Abstract
Linear double-stranded DNA (dsDNA) viruses package their genomes into preformed protein shells via nanomotors using ATP as an energy source. The central hub of the bacteriophage φ29 DNA-packaging motor contains a 3.6-nm channel for dsDNA to enter during packaging and to exit during infection. The negatively charged interior channel wall is decorated with a total of 48 positively charged lysine residues displayed as four 12-lysine rings from the 12 gp10 subunits that enclose the channel. The standard notion derived from many models is that these uniquely arranged, positively charged rings play active roles in DNA translocation through the channel. In this study, we tested this prevailing view by examining the effect of mutating these basic lysines to alanines, and assessing the impact of altering the pH environment. Unexpectedly, mutating these basic lysine residues or changing the pH to 4 or 10, which could alter the charge of lysines, did not measurably impair DNA translocation or affect the one-way traffic property of the channel. The results support our recent findings regarding the dsDNA packaging mechanism known as the “push through a one-way valve”.
Introduction
A key step in the life cycle of tailed double-stranded DNA (dsDNA) viruses is the packaging of their genomes into preformed protein shells, termed procapsids, via ATP-driven nanomotors (1, 2, 3, 4, 5, 6). This has been observed for φ29 (7, 8, 9, 10), T4 (11, 12, 13), λ (2, 14), T3 (4, 15, 16), T7 (17), ε15 (18), P22 (19, 20), SPP1 (5), P1 (21), PRD1 (22), herpes simplex virus (23, 24), human cytomegalovirus (25, 26), and adenovirus (27). The φ29 nanomotor packages its genomic DNA into a procapsid consisting of four proteins: gp7 (the scaffold protein), gp8 (the capsid protein), gp8.5 (the fiber protein), and gp10 (the connector) (28, 29). The nanomotor is composed of three components: the connector at the vertex of procapsid (30), a packaging RNA (pRNA) hexameric ring that binds to the N-terminus of the connector (9, 31, 32, 33, 34, 35), and a ring of ATPase gp16 (1) linked by the pRNA (36, 37). The connector serves as the nucleation site for procapsid assembly and determines the shape and size of the procapsid (38, 39, 40). The φ29 DNA packaging motor, which was first assembled in vitro 25 years ago (7), is the most extensively studied motor system and one of the most powerful molecular motors constructed to date (41, 42). Elucidating the molecular mechanism of this motor may aid in the development of biomimetics and synthetic nanodevices (43, 44, 45, 46) to deliver therapeutics to targets such as the heart, eyes, and other organs in the body. As a result, the φ29 DNA packaging motor has attracted great attention from scientists in the fields of virology, molecular biology, structural biology, nanotechnology, biophysics, biomaterials, nanomachines, RNA biochemistry, and therapeutics.
Although the protein sequences of many viral connectors share little homology, the global structures are morphologically similar (19, 30, 47, 48), suggesting that viral DNA packaging may utilize similar mechanisms. Extensive studies reveal that the structure of the φ29 connector is a cone-shaped ring surrounded by 12 subunits of gp10 (30, 48, 49, 50, 51, 52). The wider end, which is buried inside the procapsid, is 13.6 nm, and the narrower, protruding end is 6.8 nm. The narrowest part of the connector channel is 3.6 nm, which allows dsDNA to enter during DNA packaging and to exit during host infection. Because the highly negative charged channel wall is decorated with four lysine rings (K200, K209, K234, and K235; Fig. 1) scattered inside the channel, these basic lysine residues were proposed to interact with the negative phosphate backbone of dsDNA during DNA packaging (30). An active role for the connector and a potential function of the lysine residues in DNA packaging have also been proposed in many DNA-packaging models (30, 51, 53). Here, we focused on dissecting the role of these interior lysine residues in φ29 DNA packaging. The positively charged lysine residues were substituted with neutral residues and the pH environment was changed to either neutralize or enhance the charges. Contrary to expectations (30, 51, 53), neither the change in pH nor the exchange of the lysine residues affected the genome packaging by the active ATP-driven motor or the translocation of the dsDNA driven by electric current through membrane-embedded connectors.
Figure 1.
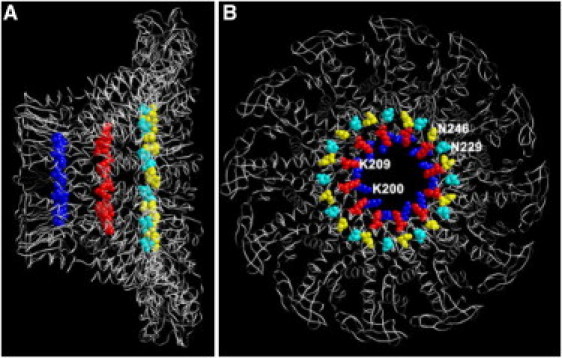
Structure of the φ29 DNA packaging motor connector, showing the location of the four lysine rings. Side view (A) and top view (B) of the φ29 connector, showing K200 (blue), K209 (red), and the border of tunnel loop N229 (cyan) and N246 (yellow). The lysine residues K234 and K235 were located in the tunnel loop of the φ29 connector, which was missing in the crystal structure, so only the boundary of loop N229 and N246 is labeled here.
Materials and Methods
Materials
The phospholipid 1,2-diphytanoyl-sn-glycerol-3-phosphocholine (DPhPC; Avanti Polar Lipids, Alabaster, AL), n-decane (Fisher Scientific, Pittsburg, PA), and chloroform (TEDIA) were used as instructed by the vendor. All other reagents were obtained from Sigma (St. Louis, MO) if not otherwise specified.
Construction of plasmids for expression of procapsids and connectors
We constructed mutant procapsids bearing the mutant connectors K200A, K209A, and K234A by mutating the DNA sequence of plasmid pARgp7-8-8.5-10 using the Stratagene (Santa Clara, CA) QuickChange site-directed mutagenesis kit with appropriate primers.
We employed a similar approach to construct the mutant connectors K200A, K209A, and K234A, using the plasmid C-His-gp10 expressing the connector with a His-tag at its C-terminus.
