Abstract
Biological membranes define not only the cell boundaries but any compartment within the cell. To some extent, the functionality of membranes is related to the elastic properties of the lipid bilayer and the mechanical and hydrophobic matching with functional membrane proteins. Supported lipid bilayers (SLBs) are valid biomimetic systems for the study of membrane biophysical properties. Here, we acquired high-resolution topographic and quantitative mechanics data of phase-separated SLBs using a recent atomic force microscopy (AFM) imaging mode based on force measurements. This technique allows us to quantitatively map at high resolution the mechanical differences of lipid phases at different loading forces. We have applied this approach to evaluate the contribution of the underlying hard support in the determination of the elastic properties of SLBs and to determine the adequate indentation range for obtaining reliable elastic moduli values. At ∼200 pN, elastic forces dominated the force-indentation response and the sample deformation was <20% of the bilayer thickness, at which the contribution of the support was found to be negligible. The obtained Young's modulus (E) of 19.3 MPa and 28.1 MPa allowed us to estimate the area stretch modulus (kA) as 106 pN/nm and 199 pN/nm and the bending stiffness (kc) as 18 kBT and 57 kBT for the liquid and gel phases, respectively.
The concept that lipid bilayers are not just a simple passive beholder of membrane proteins is now well accepted. It is important to note that membranes are heterogeneous, with local associations of lipids (and proteins) in detergent-resistant membrane (DRM) domains or rafts (1, 2).
In general, membrane dimensions and mechanical properties (i.e., bilayer thickness, bending and stretching stiffness, or membrane tension) modify the function not only of mechanosensitive proteins but of any membrane protein (3). In this framework, the mattress model is in favor of the importance of the lipid environment and provides an elastic model of lipid bilayer behavior (4). As a consequence of the established importance of bilayer compliance and lateral organization of membranes, a large number of techniques (including micropipette aspiration, surface force apparatus, biomembrane force probe or atomic force microscopy imaging, and force spectroscopy) have been employed to give insights into the structure and mechanical properties of biological membranes (5, 6, 7, 8). Among them, atomic force microscopy (AFM) (9) in particular has been used to address fundamental questions on the nanomechanics of supported membranes (10).
Here, we introduce a novel, to our knowledge, AFM-based imaging technique, PeakForce-Quantitative Nano-Mechanics (PF-QNM), to probe the structural and mechanical properties of SLBs. PF-QNM allows simultaneous imaging and quantitative mechanical mapping of the sample, both at submolecular resolution (11), and, it is important to point out, improves acquisition time and spatial resolution compared to other AFM-based techniques, such as force volume. This is achieved by oscillating the sample in the z axis at a given amplitude (tens of nanometers) and frequency (2 kHz), thus providing cycles of force-distance (FνD) curves in which the tip intermittently contacts the sample surface. Each FνD plot is thereafter analyzed to determine the mechanical properties of the sample (Fig. S1 in the Supporting Material), thus coupling topography analysis with stiffness and deformation assessment at high resolution.
The aim of this study was to probe the mechanical properties of biological membranes in the elastic regime. We present measures of the elastic properties (i.e., Young's modulus) of different lipid phases, and characterize the effect of the underlying hard substrate.
