The discovery of hepatitis c virus (hcv) (1) has led to the development of serological tests (2) for the detection of antibody to this newly discovered virus. hcv is the primary etiological agent of the parenterally transmitted non-A, non-B hepatitis (3). In Canada, groups at greater risk of acquiring hcv infection include intravenous drug users, hemodialysis patients, hemophiliacs and blood transfusion recipients (4). The screening of blood donors for anti-hcv has greatly reduced the incidence of post-transfusion hepatitis (5). One of the major problems with hcv infection is the development of chronic hepatitis in 50 to 60% of cases, which could lead to cirrhosis and hepatocellular carcinoma (6).
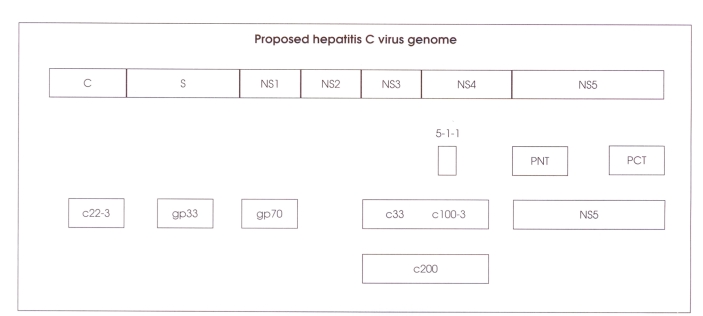
hcv is a single-stranded rna virus (9400 nucleotides) related to the family Flaviviridae (7). The 5′ end codes for core and envelope proteins followed by nonstructural proteins NS2, NS3, NS4 and NS5. There is also a noncoding region at the 5′ end (Figure 1).
Figure 1.
The entire genome is about 10,000 bases, coding for about 3000 amino acids. Only the translated portions of the genome are shown. Not shown is a 5′ conserved noncoding region of approximately 300 bases, which is located to the left of core (c) region, and a 3′ noncoding region of about 50 bases, which is at the end of NS5 region, gp Glycoprotein; PCT Polymerase carboxy terminus; PNT Polymerase amino terminus. Modified from reference 8
This paper briefly reviews the available serological and molecular diagnostic tests for the detection of hcv antibody and viral rna.
Serological assays for detection of HCV infection:
The first-generation enzyme immunoassay (eia) was developed by Chiron Corporation (California) to detect antibody against hcv in 1989 (2). Later, Abbott Laboratories (Illinois) also developed a first-generation eia test for the detection of anti-hcv under licence from Chiron Corporation. Chiron developed a recombinant immunoblot assay (riba 1.0) as a supplemental test.
The first-generation tests have become obsolete (8) because they lacked sensitivity and specificity, and were soon replaced by second- and third-generation tests. The details of the different eias are given in Table 1 and of immunoblot assays in Table 2.
TABLE 1.
Antigenic composition of second- and third-generation enzyme immunoassay tests
| Tests | Source | Date released in Canada | Antigenic composition |
|---|---|---|---|
| EIA 2.0 | Ortho | April 1991 | R; c 100–3 (NS4), c200 (NS3 + NS4), c22 (core) |
| EIA 3.0 | Ortho | July 1993 | R; c200 (NS3 + NS4), c22 (core), NS5 |
| EIA 2.0 | Abbott | April 1991 | R; c100 (NS4), 33c (NS3), HC34 (core) |
| EIA 3.0 | Abbott | September 1994 | R; c100 (NS4), HC43 (core + NS3), H34 (core), NS5 |
| IMx HCV | Abbott | September 1994 | R; Microparticles; c200 (NS3 + NS4), HC34 (core) |
| EIA | R; Probe; HC31 (NS3 + NS4), c200 (NS3 + NS4), c22 (core) | ||
| UBI HCV EIA 2.1 |
Organon | 1993 | Core (P), NS3 (R), NS4 (P), NS5 (P) |
| UBI HCV EIA 4.0 |
Organon | July 1994 | Core (P), NS3 (P), NS4 (P), NS5 (P) |
P Synthetic peptides; R Recombinant antigen
TABLE 2.