Expression and purification of mutant procapsids
The mutant plasmid DNA was transformed into Escherichia coli strain HMS174(DE3). The procapsid was expressed and induced with isopropyl-β-D-thio-galactoside (IPTG). Bacterial cells were harvested, resuspended in MMS buffer (2 mM sodium azide, 5 mM maleic acid, 100 mM NaCl, 15 mM MgCl2, pH 5.6), and then disrupted by passage through a French press. The cell lysate was clarified by centrifugation. The procapsid was isolated by applying the clarified cell lysate in 10–30% sucrose gradient in MMS buffer by centrifugation at 25,000 × rpm for 5 h in a Beckman SW28 motor. The procapsid was concentrated by spinning down the procapsid fractions and then resuspended in TMS (100 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 100 mM NaCl) buffer.
Expression and purification of connectors
Mutant connector plasmid was transformed into the E. coli strain HMS174(DE3). The connector was expressed and induced with IPTG. The His-tagged connector was purified by affinity chromatography (Novagen, Philadelphia, PA). Cells were resuspended with buffer A (15% glycerol, 500 mM NaCl, 20 mM Tris-HCl, pH 8.0) containing 5 mM imidazole, and the clarified cell lysate was loaded to a His-Bind resin column and washed with buffer A containing 60 mM imidazole. The His-tag connectors were eluted by buffer A containing 200 mM imidazole.
DNA packaging and in vitro virion assembly assay
The procedures used for DNA packaging and in vitro virion assembly assay are described elsewhere (6).
Transmission electron microscopy imaging
Transmission electron microscopy (TEM) grids (Electron Microscopy Sciences, Hatfield, PA) were coated with 400 mesh formvar and carbon, and glow-discharged before use to increase their hydrophilicity. The purified procapsid protein was diluted in TMS when necessary and negatively stained with 2% uranyl acetate. Samples were imaged on a Philips CM-100 transmission electron microscope with a 200- and 45-μm condenser and objective aperture, respectively, operated at 80 kV.
Sucrose gradient sedimentation
The procapsid samples were layered on top of a 5–20% (w/v) linear sucrose gradient in TMS and centrifuged in a Beckman SW55 rotor at 35,000 rpm for 35 min. Eleven drops per gradient fractions were collected from the bottom, and the fractions were then analyzed by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Preparation of lipid vesicles containing the reengineered connector
To prepare the gp10 reconstituted lipid vesicles (45, 46, 57, 58), lipid stock solutions of DPhPC or DOPC in chloroform were syringed into a glass vial, dried under a stream of nitrogen, and placed under vacuum overnight. To rehydrate the lipid film, 2 ml of the gp10 connector solution containing 200–300 mM sucrose were used to bud off vesicles into the solution. The lipid/connector solution was then extruded through a polycarbonate membrane filter (100 μm or 400 μm), which resulted in the formation of unilamellar lipid vesicles/connector complexes. A final molar ratio of lipid/connector was set to 4000:1 to 16000:1 for single-channel conduction assays.
Insertion of the connectors into the planar bilayer lipid membrane
We inserted the connectors into a bilayer membrane (BLM) as described previously (45, 46, 57). A planar BLM was formed in a standard BLM cell. An aperture of 100 μm or 200 μm diameter in a thin Teflon partition separated the cell into cis and trans compartments. A 0.5-μl 3% (w/v) DPhPC n-decane solution was used to prepaint the aperture twice before the cell was filled with conducting buffers (10 mM Tris/pH 7.9 or 5 mM HEPES/pH 7.9, with varied concentrations of NaCl or KCl). Directly after the planar BLM formed spontaneously, 1–2 μl of liposomes were added to the cis compartment.
Electrophysiological measurements
Current measurements were obtained with the use of two Ag/AgCl electrodes connected directly to the head-stage of an Axopatch 200B patch-clamp amplifier with an Axon DigiData 1322A analog-digital converter (Axon Instruments, Sunnyvale, CA) to detect and record the current traces across the bilayer lipid membrane. All collected data were filtered at a frequency of 2 kHz and acquired at 500-μs intervals if not otherwise specified. The PClamp 9.1 software (Axon Instruments) was used to collect the data, and the Clampfit and Origin softwares were used for data analysis (45, 46, 57, 58).
Results
Assessment of procapsids bearing mutant connectors with lysine residues 200, 209, and 234 alterations
It was previously proposed that the positive-charged lysine residues in the connector channel may interact with the negative-charged phosphate backbone of dsDNA during DNA translocation (30). To evaluate the validity of this hypothesis, we substituted lysines K200, K209, and K234 with the neutral amino acid alanine. Procapsids bearing the mutant connectors K200A, K209A, and K234A were assembled from products of coexpressed genes, including mutant gp10. The effects of the mutation on procapsid assembly and DNA packaging were tested. The morphology of the resulting mutant procapsids was examined by EM. The procapsids bearing mutant connector K234A exhibited a morphology similar to that of the wild-type (WT), implying that the change of this lysine did not affect procapsid assembly. The shape and size of most of the procapsids bearing mutant connectors K200A and K209A were also similar to those of the WT, with a prolate shape observed in side-view and a circular shape observed in the end-on view (Fig. 2). However, for these two mutants, some round and irregular particles of smaller sizes than the normal procapsids were also observed, in similarity to the gp8/gp7 connector-free particles reported previously (38, 39). These may represent particles with only capsids and scaffold proteins, but without the connectors. This notion is supported by results from an analysis of the components of these procapsids by 10% SDS-PAGE (Fig. 3). When the capsid protein was kept at a similar quantity (Fig. 3 A), procapsids bearing mutant connectors K200A and K209A had much less gp10 than the WT procapsid. The results were further substantiated by sucrose gradient analysis. The WT procapsid peak appeared in fractions 13 and 15 (Fig. 3 C), with the gp10 band clearly observed with SDS-PAGE. However, weaker gp10 bands could be observed in the peak fractions from the procapsids possessing mutant connectors K200A and K209A (Fig. 3, D and E). Therefore, mutant connectors K200A and K209A may not fold or assemble well, and/or may not interact properly with the major capsid protein, which in turn would affect the nucleation of the connector in procapsid assembly (39, 40, 54, 55, 56).
