Abstract
The incidence of methicillin-resistant Staphylococcus aureus (MRSA) has been increasing in many Canadian hospitals over the past few years. Some strains may be considered ‘epidemic’, in that they are clinically or epidemiologically significant, and have been identified in patients from multiple hospitals and geographic regions across the country. This paper describes phenotypic and genotypic characteristics of four epidemic MRSA strains in Canada and proposes standardized nomenclature.
Keywords: Methicillin-resistant Staphylococcus aureus
Résumé :
L’incidence des infections causées par des souches de Staphylococcus aureus méthicillino-résitantes (ou MRSA, pour methicillin-resistant Staphylococcus aureus) est à la hausse dans de nombreux hôpitaux canadiens depuis quelques années. Certaines souches seraient même considérées « épidémiques », en ce sens qu’elles sont importantes sur les plans clinique ou épidémiologique et qu’elles ont été signalées chez des patients de plusieurs hôpitaux de différentes régions du pays. Cet article porte sur les caractéristiques de quatre souches épidémiques de MRSA sur le plan du phénotype et du génotype signalées au Canada et propose une nomenclature standardisée.
The Canadian Hospital Epidemiology Committee (CHEC) in collaboration with the Canadian Nosocomial Infection Surveillance Program (CNISP) has been tracking methicillin-resistant Staphylococcus aureus (MRSA) in sentinel Canadian hospitals since January 1995. This surveillance program has documented a significant increase in the number of patients colonized and/or infected with MRSA in each of the past four years (1,2). Some MRSA strains appear to be able to spread more rapidly or easily within hospitals, nursing homes and other health care facilities, causing outbreaks or epidemics. Molecular typing of isolates may assist hospital epidemiologists, infection control practitioners and public health personnel in tracking the epidemiology and transmission of MRSA. Accurate identification and characterization of epidemic strains are essential to be able to apply appropriate infection control and prevention measures most effectively.
In an attempt to standardize the terminology used to describe epidemic strains of MRSA prevalent in Canadian hospitals, CNISP has characterized MRSA isolates from across the country by standard phenotypic typing methods, antibiogram determination, bacteriophage typing and various molecular typing procedures such as pulsed field gel electrophoresis (PFGE). Guidelines used for designating strains as ‘epidemic’ MRSA include the following: the strain is recognized to be clinically or epidemiologically significant (eg, associated with outbreaks of infection in health care facilities) or to possess important or unique microbiological characteristics (eg, distinct colonial morphology or phenotypic properties); the strain has been identified in patients from five or more hospital sites, or from three or more geographic regions within the country; and the strain has been characterized by standardized typing methods.
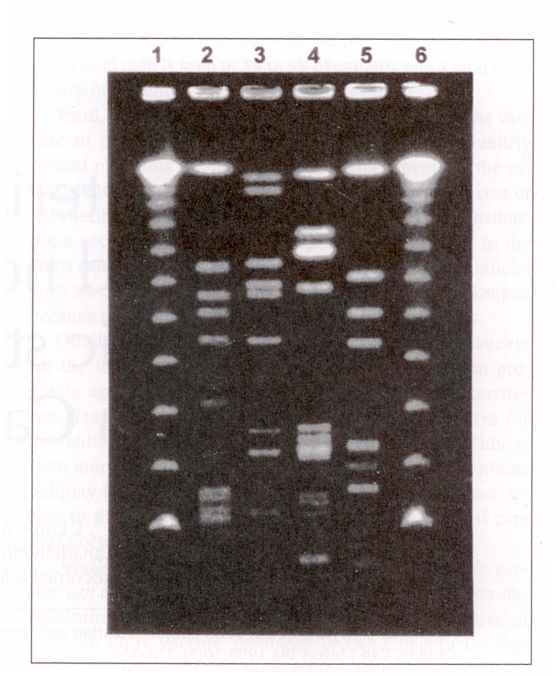
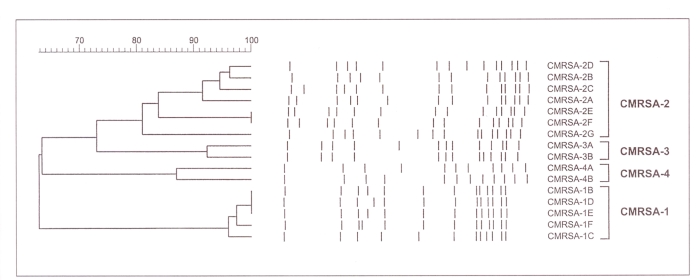
