Abstract
Induced expression of the Flock House virus in the soma of C. elegans results in the RNAi-dependent production of virus-derived, small interfering RNAs (viRNAs), which in turn silence the viral genome. We show here that the viRNA-mediated viral silencing effect is transmitted in a non-Mendelian manner to many ensuing generations. We show that the viral silencing agents, viRNAs, are transgenerationally transmitted in a template-independent manner and work in trans to silence viral genomes present in animals that are deficient in producing their own viRNAs. These results provide evidence for the transgenerational inheritance of an acquired trait, induced by the exposure of animals to a specific, biologically relevant physiological challenge. The ability to inherit such extragenic information may provide adaptive benefits to an animal.
INTRODUCTION
The inheritance of acquired traits is a topic of long-standing interest and controversy. While some of the classic Lamarckian ideas have been dismissed (Weismann, 1889), more recent studies suggest that certain traits acquired by an animal during its lifetime may be transmitted to next generations. For instance, in rats, obesity was shown to transfer from parent to offspring (Ng et al., 2010), and maternal care influenced multiple aspects of neurobiology and behavior of offspring (Champagne, 2008). Nevertheless, the controversy over the inheritance of acquired traits remains, since the genetic and mechanistic basis for these observations has remained largely unclear. We used the nematode C.elegans, to establish a robust model that can unequivocally examine whether “Lamarckian” inheritance is possible and to probe the molecular mechanisms underlying such inheritance. We chose to study the inheritance mode of a trait that is absolutely crucial for the survival of any organism, resistance to viruses and other genomic parasites. Such resistance is expected to be under strong evolutionary pressure.
One commonly employed defense strategy against viruses and other genomic parasites utilizes the process of RNA interference (RNAi), a gene silencing process that has been well characterized in C.elegans (Fire et al., 1998). An RNAi response can also be triggered by the exogenous application of heterologous double stranded RNA (dsRNA) that targets any gene of choice. Intriguingly, gene silencing effects evoked by exogenously added dsRNA can not only be observed in the treated animals, but - in a subset of cases tested - also in the progeny of the treated worms (Alcazar et al., 2008; Fire et al., 1998; Grishok et al., 2000; Vastenhouw et al., 2006). However, whether this transmission involves transgenerationally transmitted RNAs or modifications of DNA or chromatin has not been resolved (Alcazar et al., 2008; Fire et al., 1998; Grishok et al., 2000; Vastenhouw et al., 2006). Moreover, and more importantly, while these observations are technically useful, to date no biological context or function has been attributed to the transmission of RNAi-mediated gene silencing effects.
C.elegans possesses an extraordinary ability to ward off viruses. To date, no viruses have been found to hijack its genome. Nevertheless, a wild C.elegans strain has recently been discovered to be infected with a single strand RNA virus closely related to nodaviruses (Felix et al., 2011). This infected wild C. elegans strain was found to be deficient in mounting an effective RNAi response. In contrast, laboratory strains with an intact RNAi response, such as N2, could not be infected with the wild RNA virus (Felix et al., 2011). These observations indicate that the RNAi pathway attacks dsRNA intermediates that single strand RNA viruses like the natural nodaviruses generate during replication (Figure 1A). Indeed, artificially generated viral infection models have previously shown that C. elegans can fight viruses by processing the viral dsRNA trigger into virus-derived small interfering RNAs (viRNAs) to guide specific viral immunity by Argonaute-dependent RNAi (Lu et al., 2005; Schott et al., 2005; Wilkins et al., 2005). The ability to respond to specific viruses by the production of targeted viRNAs is an acquired trait, serving as an effective defense mechanism against viruses.