Figure 2.
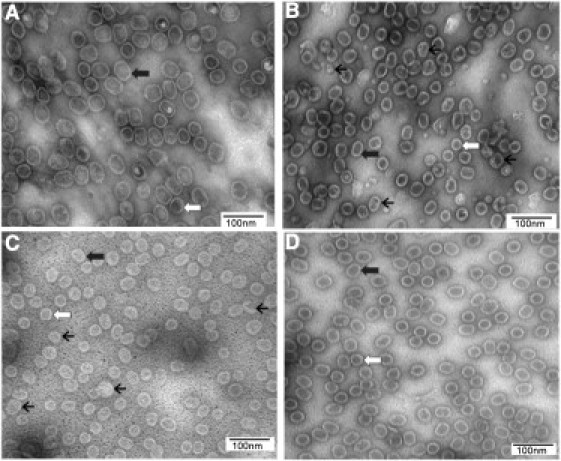
TEM micrographs of procapsids. Procapsids bearing WT (A) and mutant connectors K200A (B), K209A (C), and K234A (D) are shown. White arrow, end-on-view; thick black arrow, side view; thin black arrow, irregular particles.
Figure 3.
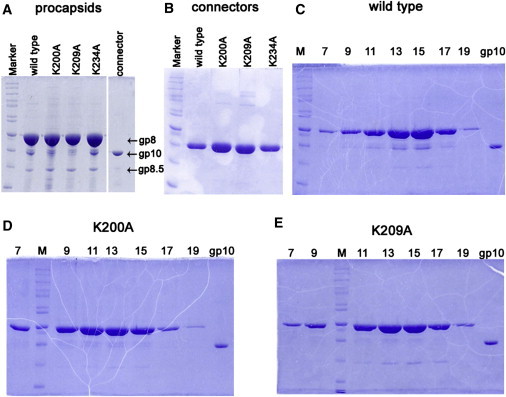
10% SDS-PAGE gel showing component profiles of different procapsids and connectors. (A) Procapsids. (B) Connectors. (C–E) Fractions 7–19 of 5–20% sucrose gradient sedimentation to demonstrate the sedimentation rate and component for WT procapsid (C), procapsid bearing mutant connector K200A (D), and procapsid bearing mutant connector K209A (E). M, molecular weight marker; gp10, the connector protein control.
Examination of the connectors' surface charges and hydrophobic profiles
The connector channel has a central hydrophobic domain sandwiched between two hydrophilic domains (45, 46, 57, 58), facilitating the incorporation of the connector channel into artificial lipid membranes (45, 46, 57, 58). Under an applied transmembrane voltage, a discrete stepwise increase of the current was observed as the connectors were incorporated into lipid membranes. Mutant connectors K200A and K209A (Fig. 3 B) were competent for insertion into the lipid membrane, but they were more difficult than the WT connector for membrane incorporation. Connectors K200A and K209A showed incorporation rates of 2 and 0.3 connectors per minute, respectively, compared with 15 connectors per minute for the WT connector (Table 1). These results indicate that compared with the WT connector, the changes of K200A and K209A alter the surface charge of the connector to some extent, impairing interaction with the lipid membrane. The other mutant connector, K234A (Fig. 3 B), had an incorporation rate of 14 connectors per minute (Table 1) and could be incorporated into the lipid membrane as easily as the WT connector, suggesting that the K234 mutation did not alter the connector's surface charges.
Table 1.
Comparison of the channel properties of connectors
| Connector | Rate (connector/min) in lipid membrane insertion | Conductance (nS) | Channel size (nm) |
|---|---|---|---|
| WT | 15 ± 8 (N = 3) | 4.3 ± 0.2 (N = 46) | 3.6 |
| K234A | 14 ± 9 (N = 5) | 2.9 ± 0.3 (N = 100) | 3.0 |
| 4.2 ± 0.2 (N = 19) | 3.6 | ||
| K200A | 2 ± 1 (N = 3) | 4.7 ± 0.5 (N = 125) | 3.7 |
| K209A | 0.3 ± 0.2 (N = 3) | 4.3 ± 0.6 (N = 34) | 3.6 |
Evaluation of the connectors' channel size by single-pore conductance assays
The channel sizes of the lysine mutant connectors can be deduced from single-channel conductance assays. Upon insertion of the connector into lipid membranes, discrete current jumps can be observed under an applied voltage representing the open-pore current amplitude. When nonelectrolyte polymers, such as DNA molecules, pass through the channel, the capacity of the electrolyte ion passage is reduced, resulting in transient current blockade events. Because the diameter of dsDNA is 2 nm and the size of the narrowest region of the WT connector channel is 3.6 nm, one can use the ratio of the cross-sectional areas (represented by the ratio of the open-pore current and the current during the DNA blockade) as a parameter to estimate the channel size at the narrowest point. As shown in Fig. 4, the conductance measurements revealed the diameters of mutant connectors K200A and K209A to be 3.7 nm and 3.6 nm, respectively, in similarity to the WT connector (Figs. 4, F and G, and 5; Table 1). Of interest, for mutant connector K234A, two different pore sizes were observed: 15% of the channels had a diameter similar to that of the WT connector (3.6 nm; Fig. 4 H), whereas ∼85% had a smaller diameter (∼3.0 nm; Figs. 4 H and 5; Table 1).
Figure 4.