Based on these criteria, CHEC and CHISP propose that the four MRSA strains described below be designated epidemic strains of MRSA. Suggested nomenclature to be used when referring to these strains are Canadian MRSA strain (CMRSA)-1, CMRSA-2, CMRSA-3, etc. A representative PFGE DNA pattern, using standard typing methods (3) for each of these strains, is shown in Figure 1. A dendrogram of epidemic MRSA subtypes is shown in Figure 2. PFGE DNA profiles were digitized into the GelCompar computer software program (GelCompar, version 4.1, Kortrijk, Belgium) for analysis. DNA fragments on each gel were normalized using the molecular weight standards run on each gel to allow comparisons between different gels. A 2.0% tolerance was selected for use during comparisons of DNA profiles. Cluster analysis was performed by the unweighted pair group method using arithmetic averages (UPGMA), and DNA relatedness was calculated based on the Dice coefficient. Determination of genetic relatedness of isolates was based on criteria recommended by Bannerman et al (3) and Tenover et al (4). Isolates were considered to be genetically related if their macrorestriction DNA patterns differed by six bands or less and the Dice coefficient of correlation was 80% or greater.
Figure 1.
Representative DNA profiles obtained by pulsed field gel electrophoresis of SmaI digests from Canadian epidemic strains of methicillin-resistant Staphylococcus aureus (MRSA). Lanes 1 and 6, lambda DNA concatamers; lane 2, Canadian MRSA strain (CMRSA)-1; lane 3, CMRSA-2; lane 4, CMRSA-3; lane 5, CMRSA-4
Figure 2.
Dendrogram of SmaI digested DNA profiles obtained by pulsed field gel electrophoresis of Canadian epidemic methicillin-resistant Staphylococcus aureus (CMRSA) strains (CMRSA-1, CMRSA-2, CMRSA-3, CMRSA-4) with representative subtype variations
Each of these epidemic strains have been associated with significant infections, including bacteremia and pneumonia. In surveillance conducted by CNISP, approximately half of all patients with MRSA were thought to be infected with MRSA. Clinical isolates of the four epidemic strains were reported most often from skin and soft tissue infections, surgical wounds, sputum, urine, and blood cultures; strain CMRSA-4 was more likely to be isolated from sputum than were the other strains. In colonized patients, all strains were commonly recovered from nasal mucosa, skin, and rectal or perineal sites. All of these MRSA strains were typically resistant to erythromycin, clindamycin (Dalacin C, Pharmacia & Upjohn, Mississauga, Ontario) and ciprofloxacin (Cipro, Bayer Inc, Etobicoke, Ontario). CMRSA-1 and CMRSA-3 were generally resistant to trimethoprim/sulphamethoxazole, whereas CMRSA-2 and CMRSA-4 were usually susceptible to this agent. No strains with reduced susceptibility to vancomycin have been detected thus far.
CMRSA strains are available upon request from the Laboratory Centre for Disease Control, Health Canada.
DESCRIPTION OF CANADIAN EPIDEMIC STRAINS OF MRSA
CMRSA-1:
The strain designated as CMRSA-1 has been most widely found in the province of Ontario, where it accounts for the majority of strains identified in the province’s hospitals; it has commonly been referred to as the ‘Ontario epidemic strain’. However, this strain has also been isolated sporadically from patients hospitalized in British Columbia, Alberta, Manitoba and Newfoundland. In addition, the strain has been identified in the United States and some European countries. Laboratory identification of this strain is often problematic because the organism’s colonial morphology may resemble that of coagulase-negative staphylococci and because the isolate may have weak or negative test results with rapid slide agglutination assays (5,6). Detection of coagulase activity may also be weak or delayed, often requiring overnight incubation. Most CMRSA-1 isolates have been determined to be a phage-type 95, although some isolates are nontypeable and, in about 30% of isolates, other phage types have been identified (7). SmaI macrorestriction of chromosomal DNA followed by PFGE has identified at least five closely related DNA profiles (Figures 1,2).