Antigenic composition of second- and third-generation immunoblot assays
| Tests | Source | Date released in Canada | Antigenic composition |
|---|---|---|---|
| RIBA 2.0 | Ortho | April 1994 | R; 5-1-1 (NS4), c100 (NS4), c22 (core), c33 (NS3) |
| RIBA 3.0 | Ortho | June 1994 | R; c33 (NS3), NS5 (P); c100 (NS4), c22 (core) |
| Matrix 1.0 | Abbott | April 1993 | R; H34 (core), NS3, NS4 (yeast), NS4 (Escherichia coli) |
| Matrix 2.0 | Abbott | Not released | R; H34 (core), NS3, NS4 (yeast), NS4 (E coli), NS5 |
| Liatek III | Organon | 1993 | C1 (P), C2 (P), E2/NS1 (P), NS3 (R), NS4 (P), NS5 (P) |
P Synthetic peptides; R Recombinant antigen
Third-generation assays are more sensitive and specific for the detection of anti-hcv, and they should be the test of choice. In Canada anti-hcv test kits are available mainly from Ortho Diagnostics (New Jersey), Abbott Laboratories and Organon Teknika (The Netherlands).
The diagnosis of hcv infection depends mainly on detecting circulating antibodies to this virus, eia 2.0 detects anti-hcv in approximately 90% of cases (9). The third-generation eia 3.0 is more sensitive than eia 2.0 and the predictive positive values are 0.52 versus 0.23 (10). eia 3.0 detects antibody earlier in the course of infection (11), five to six weeks after the onset of hepatitis in 80% of patients.
Some limitations have been observed with eia tests in that they could not differentiate among acute, chronic and past infection. In some acute cases there could be a long interval before seroconversion. In low risk groups such as blood donors, even the third-generation eias produce false positive results.
Most of the false positive eia results could be resolved by supplemental testing with immunoblot assays, riba 3.0 is more sensitive and specific than riba 2.0, and 60 to 95% of the indeterminates could be resolved by the later test (10,12–14). riba 3.0 contains two recombinant (c33[NS3], NS5) and two synthetic (c22[core], c100[NS4]) antigens. The concentration of antigens was optimized to improve sensitivity and specificity. Improvement was observed with riba 3.0 and was obtained mostly by the synthetic peptides but was not due to NS5 antigen (10). A comparative study (12) of immunoblot assays (riba 3.0, Matrix 1, LiaTek III) showed a poor correlation among the three tests.
Molecular assays for the detection of HCV infection:
The diagnosis of chronic or acute infection is still restricted due to the lack of a test for the detection of viral antigen. However, recent developments in the polymerase chain reaction (pcr) technique have made it possible to detect hcv rna in serum or plasma (15–17).
Primers specific for 5′ untranslated region (utr) are the most sensitive because this region is highly conserved (15). hcv rna could be detected many weeks before the appearance of anti-hcv (6) and in some cases this may be the only evidence of hcv infection. Nested pcr is the most sensitive technique for the detection of hcv rna. In our study (17) 67% of the riba reactive samples were positive for hcv rna by nested pcr with primers from the 5′ utr. On the other hand, 100% of riba reactive samples from high risk groups such as transfused patients, hemodialysis patients and intravenous drug users were pcr positive. A commercial pcr kit is available for the fast detection of hcv rna (Amplicor hcv) from Roche Diagnostic Systems (New Jersey). The Amplicor hcv test uses 5′ utr specific primers, the antisense primer is biotinylated at the 5′ end, and a thermostable polymerase from Thermus thermophilus is used for both reverse transcription and pcr steps. The amplified product is detected by hybridization to a specific nucleic acid probe (18). We have tried this kit and our prelimary data indicate that it is a useful assay for the detection of hcv rna.
The other assay that is commercially available for the detection of hcv rna is called ‘branched-dna signal amplification assay’ (Chiron). In this case, hcv rna is hybridized directly to synthetic oligonucleotides from the highly conserved 5′ utr and core gene of hcv immobilized on a microwell plate. Synthetic bdna amplifier molecules and multiple copies of an alkaline-phosphatase linked probe are hybridized to the immobilized complex. The complex is then incubated with a chemi-luminescent substance (dioxetane) and the light emission is measured. This technique has an added advantage that it can quantitate hcv rna (19). The disadvantage of this technique is that it is less sensitive than pcr (limitation 300,000 rna molecules).