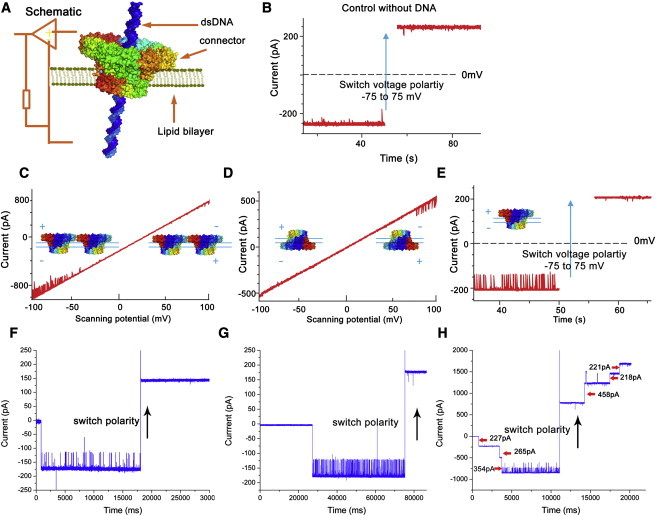
DNA translocation through the connector channel demonstrated as current blockage. (A) Schematic of the electrophysiological setup of membrane-embedded connector under an applied voltage. (B) Control experiment in the absence of DNA with the WT connector at −75 mV. (C and D) One-way traffic in DNA translocation through the WT connector channel under a ramping potential from −100 mV to +100 mV (2.2 mV/s) with different connector orientation. (E) One-way traffic in DNA translocation through the WT connector (as control) by switching polarity at −75 mV. (F–H) One-way traffic in DNA translocation through the K200A verified by switching polarity at −40 mV (F), K209A verified by switching polarity at −40 mV (G), and K234A verified by switching polarity at −75 mV (H). DNA was added in both chambers. K234A (H) had two sizes of pores. The larger one was similar to the WT connector, and the smaller one did not allow DNA to pass.
Figure 5.
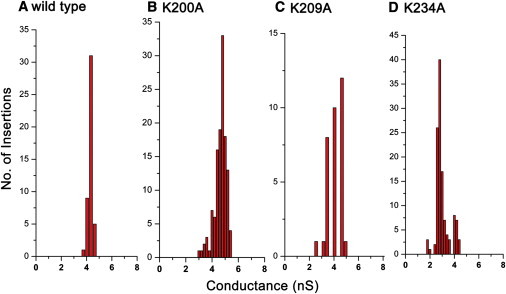
Histogram of conductance measurements for single connector insertions: (A) WT connector, (B) K200A, (C) K209A, and (D) K234A. Measurements were made under specific voltages of −75 mV (A) and −40 mV (B–D) with 1 M NaCl, 5 mM Tris, pH 7.8.
Measurement of DNA translocation through the connector channel by a single-pore conductance assay
To measure DNA translocation, we inserted each connector into a lipid BLM. Single-channel conductance assays using the connectors showed a robust ability to reliably and precisely assess the transportation of ions and DNA (45, 46, 57, 58). A standard horizontal BLM chamber containing a thin Teflon film with an aperture of 100–200 μm diameter was used as a partition to separate the chamber into two compartments. A pair of Ag/AgCl electrodes were connected directly to both compartments and used to measure the current traces across the motor channel incorporated into the lipid BLM (Figure 4, Figure 5, Figure 6). Previous studies showed that when dsDNA passes through the channel, it blocks the channel, resulting in a reduction of current and a change in the current signature (Fig. 4, E–H). The current-blockage events are used as an indicator of dsDNA translocation through the channel (45, 46, 57, 58). In this study, conductance assays revealed that dsDNA readily translocated through the mutant connectors K200A and K209A (Fig. 4, F and G), and displayed a one-way DNA translocation property from the narrower end to the wider end, in similarity to the WT connector (46). The pores of connector K234A were of two different sizes. The larger pores displayed properties similar to those of the WT connector with respect to DNA translocation capacity and one-way traffic property. In contrast, the connector with the smaller pore size did not allow dsDNA to translocate (Fig. 4 H).
Figure 6.
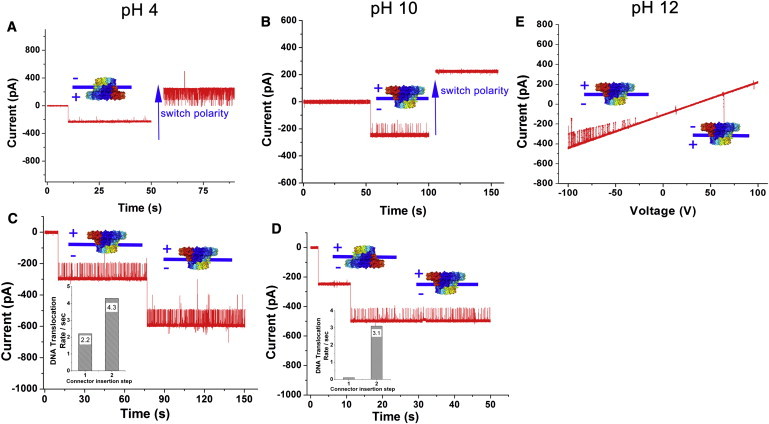
DsDNA translocation and the one-way traffic property of connectors at different pH values. Switching the electrode polarity at pH 4 (A) and pH 10 (B). Comparison of the DNA translocation rate at pH 4 (C) and pH 10 (D). (E) Under a ramping potential from −100 mV to +100 mV (2.2 mV/s) at pH 12.
Assessment of the pH environment in DNA translocation
Lysine has one pKa3 of 10.28 and is positively charged by protonation at neutral pH. In previous studies, we found that DNA translocation through the WT connector was not affected by extremely acid (pH 2) or basic (pH 12) environments (45, 46, 57). In this work, to determine whether the positively charged lysine residues in the channel wall interact with the negatively charged DNA backbone, we investigated DNA translocation through the WT connector under various pH environments ranging from acidic (pH 4) to basic (pH 12). At pH 10 and pH 12 (close to or higher than the pKa3 of the lysine side chain), the positive charge of the lysine would be expected to disappear. Surprisingly, however, dsDNA could pass through the WT connector and still display a one-way traffic property (Fig. 6, B, D, and E). Although in an acidic environment (pH 4) the negatively charged residues glutamate and aspartate would be expected to protonate, the DNA translocation remained unaffected (Fig. 6, A and C). These results suggest that the charge state of the lysine residues do not play a key role in DNA translocation under these in vitro conditions.