CMRSA-2:
This strain is the most widespread of current Canadian strains, although CMRSA-1 and CMRSA-3 are more frequently encountered in some parts of the country. In the CNISP surveillance in 1995, CMRSA-2 was reported from 12 of 16 sentinel hospital sites and from all areas of the country. The strain has also been reported in the United States and Argentina. This strain has the typical colonial morphology of S aureus in culture, and it is reliably positive when tested with rapid agglutination assays. CMRSA-2 exhibits variable phage-typing results: The strain generally reacts with Group III phages, but it may also be nontypeable. PFGE with SmaI yields a variety of closely related DNA profiles, with as many as 15 variants identified to date (Figures 1,2).
CMRSA-3:
CMRSA-3 was first identified in Canada in 1993 in a patient transferred directly from a hospital in the Punjab region of India to a hospital in British Columbia. Therefore, the strain has also been referred to as the ‘Punjabi strain’. Since then, it has spread to other hospitals in British Columbia and in Manitoba (8). Sporadic cases with CMRSA-3 have also been reported in Alberta and in one patient transferred directly to an Ontario hospital from the Punjab. The colonial morphology of CMRSA-3 is typical of that for S aureus, and the strain has the expected positive reaction with rapid agglutination tests. Variable phage types are associated with CMRSA-3, although most isolates appear to react with group III phages. A representative DNA profile is shown in Figure 1.
CMRSA-4:
Most CMRSA-4 isolates have been recovered from patients in Ontario, particularly from a small number of Toronto hospitals. This strain has also been identified in Alberta, Quebec, Nova Scotia and Newfoundland. The strain has the typical colonial morphology of S aureus and gives a positive reaction with rapid agglutination assays. CMRSA-4 has been associated with many variable phage types. DNA macrorestriction with SmaI has identified two predominant closely related molecular subtypes (Figures 1,2).
SUMMARY
We hope that standardized nomenclature for identifying epidemic MRSA strains prevalent in Canadian hospitals will be helpful to physicians and infection control practitioners attempting to understand and control the spread of the organism in health care facilities. It is anticipated that, as MRSA continues to evolve in Canadian health care facilities, other strains may be recognized as ‘epidemic’; as these strains become better characterized, they may be added to those designated above. We invite laboratory physicians and infection control personnel to submit strains that may warrant characterization and designation as a Canadian epidemic strain to the Laboratory Centre for Disease Control, Health Canada, Winnipeg, Manitoba.
REFERENCES
- 1.Simor A, Ofner-Agostini M, Paton S. The Canadian Nosocomial Infection Surveillance Program: Results of the first 18 months of surveillance for methicillin-resistant Staphylococcus aureus in Canadian hospitals. Can Commun Dis Rep. 1997;23:41–5. [PubMed] [Google Scholar]
- 2.Simor AE, Ofner-Agostini M, Onno S, et al. The evolution of methicillin-resistant Staphylococcus aureus in Canadian hospitals: four years of national surveillance. Ninth Annual Scientific Meeting, The Society for Healthcare Epidemiology of America; San Francisco. April 18–20, 1999; (Abst 61) [Google Scholar]
- 3.Bannerman TL, Hancock GA, Tenover FC, Miller JM. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J Clin Microbiol. 1995;33:551–5. doi: 10.1128/jcm.33.3.551-555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGeer A, Low DE, Conly J, Campbell I, Willey B, Simor A. The rapid emergence of a new strain of MRSA in Ontario: Laboratory and infection control implications. LPTP Newsl. 1996;190:1–4. [Google Scholar]
- 6.McGeer A, Low D, Conly J, et al. Methicillin-resistant Staphylococcus aureus in Ontario. Can Commun Dis Rep. 1997;23:45–6. [PubMed] [Google Scholar]
- 7.Preston M, Borczyk A, Jamieson F. Epidemic methicillin-resistant Staphylococcus aureus strain – Ontario. Can Commun Dis Rep. 1998;24:47–9. [PubMed] [Google Scholar]
- 8.Roman RS, Smith J, Walker M, et al. Rapid geographic spread of a methicillin-resistant Staphylococcus aureus strain. Clin Infect Dis. 1997;25:698–705. doi: 10.1086/513758. [DOI] [PubMed] [Google Scholar]