Nanomechanical mapping of SLBs was performed on DOPC/DPPC (1:1, mol/mol) membranes (Fig. 1), which is one of the best-characterized SLBs and is commonly used as a straightforward model membrane for AFM studies (12, 13). DOPC/DPPC bilayers display phase separation at room temperature between liquid (Lα) and gel (Lβ) phase, as a consequence of the different transition temperatures of DOPC and DPPC (−20°C and 41°C, respectively) (14). The presence of two segregated domains was readily detected by means of AFM topography with heights for liquid and gel phases of 4.1 ± 0.2 nm and 5.3 ± 0.4 nm, respectively, over the mica support (see Fig. S2), which is in agreement with previous observations (12). It is worthy of note that DOPC has two 18C with one unsaturation (18:1) and DPPC has two 16C fully saturated hydrocarbon chains. These structural differences account not only for the different transition temperatures, more densely packed and gel-like ordered structure for DPPC, and liquidlike behavior for DOPC at room temperature, but also for other physicochemical properties, including differences in breakthrough forces, response to detergent addition, etc. (6, 12). Indeed, using PF-QNM AFM imaging, we observed that the gel phase was systematically stiffer than the liquid phase (Figure 1, Figure 2). Given the thinness of lipid bilayers, the contribution of the underlying hard mica substrate must be a matter of careful consideration. To address this sensible question, we employed PF-QNM at different loading forces (100 pN, 200 pN, 300 pN, 400 pN, and 550 pN). The Young's modulus of SLBs was determined by fitting the Hertz model to retracting curves at each applied force (Fig. S1). The resulting topography and nanomechanical maps document force-dependent height (nm), stiffness (MPa), and deformation (nm) behavior of the DOPC/DPPC SLBs (Fig. 1).
Figure 1.
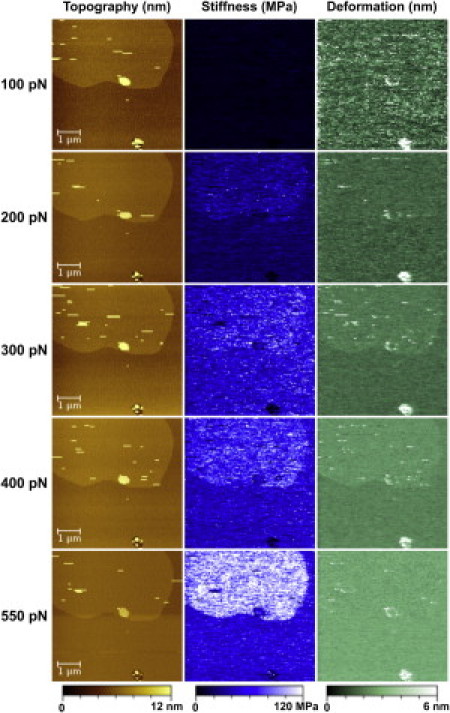
PF-QNM images showing the topography (nm) (left), stiffness (MPa) (middle), and deformation (nm) (right) at different peak loading forces (100 pN, 200 pN, 300 pN, 400 pN, and 550 pN). The false color scale is 12 nm for height, 120 MPa for apparent stiffness, and 6 nm for deformation.
Figure 2.
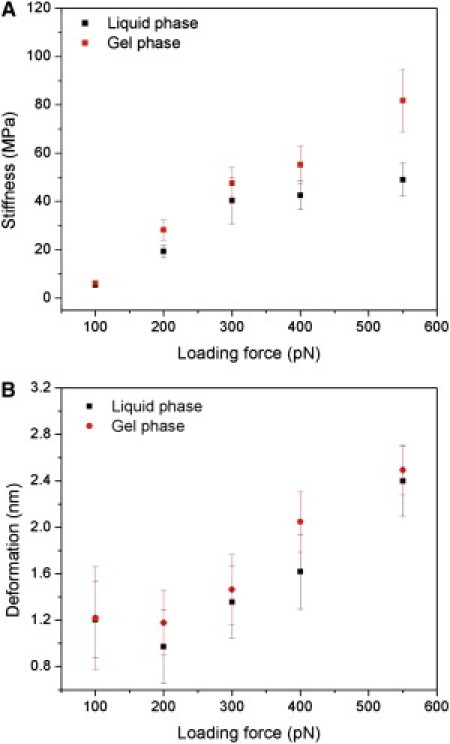
Graphical representation of stiffness (A) and deformation (B) as functions of the peak loading force for both fluid and gel phases. Data are shown as the mean ± SD.