Study of DNA packaging activity and virus assembly with the ATP-driven active motor bearing lysine-mutant connectors
The DNA packaging activities and viral assembly activities of procapsids bearing lysine-mutant connectors were assessed in vitro by means of DNase protection and virion production assays. DNA packaged into the procapsid is protected from DNase degradation. In contrast to the WT procapsid, procapsids bearing mutant connector K200A or K209A could package DNA, albeit with slight reduction in activity (Fig. 7 B). The procapsid bearing mutant connector K234A displayed activity similar to that of the WT procapsid (Fig. 7 B).
Figure 7.
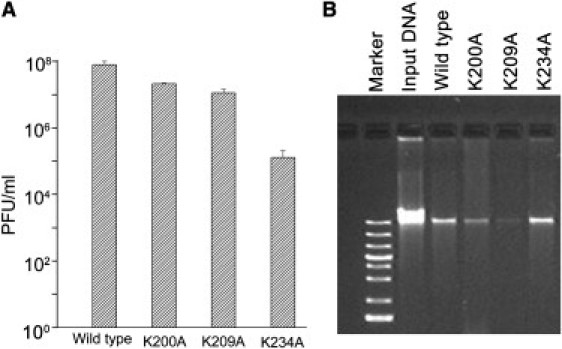
Virus assembly (A) and DNA packaging (B) activity of procapsids bearing different connectors.
After DNA packaging, the neck and tail proteins were incubated with a DNA packaging reaction to produce active viruses (6). Although the DNA packaging activity of the procapsid containing mutant connector K234A was similar to the WT procapsid, surprisingly, its phage assembly activity was ∼600-fold lower than that of the WT procapsid (Fig. 7 A). This result suggests that this particular mutation has adverse effects on viral production, which may involve the DNA ejection step (i.e., ejection of genomic DNA into the host cell). On the other hand, the K200A and K209A mutations caused slight reductions (by ∼4- and ∼7-fold, respectively) in the virus assembly activities(Fig. 7 A). A comparison of the procapsids' specific activities (i.e., the yield of viral production divided by the number of procapsids) revealed that the reduction of DNA packaging activity and virus assembly activity of procapsids bearing mutant connector K200A or K209A was due to the presence of abnormal procapsids that lacked connectors (Figure 2, Figure 3) (38, 39). These results support the conclusion that these lysine residues were not essential for DNA translocation through the channel embedded in the procapsid.
Discussion
The packaging of DNA has been extensively investigated in a number of viral systems, but the mechanism of motor action remains to be determined. Many packaging models have been proposed, including 1), DNA compression and relaxation (3, 59, 60); 2), force of osmotic pressure (61); 3), ratchet mechanism (62); 4), Brownian motion (63); 5), fivefold/sixfold mismatch connector rotating thread (64); 6), supercoiled DNA wrapping (53); 7), the sequential action of motor components (65, 66); 8), electro-dipole within the central channel (30); 9), the connector contraction hypothesis (4); and 10), the connector rotation model (30). However, none of these models are conclusively supported by experimental data. Some of these models have been validated in one viral system but refuted in other systems. The fivefold/sixfold mismatch connector rotation model, first proposed in 1978, was in vogue because it could account for the continual work of the motor (64). A mechanical motor prototype was also prevalent (52) based on the mismatch between the fivefold symmetry of capsid at the vertex and sixfold symmetry of the connector, which is a 12-subunit dodecamer. Years later, another model was proposed in which the connector revolves during DNA packaging (30). In this model, the positively charged lysine residues inside the highly negatively charged connector channel were thought to interact favorably with the negatively charged phosphate backbone of DNA in the process of driving the DNA into the procapsid. However, both the proposed popular connector rotation model (30) and the fivefold/sixfold mismatch connector rotating thread model (52) were invalidated by single-molecule studies using fluorescent labeling of the connector (67) and connector cross-linking experiments (68, 69).
In this study, we have shown that the interior channel lysine residues K200, K209, and K234 are not essential for φ29 DNA translocation. Our results also reveal that the K234A alteration did not affect the mutant connector's role as a nucleation site to assemble the procapsid or alter the DNA packaging activity of the procapsid. However, of interest, the virus assembly activity of this mutant procapsid was decreased by ∼600-fold. This is an indication that although mutation of this lysine ring did not overtly impair the entry of DNA, it did disrupt the exit of DNA during the host infection process. This result concurs with our recent finding that channel gating plays a role in regulating DNA traffic (58). These experimental data are actually consistent with the recent finding (70) that the loop containing lysine K234 and K235 has no active role in the translocation of DNA. Rather, it appears to retain the packaged DNA in the procapsid. Deletion of this loop leads to DNA leaking out before virion assembly occurs (70), and low infectivity.
The conclusion that channel lysine rings do not play an active role in DNA translocation is also supported by our pH manipulations. Translocation was retained even in high pH (basic) environments (Fig. 6), where the lysine residues would exist in a deprotonated state. Furthermore, the DNA translocation was also not affected at low pH, a state that would be predicted to reduce the overall net negative charge of the channel (Fig. 6). Recently, however, we demonstrated that the connector channel operates as a one-way valve that allows dsDNA to travel in only one direction from the narrow external end (N-terminus) toward the wider internal end (C-terminus) of the channel in DNA packaging (46). Here we found that this one-way traffic property was retained within the full spectrum of pH values. This suggests that the charge states of lysine residues are not essential for DNA translocation. The in vitro observation of one-way DNA translocation through the membrane-inserted connector has not been tested in the context of the virion or in vivo. Recently, Grimes et al. (70) reported that DNA exited procapsids that bore connectors lacking the channel loops. It is speculated that the channel loops may be involved in the one-way valve function.