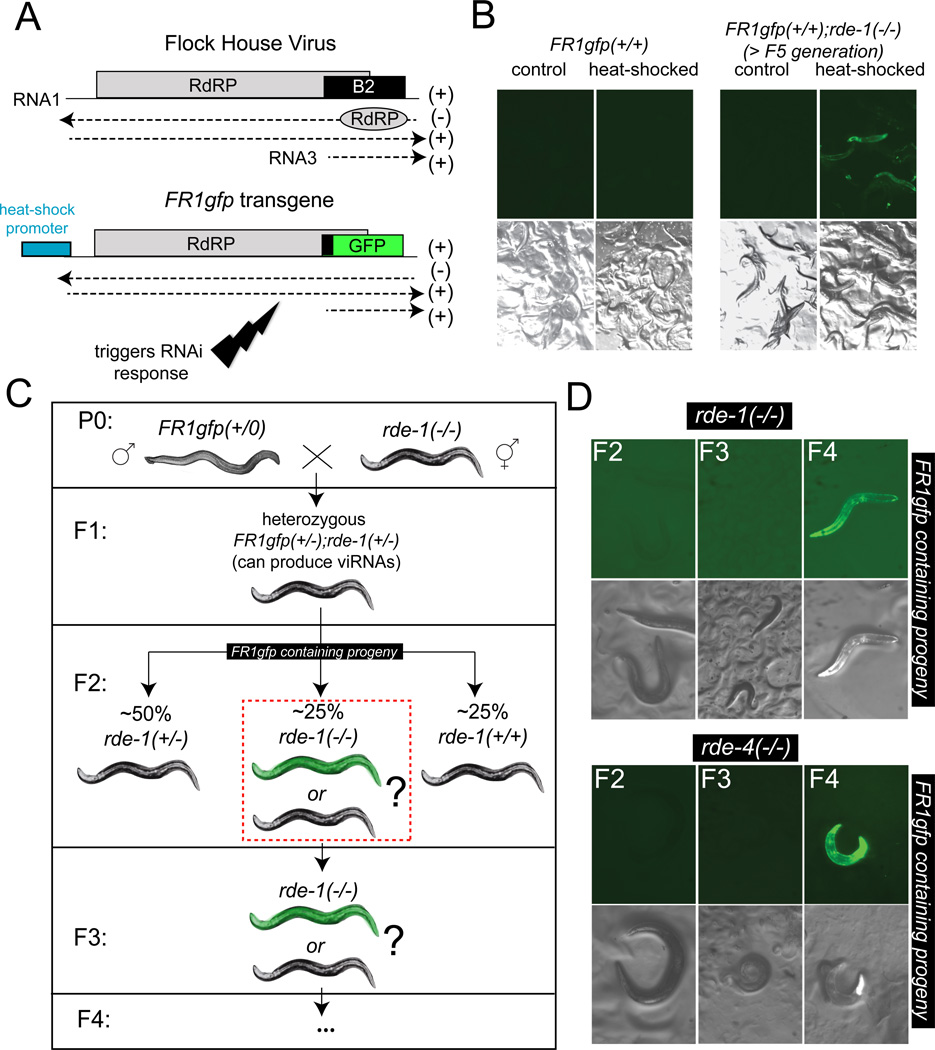
Figure 1. Inheritance of an antiviral RNAi response.
(A) Schematic presentation of the FHV genome and the FR1gfp transgene used in this study (Lu et al., 2009). The RNA2 transcript that produces the capsid is not shown.
(B) Replication of FHV was monitored by recording the expression of GFP in rde-1 and rde-4 mutant worms containing the FR1gfp transgene 48 hours after heat-shock induction of viral replication. The animals were RNAi-deficient for more than 5 generations.
(C) A scheme depicting the cross that tests the requirement for rde-1 and rde-4 in generating virus-silencing viRNAs. Animals shown in the F2 and later generation were all selected to contain the FR1gfp transgene as assessed by the co-injection marker rol-6. rde-1(−/−) animals were scored in the F2 generation for whether they express or do not express GFP after heat-shock induction. Since rde-1(−/−) animals (or rde-4 mutant animals) are not capable of initiating an RNAi response, the FR1gfp; rde-1(−/−) F2 generation is either not able to protect itself against viral propagation (hence heat-shocked animals would be GFP(+) as indicated schematically with a green animal); or the F2 generation inherits antiviral viRNAs that were produced by earlier generations and can therefore protect itself against the virus (hence heat-shocked animals would be GFP(−) as indicated schematically with a dark animal). As shown in panel D, the latter is the case.
(D) DIC and GFP images of heat-shocked rde-1(−/−) and rde-4(−/−) worms that have been homozygous mutant for rde-1 or rde-4 for several generations (as indicated in panel C) and contain FR1gfp as assessed by the rol-6 transgene marker. The rde-1 and rde-4 genotypes were assessed through a linked unc marker.
We describe here how we have tested whether this acquired trait is transmitted transgenerationally. We show that an episode of viral expression is memorized in the form of small viRNA molecules that are transmitted through many ensuing generations in the absence of the genetic template and even in the absence of a functional small RNA-generating machinery. These inherited viRNA molecules protect ensuing generations from the virus by silencing the expression of the viral genome. We therefore provide here evidence for transgenerational transmittance of extrachromosomal information, and suggest a biologically relevant context in which such extrachromosomal information provides a benefit of potential evolutionary relevance to an organism.