Quantitation of HCV RNA:
In of hcv rna positive cases, quantitation may provide prognostic information; lower hcv rna levels appear to be associated with less symptomatic disease and with improved response to interferon therapy. Higher viral titres are associated with prolonged infection and are less responsive to treatment. Clinical studies indicate a long term response rate of about 25%.
Genotyping of HCV isolates:
Several distinct hcv genotypes have been described on the basis of complete or partial sequence analysis (20,21). Recently, a direct pcr for hcv genotyping was introduced (22,23) that eliminates the time consuming procedure of sequencing. The genotyping of hcv is important for the epidemiological study of hcv infection and for interferon treatment. Patients infected with different genotypes of hcv respond differently to interferon therapy (24). The subtyping of Canadian isolates (25) has shown that 60.5% are type I, 10.9% type II, 6.7% type III, 5.9% type IV, 10.9% not typeable and 5% type I + II. A commercial test for hcv subtyping has been developed by Innogenetics NV Belgium (26). In this assay, oligonucleotides derived from the 5′ utr act as specific probes for each genotype. The specific probes are immobilized as parallel lines on nitrocellulose strips and then hybridized with the amplified pcr products (26). The subtyping of Canadian hcv isolates by this line probe assay showed a good correlation with other methods we have used for genotyping (27).
The Laboratory for Viral Hepatitis in the National Laboratory for Special Pathogens, Bureau of Microbiology, Laboratory Centre for Disease Control provides reference service for hcv testing. This includes confirmation of eia-positive samples for anti-hcv by riba 3.0, testing for hcv rna by pcr and genotyping. In addition, we provide a proficiency panel for hcv testing to all provincial public health laboratories and large hospital centres.
REFERENCES
- 1.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–62. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Kuo G, Choo QL, Alter HJ, et al. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362–4. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- 3.Esteban JI, Gonzalez A, Hernadez JM, et al. Evaluation of antibodies to hepatitis C virus in a study of transfusion-associated hepatitis. N Engl J Med. 1990;323:1107–12. doi: 10.1056/NEJM199010183231605. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhary RK, Mo T. Antibody to hepatitis C virus in risk groups in Canada. Can J Infect Dis. 1992;3:27–9. doi: 10.1155/1992/710476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson KE, Ahmed F, Ness P, Stambolis V, Yawn D, McAllister H., Jr Comparison of first- and second-generation ELISA screening test in detecting HCV infection in transfused cardiac surgery patients. Program and Abstracts of the International Symposium on Viral Hepatitis and Liver Diseases; Tokyo, Japan. May 10–14, 1994. [Google Scholar]
- 6.Alter MJ. The detection, transmission and outcome of hepatitis virus infection. Infect Agents Dis. 1993;2:155–6. [PubMed] [Google Scholar]
- 7.Van Doom L-J. Molecular biology of the hepatitis C virus. J Med Virol. 1994;45:345–56. doi: 10.1002/jmv.1890430406. [DOI] [PubMed] [Google Scholar]
- 8.Alter HJ. New kit on the block: Evaluation of second-generation assays for detection of antibody to the hepatitis C virus. Hepatology. 1992;15:350–3. doi: 10.1002/hep.1840150228. [DOI] [PubMed] [Google Scholar]
- 9.Alter MJ, Margolis HS, Krawczynski K, et al. The natural history of community-acquired hepatitis C in the USA. N Engl J Med. 1992;327:1899–905. doi: 10.1056/NEJM199212313272702. [DOI] [PubMed] [Google Scholar]
- 10.Uyttendaele S, Claeys H, Mertens W, Verhaert H, Vermylen C. Evaluation of third-generation screening and confirmatory assays for HCV antibodies. Vox Sang. 1994;66:122–9. doi: 10.1111/j.1423-0410.1994.tb00293.x. [DOI] [PubMed] [Google Scholar]