As documented in many phage systems (10, 43, 71), the ATPase, or terminase, plays a key role in transporting the DNA into the procapsid. What, then, is the role of the connector in DNA translocation? The results obtained here are consistent with the “push through a one-way valve” model (Fig. 8) recently proposed by Schwartz and co-workers (72), which suggests separate roles for the connector and ATPase gp16. In this model, dsDNA packaging occurs through a combination of two separate tasks. In the first task, an active pushing mechanism is provided by the ATPase, gp16 (1, 72), bound to either pRNA (the fulcrum) (36, 37) (Fig. 8) of φ29, or, in the case of other dsDNA phage systems, the large subunit of the two terminases grips the motor component to push the dsDNA coupled with ATP hydrolysis. In the second task, the direction of DNA migration is controlled by a one-way valve action (46). This mechanism is consistent with and supported by the recent finding in a T4 phage system that the dsDNA was crunched and compressed when DNA entry was blocked at the front end (3, 11, 12, 60). Although the authors concluded that the force for compression was due to the torsion force from the coiled DNA at the external end, this does not contradict our results because the compression can also be attributed to ATPase pushing. In fact, their result supports our report in 1998 that the function of ATPase gp16 was similar to that of the DNA-tracking motors of the AAA+ (ATPase Associated with diverse cellular Activities) family, which twist or rotate the dsDNA (32). Furthermore, it was reported that in the T4 system, both ends of the packaged DNA were outside the procapsid (11, 12). If the motor generates a pulling rather than a pushing function, it is difficult to envisage how the initiation of DNA translocation could begin when both ends are located outside the capsid (11, 12). The “push through a one-way valve” model is also consistent with the supercoiled DNA wrapping model (53), which shows the coiling of DNA, and the ratchet model (62), which specifies the direction control of DNA translocation.
Figure 8.
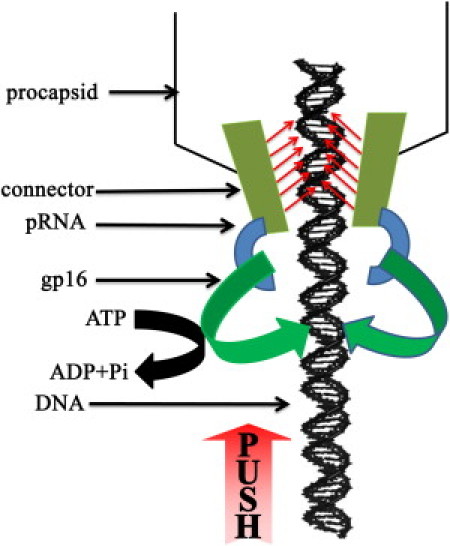
New model of the “pushing through a one-way valve” mechanism of the φ29 DNA packaging motor. ATPase gp16 pushes DNA into the procapsid using the energy from ATP hydrolysis. The connector channel exercises a unidirectional one-way traffic control to prevent DNA from sliding out during DNA packaging.
Acknowledgments
We thank Jose Carrascosa for inspiring communications concerning the connector structure and function, Daniel Binzel and Zhanxi Hao for proofreading, and Dr. Eric Smith for insightful comments on the manuscript.
P.G. is a cofounder of Kylin Therapeutics, Inc., and Biomotor and Nucleic Acid Nanotechnology Development Corp., Ltd.
This work was supported by National Institutes of Health grant EB012135.
Editor: Robert Nakamoto.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Guo P., Peterson C., Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage ϕ29. J. Mol. Biol. 1987;197:229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- 2.Hwang Y., Catalano C.E., Feiss M. Kinetic and mutational dissection of the two ATPase activities of terminase, the DNA packaging enzyme of bacteriophage χ. Biochemistry. 1996;35:2796–2803. doi: 10.1021/bi952322z. [DOI] [PubMed] [Google Scholar]
- 3.Sabanayagam C.R., Oram M., et al. Black L.W. Viral DNA packaging studied by fluorescence correlation spectroscopy. Biophys. J. 2007;93:L17–L19. doi: 10.1529/biophysj.107.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morita M., Tasaka M., Fujisawa H. Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme: importance of the C-terminal region in prohead binding. J. Mol. Biol. 1995;245:635–644. doi: 10.1006/jmbi.1994.0052. [DOI] [PubMed] [Google Scholar]
- 5.Oliveira L., Cuervo A., Tavares P. Direct interaction of the bacteriophage SPP1 packaging ATPase with the portal protein. J. Biol. Chem. 2010;285:7366–7373. doi: 10.1074/jbc.M109.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee C.S., Guo P. In vitro assembly of infectious virions of double-stranded DNA phage ϕ29 from cloned gene products and synthetic nucleic acids. J. Virol. 1995;69:5018–5023. doi: 10.1128/jvi.69.8.5018-5023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo P., Grimes S., Anderson D. A defined system for in vitro packaging of DNA-gp3 of the Bacillus subtilis bacteriophage φ29. Proc. Natl. Acad. Sci. USA. 1986;83:3505–3509. doi: 10.1073/pnas.83.10.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang L.P., Guo P. Use of acetone to attain highly active and soluble DNA packaging protein Gp16 of φ29 for ATPase assay. Virology. 2003;312:449–457. doi: 10.1016/s0042-6822(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 9.Guo P.X., Erickson S., Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage φ29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H., Schwartz C., et al. Guo P. AAA+ hexameric viral DNA packaging motor using a “push through a one-way valve” mechanism. Adv. Virus Res. 2011 doi: 10.1016/B978-0-12-394438-2.00009-8. In press. [DOI] [PubMed] [Google Scholar]
- 11.Ray K., Sabanayagam C.R., et al. Black L.W. DNA crunching by a viral packaging motor: compression of a procapsid-portal stalled Y-DNA substrate. Virology. 2010;398:224–232. doi: 10.1016/j.virol.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray K., Ma J., et al. Black L.W. Single-molecule and FRET fluorescence correlation spectroscopy analyses of phage DNA packaging: colocalization of packaged phage T4 DNA ends within the capsid. J. Mol. Biol. 2010;395:1102–1113. doi: 10.1016/j.jmb.2009.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray K., Oram M., et al. Black L.W. Portal control of viral prohead expansion and DNA packaging. Virology. 2009;391:44–50. doi: 10.1016/j.virol.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Q., Catalano C.E., Maluf N.K. Kinetic analysis of the genome packaging reaction in bacteriophage λ. Biochemistry. 2009;48:10705–10715. doi: 10.1021/bi901016n. [DOI] [PubMed] [Google Scholar]