As expected, the obtained average stiffness values increased with increasing loading force, with the gel phase stiffer than the fluid phase at all loads (Fig. 2 A). The degree of deformation followed a similar trend (Fig. 2 B), except at very low loading forces (100 pN), where a high apparent deformation (1.2 nm) was observed as a result of long-range electrostatic forces that dominated the interaction over elastic forces (Table S1).
The force-indentation relationship of a parabolic tip indenting a thin layer has been developed and validated on thin layers of soft gels (15). According to this theoretical approach, the overestimation of E is small (<25%) when the indentation is <20% of the sample thickness. Thus, at forces of ∼200 pN, where the deformation is ∼1 nm, PF-QNM mapping provides reliable values of the actual bilayer elastic modulus of 19.3 MPa and 28.1 MPa for the liquid and gel phases, respectively (Table S1). Even though it is difficult to asses the validity of continuous models at the nanometer scale, and possible nonlinear strain hardening may occur, our values were also confirmed by determining E on supported lipid vesicles (Fig. S3). Moreover, our results show that gel-phase lipids are stiffer than fluid-phase lipids, though the former are thicker than the latter.
The measured Young's modulus on the gel- and fluid-phase bilayers allowed us to calculate the area stretch modulus (kA) and bending stiffness (kc), by invoking thin shell theory
where ν is the Poisson ratio, assumed as 0.5, and h is the bilayer thickness (7). From our results, we estimated kA at 106 pN/nm and 199 pN/nm and kc at 18 kBT and 57 kBT for the liquid and gel phases, respectively. Previous estimations from micropipette aspiration and AFM-based methods were in quantitative agreement with our results (7, 8, 16). However, we found a considerably high bending stiffness in the case of the SLB in the gel phase, which to our knowledge has not been calculated before. The measured thickness and mechanical properties between the two phases allow us to estimate the energetic cost due to bilayer deformation to be 0.81 kBT and 0.36 kBT for DOPC and DPPC, respectively (see Supporting Material), which is in conceptual agreement with previous works (13, 17). Thus, even though the energetic cost of mixing is low, it is sufficient for phase separation.
The improved time and lateral resolution of PF-QNM offered us more physical insight about SLB mechanical properties (Fig. 3). The stiffer gel-lipid phase can be understood by the lower mobility and tighter order of DPPC lipid tails. According to this interpretation, one could suggest that the edges between DOPC and DPPC would have intermediate stiffness, as lipids might be more disordered and mobile at phase edges. We observed slight changes in mechanics at phase edges on the order of ∼10 nm, suggesting that the lateral range of this effect is short.
Figure 3.
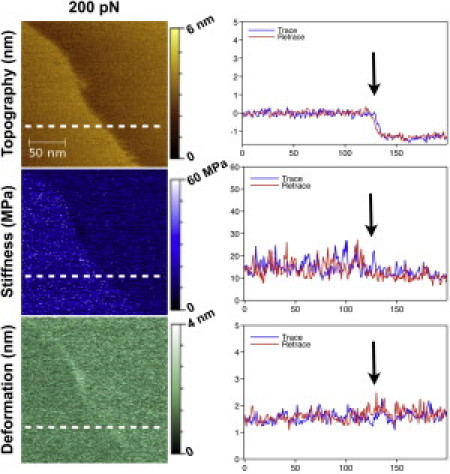
PF-QNM images at the lipid phase boundary showing the topography (nm), stiffness (MPa), and deformation (nm) at 200 pN peak loading force. The false color scale (from top to bottom) is 6 nm, 60 MPa, and 4 nm. Cross section of the trace (blue line) and retrace (red line) along the white dashed line for topography, stiffness, and deformation (average of three consecutive lines).
Our approach illustrates the suitability of PF-QNM AFM for the nanomechanical mapping of membrane models at high resolution and sufficient sensitivity to detect the properties of lipid phases. Our results suggest that at moderate indentations, determination of the stiffness is actually not perturbed by the solid support, thus providing reliable values of the Young's modulus. However, relative differences between lipid phases become more obvious at high applied forces. Accordingly, we propose that the high spatial resolution and sensitivity of the applied technique might be used to mechanically detect DRMs or rafts on cell surfaces before they can be directly visualized.
