N. Analysing the role of endogenous matrix molecules in the development of osteoarthritis. Int J Exp Pathol. 2009;90:463–479. doi: 10.1111/j.1365-2613.2009.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hynes . Fibronectins. New York: Springer; 1990. pp. 113–175. [Google Scholar]
- 17.Homandberg GA, Hui F, Wen C, Purple C, Bewsey K, Koepp H, Huch K, Harris A. Fibronectin-fragment-induced cartilage chondrolysis is associated with release of catabolic cytokines. Biochem J. 1997;321:751–757. doi: 10.1042/bj3210751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasuda T, Shimizu M, Nakagawa T, Julovi SM, Nakamura T. Matrix metalloproteinase production by COOH-terminal heparin-binding fibronectin fragment in rheumatoid synovial cells. Lab Invest. 2003;83:153–162. doi: 10.1097/01.lab.0000056999.08437.b2. [DOI] [PubMed] [Google Scholar]
- 19.Su SL, Tsai CD, Lee CH, Salter DM, Lee HS. Expression and regulation of Toll-like receptor 2 by IL-1beta and fibronectin fragments in human articular chondrocytes. Osteoarthritis Cartilage. 2005;13:879–886. doi: 10.1016/j.joca.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q, Hui W, Litherland GJ, Barter MJ, Davidson R, Darrah C, Donell ST, Clark IM, Cawston TE, Robinson JH, Rowan AD, Young DA. Differential Toll-like receptor-dependent collagenase expression in chondrocytes. Ann Rheum Dis. 2008;67:1633–1641. doi: 10.1136/ard.2007.079574. [DOI] [PubMed] [Google Scholar]
- 21.Gondokaryono SP, Ushio H, Niyonsaba F, Hara M, Takenaka H, Jayawardena ST, Ikeda S, Okumura K, Ogawa H. The extra domain A of fibronectin stimulates murine mast cells via Toll-like receptor 4. J Leukoc Biol. 2007;82:657–665. doi: 10.1189/jlb.1206730. [DOI] [PubMed] [Google Scholar]
- 22.Lefebvre JS, Lévesque T, Picard S, Paré G, Gravel A, Flamand L, Borgeat P. The extra domain A of fibronectin primes leukotriene biosynthesis and stimulates neutrophil migration through Toll-like receptor 4 activation. Arthritis Rheum. 2011;63:1527–1533. doi: 10.1002/art.30308. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto G, Shimoda M, Okada Y. ADAMTS4 (aggrecanase-1) interaction with the C-terminal domain of fibronectin inhibits proteolysis of aggrecan. J Biol Chem. 2004;279:32483–32491. doi: 10.1074/jbc.M314216200. [DOI] [PubMed] [Google Scholar]
- 24.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda K, Akira S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipolpolysaccharide: evidence for TLR4 as the LPS gene product. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 25.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 26.Farndale RW, Buttle DJ, Barrett AJ. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta. 1986;883:173–177. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]
- 27.Bury AF. Analysis of protein and peptide mixtures: evaluation of three SDS-PAGE buffer systems. J Chromatogr. 1981;213:491–500. [Google Scholar]
- 28.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1980;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 29.Gendron C, Kashiwagi M, Hughes C, Caterson B, Nagase H. TIMP-3 inhibits aggrecanase-mediated glycosaminoglycan release from cartilage explants stimulated by catabolic factors. FEBS Lett. 2003;555:431–436. doi: 10.1016/s0014-5793(03)01295-x. [DOI] [PubMed] [Google Scholar]
- 30.Weiss RE, Reddi AH. Isolation and characterization of rat plasma fibronectin. Biochem J. 1981;197:529–534. doi: 10.1042/bj1970529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Midwood K, Sacre S, Piccinini AM, Inglis J, Trebaul A, Chan E, Drexler S, Sofat N, Kashiwagi M, Orend G, Brennan F, Foxwell B. Tenascin-C is an endogenous activator of TLR4 that is essential for maintaining inflammation in arthritic joint disease. Nat Med. 2009;15:774–780. doi: 10.1038/nm.1987. [DOI] [PubMed] [Google Scholar]
- 32.Wait R, Gianazza E, Eberini I, Sironi L, Dunn MJ, Gemeiner M, Miller I. Proteins of rat serum, urine, and cerebrospinal fluid. VI. Further protein identifications and interstrain comparison. Electrophoresis. 2001;22:3043–3052. doi: 10.1002/1522-2683(200108)22:14<3043::AID-ELPS3043>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 33.Sharma A, Askari JA, Humphries MJ, Jones EY, Stuart DI. Crystal structure of a heparin- and integrin-binding segment of human fibronectin. EMBO J. 1999;18:1468–1479. doi: 10.1093/emboj/18.6.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Neill LA. Primer: Toll-like receptor signalling pathways – what do rheumatologists need to know? Nat Clin Pract Rheumatol. 2008;4:319–327. doi: 10.1038/ncprheum0802. [DOI] [PubMed] [Google Scholar]
- 35.Gruber H, Vincent TL, Hermansson M, Bolton M, Wait R, Saklatvala J. Induction of interleukin-1 in articular cartilage by explantation and cutting. Arthritis Rheum. 2004;50:2539–2546. doi: 10.1002/art.20369. [DOI] [PubMed] [Google Scholar]
- 36.Parks WC, Wilson CL, Lopez-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol. 2004;4:617–629. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 37.Xie DL, Meyers R, Homandberg GA. Fibronectin fragments in osteoarthritic synovial fluid. J Rheumatol. 1992;19:1448–1452. [PubMed] [Google Scholar]
- 38.Peters JH, Carsons S, Yoshida M, Ko F, McDougall S, Loredo GA, Hahn TJ. Electrophoretic characterization of species of fibronectin bearing sequences from the N-terminal heparin-binding domain in synovial fluid samples from patients with osteoarthritis and rheumatoid arthritis. Arthritis Res Ther. 2003;5:R329–R339. doi: 10.1186/ar1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okamura Y, Watari M, Jerud ES, Young DW, Ishizaka ST, Rose J, Chow JC, Strauss JF., 3rd The extra domain A of fibronectin activates Toll-like receptor 4. J Biol Chem. 2001;276:10229–10233. doi: 10.1074/jbc.M100099200. [DOI] [PubMed] [Google Scholar]
- 40.Taylor KR, Yamasaki K, Radek KA, Nardo A, Goodarzi H, Golenbock, Beutler B, Gallo RL. Recognition of hyaluronan released in sterile injury involves a unique receptor complex dependent on Toll-like receptor 4, CD44 and MD-2. J Biol Chem. 2007;282:18265–18275. doi: 10.1074/jbc.M606352200. [DOI] [PubMed] [Google Scholar]
- 41.Ionita MG, Arslan F, de Kleijn DP, Pasterkamp G. Endogenous inflammatory molecules engage Toll-like receptors in cardiovascular disease. J Innate Immun. 2010;2:307–315. doi: 10.1159/000314270. [DOI] [PubMed] [Google Scholar]