- 15.Serwer P., Wright E.T., et al. Jiang W. DNA packaging-associated hyper-capsid expansion of bacteriophage t3. J. Mol. Biol. 2010;397:361–374. doi: 10.1016/j.jmb.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang P.A., Wright E.T., et al. Jiang W. Visualization of bacteriophage T3 capsids with DNA incompletely packaged in vivo. J. Mol. Biol. 2008;384:1384–1399. doi: 10.1016/j.jmb.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerritelli M.E., Conway J.F., et al. Steven A.C. Molecular mechanisms in bacteriophage T7 procapsid assembly, maturation, and DNA containment. Adv. Protein Chem. 2003;64:301–323. doi: 10.1016/s0065-3233(03)01008-8. [DOI] [PubMed] [Google Scholar]
- 18.Jiang W., Chang J., et al. Chiu W. Structure of ε15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olia A.S., Prevelige P.E., Jr., et al. Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat. Struct. Mol. Biol. 2011;18:597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang J.T., Lander G.C., et al. Johnson J.E. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in p22. Structure. 2011;19:496–502. doi: 10.1016/j.str.2011.02.010. Erratum in Structure.2011. 19:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J., Chen C.Y., et al. Margolin W. Visualization of bacteriophage P1 infection by cryo-electron tomography of tiny Escherichia coli. Virology. 2011;417:304–311. doi: 10.1016/j.virol.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrescia N.G., Cockburn J.J., et al. Bamford J.K. Insights into assembly from structural analysis of bacteriophage PRD1. Nature. 2004;432:68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- 23.Salmon B., Baines J.D. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of U(L)15-encoded proteins with B capsids requires at least the U(L)6, U(L)17, and U(L)28 genes. J. Virol. 1998;72:3045–3050. doi: 10.1128/jvi.72.4.3045-3050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheaffer A.K., Newcomb W.W., et al. Tenney D.J. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 2001;75:687–698. doi: 10.1128/JVI.75.2.687-698.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheffczik H., Savva C.G., et al. Bogner E. The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure. Nucleic Acids Res. 2002;30:1695–1703. doi: 10.1093/nar/30.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Champier G., Hantz S., et al. Alain S. New functional domains of human cytomegalovirus pUL89 predicted by sequence analysis and three-dimensional modelling of the catalytic site DEXDc. Antivir. Ther. (Lond.) 2007;12:217–232. [PubMed] [Google Scholar]
- 27.Ostapchuk P., Hearing P. Control of adenovirus packaging. J. Cell. Biochem. 2005;96:25–35. doi: 10.1002/jcb.20523. [DOI] [PubMed] [Google Scholar]
- 28.Peterson C., Simon M., et al. Anderson D. Composition and mass of the bacteriophage φ29 prohead and virion. J. Struct. Biol. 2001;135:18–25. doi: 10.1006/jsbi.2001.4375. [DOI] [PubMed] [Google Scholar]
- 29.Meijer W.J., Horcajadas J.A., Salas M. φ29 family of phages. Microbiol. Mol. Biol. Rev. 2001;65:261–287. doi: 10.1128/MMBR.65.2.261-287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guasch A., Pous J., et al. Coll M. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage φ29 connector particle. J. Mol. Biol. 2002;315:663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- 31.Shu D., Zhang H., et al. Guo P. Counting of six pRNAs of φ29 DNA-packaging motor with customized single-molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo P., Zhang C., et al. Trottier M. Inter-RNA interaction of phage φ29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 33.Chen C., Zhang C., Guo P. Sequence requirement for hand-in-hand interaction in formation of RNA dimers and hexamers to gear φ29 DNA translocation motor. RNA. 1999;5:805–818. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao F., Moll W.D., et al. Guo P. Binding of pRNA to the N-terminal 14 amino acids of connector protein of bacteriophage φ29. Nucleic Acids Res. 2005;33:2640–2649. doi: 10.1093/nar/gki554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atz R., Ma S., et al. Grimes S. Alanine scanning and Fe-BABE probing of the bacteriophage ø29 prohead RNA-connector interaction. J. Mol. Biol. 2007;369:239–248. doi: 10.1016/j.jmb.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee T.J., Guo P. Interaction of gp16 with pRNA and DNA for genome packaging by the motor of bacterial virus φ29. J. Mol. Biol. 2006;356:589–599. doi: 10.1016/j.jmb.2005.10.045. [DOI] [PubMed] [Google Scholar]
- 37.Zhao W., Morais M.C., et al. Grimes S. Role of the CCA bulge of prohead RNA of bacteriophage ø29 in DNA packaging. J. Mol. Biol. 2008;383:520–528. doi: 10.1016/j.jmb.2008.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo P.X., Erickson S., et al. Anderson D. Regulation of the phage ϕ29 prohead shape and size by the portal vertex. Virology. 1991;183:366–373. doi: 10.1016/0042-6822(91)90149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee C.S., Guo P. Sequential interactions of structural proteins in phage φ29 procapsid assembly. J. Virol. 1995;69:5024–5032. doi: 10.1128/jvi.69.8.5024-5032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu C.Y., Prevelige P.E., Jr. In vitro incorporation of the phage φ29 connector complex. Virology. 2009;394:149–153. doi: 10.1016/j.virol.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aathavan K., Politzer A.T., et al. Bustamante C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature. 2009;461:669–673. doi: 10.1038/nature08443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith D.E., Tans S.J., et al. Bustamante C. The bacteriophage straight φ29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 43.Guo P.X., Lee T.J. Viral nanomotors for packaging of dsDNA and dsRNA. Mol. Microbiol. 