- 11.Alter MJ. Review of serologic testing for hepatitis C virus infection and risk of posttransfusion hepatitis C. Arch Pathol Lab Med. 1994;118:342–5. [PubMed] [Google Scholar]
- 12.Chaudhary RK, Jacobsen H. Performance of third-generation confirmatory tests for detection of antibody to hepatitis C virus. J Clin Microbiol. doi: 10.1128/jcm.32.10.2606-2608.1994. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joller-Jemelka HI, Wick AN, Opitz R. Improved detection of antibodies to hepatitis C virus by use of a new recombinant immunoblot assay. Eur J Clin Microbiol Infect Dis. 1993;12:398–400. doi: 10.1007/BF01964444. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Samaniego J, Enriquez A, Soriano V, Gutierrez M, Baquero M, Munoz F. Third-generation recombinant immunoblot assay to confirm hepatitis C virus-indeterminate serological samples. Vox Sang. 1993;64:191–2. doi: 10.1111/j.1423-0410.1993.tb05384.x. [DOI] [PubMed] [Google Scholar]
- 15.Okamato H, Okada S, Sugiyama Y, et al. Detection of hepatitis C virus RNA by a two-stage polymerase chain reaction with two pairs of primers deduced from 5′-noncoding region. Jpn J Exp Med. 1990;60:215–22. [PubMed] [Google Scholar]
- 16.Lin H-H, Kao J-H, Leu J-H, et al. Comparison of three different immuno assays and PCR for the detection of hepatitis C virus infection in pregnant women in Taiwan. Vox Sang. 1993;65:117–21. doi: 10.1111/j.1423-0410.1993.tb02127.x. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhary RK, Andonov A, MacLean C. Detection of hepatitis C virus infection with recombinant immunoblot assay, synthetic immunoblot assay and polymerase chain reaction. J Clin Lab Anal. 1993;7:164–7. doi: 10.1002/jcla.1860070306. [DOI] [PubMed] [Google Scholar]
- 18.Young KK, Resnick RM, Myers TW. Detection of hepatitis C virus RNA by a combined reverse transcription-polymerase chain reaction assay. J Clin Microbiol. 1993;31:882–6. doi: 10.1128/jcm.31.4.882-886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau JXN, Davis GL, Kniffen J, et al. Significance of serum hepatitis C virus RNA levels in chronic hepatitis C. Lancet. 1993;341:1501–4. doi: 10.1016/0140-6736(93)90635-t. [DOI] [PubMed] [Google Scholar]
- 20.Bukh J, Purcell R, Miller R. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative El gene of isolates collected worldwide. Proc Natl Acad Sci USA. 1993;90:8234–8. doi: 10.1073/pnas.90.17.8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan S, McOmish F, Holmes E, et al. Analysis of new hepatitis C virus type and its phylogenetic relationship to existing variants. J Gen Virol. 1992;73:1131–41. doi: 10.1099/0022-1317-73-5-1131. [DOI] [PubMed] [Google Scholar]
- 22.Chyma K, Tsubota A, Arase Y, et al. Genotyping subtyping of hepatitis C virus. Gastroenterology. 1993;8:150–6. doi: 10.1111/j.1440-1746.1993.tb01507.x. [DOI] [PubMed] [Google Scholar]
- 23.Okamoto H, Sugiyama Y, Okada S, et al. Typing hepatitis C virus by polymerase chain rection with type specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol. 1992;73:673–89. doi: 10.1099/0022-1317-73-3-673. [DOI] [PubMed] [Google Scholar]
- 24.Tsubota A, Chayama K, Arase Y, et al. Factors useful in predicting the response to interferon therapy in chronic hepatitis C. J Gastroenterol Hepatol. 1993;8:535–9. doi: 10.1111/j.1440-1746.1993.tb01648.x. [DOI] [PubMed] [Google Scholar]
- 25.Andonov A, Chaudhary RK. Genotyping of Canadian isolates by PCR. J Clin Microbiol. 1994;32:2031–4. doi: 10.1128/jcm.32.8.2031-2034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stuyver L, Rossan R, Wyseur A, et al. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J Gen Virol. 1993;74:1093–102. doi: 10.1099/0022-1317-74-6-1093. [DOI] [PubMed] [Google Scholar]
- 27.Andonov A, Chaudhary RK. Subtyping of hepatitis C virus isolates with a line probe assay by hybridization. J Clin Microbiol. doi: 10.1128/jcm.33.1.254-256.1995. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]