Acknowledgments
The authors thank Pierre Sens for fruitful discussion.
This study was supported by a Fondation Pierre-Gilles de Gennes Fellowship (to L.P.) and a European Community Marie Curie Intra-European Fellowship for Career Development (to F.R.).
Editor: Peter Hinterdorfer.
Footnotes
Materials and Methods, two figures, a table, and references are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(11)05352-5.
Supporting Material
References and Footnotes
- 1.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 2.Engelman D.M. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 3.Phillips R., Ursell T., et al. Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jensen M.O., Mouritsen O.G. Lipids do influence protein function-the hydrophobic matching hypothesis revisited. Biochim. Biophys. Acta. 2004;1666:205–226. doi: 10.1016/j.bbamem.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Dufrêne Y.F., Boland T., et al. Lee G.U. Characterization of the physical properties of model biomembranes at the nanometer scale with the atomic force microscope. Faraday Discuss. 1998;111:79–94. doi: 10.1039/a807637e. discussion 111:137–157. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Manyes S., Sanz F. Nanomechanics of lipid bilayers by force spectroscopy with AFM: a perspective. Biochim. Biophys. Acta. 2010;1798:741–749. doi: 10.1016/j.bbamem.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 7.Rawicz W., Olbrich K.C., et al. Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das C., Sheikh K.H., et al. Connell S.D. Nanoscale mechanical probing of supported lipid bilayers with atomic force microscopy. Phys. Rev. E. 2010;82:041920. doi: 10.1103/PhysRevE.82.041920. [DOI] [PubMed] [Google Scholar]
- 9.Binnig G., Quate C.F., Gerber C. Atomic force microscope. Phys. Rev. Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 10.Gonçalves R.P., Agnus G., et al. Scheuring S. Two-chamber AFM: probing membrane proteins separating two aqueous compartments. Nat. Methods. 2006;3:1007–1012. doi: 10.1038/nmeth965. [DOI] [PubMed] [Google Scholar]
- 11.Rico F., Su C., Scheuring S. Mechanical mapping of single membrane proteins at submolecular resolution. Nano Lett. 2011;11:3983–3986. doi: 10.1021/nl202351t. [DOI] [PubMed] [Google Scholar]
- 12.Milhiet P.E., Gubellini F., et al. Lévy D. High-resolution AFM of membrane proteins directly incorporated at high density in planar lipid bilayer. Biophys. J. 2006;91:3268–3275. doi: 10.1529/biophysj.106.087791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leonenko Z.V., Finot E., et al. Cramb D.T. Investigation of temperature-induced phase transitions in DOPC and DPPC phospholipid bilayers using temperature-controlled scanning force microscopy. Biophys. J. 2004;86:3783–3793. doi: 10.1529/biophysj.103.036681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsh D. 2nd ed. Taylor and Francis; New York: 2012. Handbook of Lipid Bilayers. [Google Scholar]
- 15.Dimitriadis E.K., Horkay F., et al. Chadwick R.S. Determination of elastic moduli of thin layers of soft material using the atomic force microscope. Biophys. J. 2002;82:2798–2810. doi: 10.1016/S0006-3495(02)75620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dieluweit S., Csiszár A., et al. Merkel R. Mechanical properties of bare and protein-coated giant unilamellar phospholipid vesicles. A comparative study of micropipet aspiration and atomic force microscopy. Langmuir. 2010;26:11041–11049. doi: 10.1021/la1005242. [DOI] [PubMed] [Google Scholar]
- 17.Wallace E.J., Hooper N.M., Olmsted P.D. Effect of hydrophobic mismatch on phase behavior of lipid membranes. Biophys. J. 2006;90:4104–4118. doi: 10.1529/biophysj.105.062778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