2007;64:886–903. doi: 10.1111/j.1365-2958.2007.05706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee T.J., Schwartz C., Guo P. Construction of bacteriophage φ29 DNA packaging motor and its applications in nanotechnology and therapy. Ann. Biomed. Eng. 2009;37:2064–2081. doi: 10.1007/s10439-009-9723-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wendell D., Jing P., et al. Guo P. Translocation of double-stranded DNA through membrane-adapted φ29 motor protein nanopores. Nat. Nanotechnol. 2009;4:765–772. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jing P., Haque F., et al. Guo P. One-way traffic of a viral motor channel for double-stranded DNA translocation. Nano Lett. 2010;10:3620–3627. doi: 10.1021/nl101939e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lebedev A.A., Krause M.H., et al. Antson A.A. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simpson A.A., Leiman P.G., et al. Rossmann M.G. Structure determination of the head-tail connector of bacteriophage φ29. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1260–1269. doi: 10.1107/s0907444901010435. [DOI] [PubMed] [Google Scholar]
- 49.Jiménez J., Santisteban A., et al. Carrascosa J.L. Computer graphic display method for visualizing three-dimensional biological structures. Science. 1986;232:1113–1115. doi: 10.1126/science.3754654. [DOI] [PubMed] [Google Scholar]
- 50.Guasch A., Pous J., et al. Coll M. Crystallographic analysis reveals the 12-fold symmetry of the bacteriophage φ29 connector particle. J. Mol. Biol. 1998;281:219–225. doi: 10.1006/jmbi.1998.1928. [DOI] [PubMed] [Google Scholar]
- 51.Simpson A.A., Tao Y., et al. Rossmann M.G. Structure of the bacteriophage φ29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Badasso M.O., Leiman P.G., et al. Anderson D. Purification, crystallization and initial X-ray analysis of the head-tail connector of bacteriophage φ29. Acta Crystallogr. D Biol. Crystallogr. 2000;56:1187–1190. doi: 10.1107/s0907444900009239. [DOI] [PubMed] [Google Scholar]
- 53.Grimes S., Anderson D. The bacteriophage φ29 packaging proteins supercoil the DNA ends. J. Mol. Biol. 1997;266:901–914. doi: 10.1006/jmbi.1996.0843. [DOI] [PubMed] [Google Scholar]
- 54.Moore S.D., Prevelige P.E., Jr. Structural transformations accompanying the assembly of bacteriophage P22 portal protein rings in vitro. J. Biol. Chem. 2001;276:6779–6788. doi: 10.1074/jbc.M007702200. [DOI] [PubMed] [Google Scholar]
- 55.Tuma R., Tsuruta H., et al. Prevelige P.E. Detection of intermediates and kinetic control during assembly of bacteriophage P22 procapsid. J. Mol. Biol. 2008;381:1395–1406. doi: 10.1016/j.jmb.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodríguez-Casado A., Moore S.D., et al. Thomas G.J., Jr. Structure of bacteriophage P22 portal protein in relation to assembly: investigation by Raman spectroscopy. Biochemistry. 2001;40:13583–13591. doi: 10.1021/bi0110488. [DOI] [PubMed] [Google Scholar]
- 57.Jing P., Haque F., et al. Guo P. Robust properties of membrane-embedded connector channel of bacterial virus φ29 DNA packaging motor. Mol. Biosyst. 2010;6:1844–1852. doi: 10.1039/c003010d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geng J., Fang H., et al. Guo P. Three reversible and controllable discrete steps of channel gating of a viral DNA packaging motor. Biomaterials. 2011;32:8234–8242. doi: 10.1016/j.biomaterials.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khan S.A., Hayes S.J., et al. Serwer P. Specific single-stranded breaks in mature bacteriophage T7 DNA. Virology. 1995;211:329–331. doi: 10.1006/viro.1995.1411. [DOI] [PubMed] [Google Scholar]
- 60.Oram M., Sabanayagam C., Black L.W. Modulation of the packaging reaction of bacteriophage t4 terminase by DNA structure. J. Mol. Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serwer P. The source of energy for bacteriophage DNA packaging: an osmotic pump explains the data. Biopolymers. 1988;27:165–169. doi: 10.1002/bip.360270113. [DOI] [PubMed] [Google Scholar]
- 62.Fujisawa H., Morita M. Phage DNA packaging. Genes Cells. 1997;2:537–545. doi: 10.1046/j.1365-2443.1997.1450343.x. [DOI] [PubMed] [Google Scholar]
- 63.Astumian R.D. Thermodynamics and kinetics of a Brownian motor. Science. 1997;276:917–922. doi: 10.1126/science.276.5314.917. [DOI] [PubMed] [Google Scholar]
- 64.Hendrix R.W. Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl. Acad. Sci. USA. 1978;75:4779–4783. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen C., Guo P. Sequential action of six virus-encoded DNA-packaging RNAs during phage φ29 genomic DNA translocation. J. Virol. 1997;71:3864–3871. doi: 10.1128/jvi.71.5.3864-3871.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moffitt J.R., Chemla Y.R., et al. Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hugel T., Michaelis J., et al. Bustamante C. Experimental test of connector rotation during DNA packaging into bacteriophage φ29 capsids. PLoS Biol. 2007;5:e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baumann R.G., Mullaney J., Black L.W. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol. Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- 69.Maluf N.K., Feiss M. Virus DNA translocation: progress towards a first ascent of mount pretty difficult. Mol. Microbiol. 2006;61:1–4. doi: 10.1111/j.1365-2958.2006.05214.x. [DOI] [PubMed] [Google Scholar]
- 70.Grimes S., Ma S., et al. Jardine P.J. Role of φ29 connector channel loops in late-stage DNA packaging. J. Mol. Biol. 2011;410:50–59. doi: 10.1016/j.jmb.2011.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Casjens S.R. The DNA-packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 72.Schwartz C., Fang H., et al. Guo P. Sequential action of ATPase, ATP, ADP, Pi and dsDNA in procapsid-free system to enlighten mechanism in viral dsDNA packaging. Nucleic Acids Res. 2011;Nov. 22 doi: 10.1093/nar/gkr841. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]