RESULTS
A reporter-based system to visualize RNAi-mediated viral silencing in C.elegans
To test the hypothesis of transgenerational inheritance of an acquired trait in a well-controlled setting, we utilized a previously established model to monitor viral propagation, namely, a transgenic worm that supports autonomous replication of the Flock House Virus (FHV) (Lu et al., 2005; Lu et al., 2009). FHV is a plus-strand RNA nodavirus with broad host specificity that is very similar to the virus that was recently identified in a wild C.elegans strain (Felix et al., 2011). To bypass the initial steps of infection (cuticle penetration and cell entry), worms were engineered to carry a chromosomally integrated DNA, called FR1gfp, corresponding to the RNA1 and parts of the RNA3 gene of FHV. The RNA1 of all known nodaviruses encodes the viral RNA-dependent RNA polymerase (RdRP) (Felix et al., 2011). The subgenomic RNA3 transcript encodes the B2 protein, a viral suppressor of RNAi essential for FHV propagation in D. melanogaster (Li et al., 2002), and was largely replaced in FR1gfp by gfp as previously described (Figure 1A) (Lu et al., 2009). Transcription of the viral RNA in the FR1gfp transgene is initially triggered by a heat-inducible promoter (Figure 1A) (Lu et al., 2009) and ensuing viral replication (including the replication of the subgenomic gfp) is then carried out autonomously by the RdRP (Lu et al., 2009). Generally, RdRP-catalyzed replication of FHV progresses at a vigorous rate, reaching levels as high as those of rRNAs (Ball et al., 1994; Johnson and Ball, 1997). Transgenic worms that were heat-induced to initiate the expression of the virus show robust silencing of the B2-deficient FHV mutant virus because the RNAi pathway generates viRNAs that target viral RNA for degradation (Figure 1B) (Lu et al., 2005; Lu et al., 2009). Worms mutant in individual components of the RNAi pathway do, however, display robust virus production as monitored by strong GFP expression throughout the animal (Figure 1B; Table 1), as previously reported (Lu et al., 2009).
Table 1.
Viral silencing in RNAi deficient mutants
| Genotype | Generation | GFP/Virus(+) animals after heat shock *** |
Total number of animals examined |
|---|---|---|---|
| wild type | any | 0% | >100 |
| rde-1 (ne300) | rde-1(−/−) P0* | 100% | 50 |
| rde-1(+/−) F1 cross progeny ** | 0% | 250 (5 experiments) | |
| rde-1(−/−) F2 | 0% | 250 (5 experiments) | |
| rde-1(−/−) F3 | 0% | 250 (5 experiments) | |
| rde-1(−/−) F4 | 0.71% | 882 (5 experiments) | |
| rde-1(−/−) F5 | 10.45% | 908 (5 experiments) | |
| rde-4 (ne299) | rde-4(−/−) P0* | 4.9% | (n=102) |
| rde-4(+/−) F1 cross progeny ** | 0% | 250 (5 experiments) | |
| rde-4(−/−) F2 | 0% | 250 (5 experiments) | |
| rde-4(−/−) F3 | 0% | 267 (5 experiments) | |
| rde-4(−/−) F4 | 3.4% | 441 (5 experiments) | |
| rde-4(−/−) F5 | 3.6% | 307 (5 experiments) |
All animals contain the FR1gfp array in the background. Animals were scored in a binary manner as either producing (GFP/Virus(+)) or not producing (GFP/Virus(−)) any GFP signal. Production of GFP is usually observed in many tissue types. See Supplementary Figure 1 for further information.
Animals were homozygous for rde-1 or rde-4 for at least 10 generations.
Cross of wild type with rde-1(−/−) or rde-4(−/−), respectively.
Numbers are average over several experiments. Individual experiments and animals are shown in Supplementary Figure 1.
Transgenerational inheritance of the antiviral response
To test whether C.elegans can remember an episode of viral propagation and pass this memory to its progeny, perhaps in the form of viRNAs, we performed a set of genetic crosses (Figure 1C). Specifically, we generated animals that are heterozygous for two different RNAi-defective mutants, rde-1 or rde-4 (both coding for RNA binding proteins required for the initiation of an RNAi response (Aoki et al., 2007; Parrish and Fire, 2001; Steiner et al., 2009)), and which contain the heat-inducible FR1gfp viral transgene in the genetic background (see Experimental Procedures for how the genotype of mutants and transgene was determined and how virus expression was induced). Due to the presence of one functional copy of rde-1 or rde-4, these heterozygous animals display a robust antiviral silencing response and therefore display no GFP fluorescence upon heat-induction of virus expression (Table 1; Figure 1C). Throughout this paper, we call this phenotype "GFP/Virus(−)". Self-fertilization of these heterozygous animals results in the generation of rde-1 or rde-4 homozygous animals. Such homozygous mutant progeny are expected to be unable to silence the virus and therefore should be GFP/Virus(+) upon heat-shock induction of viral expression. However, we find that homozygous mutant offspring of rde-1 and rde-4 heterozygotes show robust viral silencing upon heat-shock induction, as evidenced by animals being GFP/Virus(−) (Figure 1C,D; Table 1). This observation can not be simply explained by maternal deposition of wildtype rde-1 or rde-4 activity from the heterozygous parents, because it has been explicitly demonstrated that neither gene activity is maternally transmitted (Blanchard et al., 2011). Moreover, the silencing effect persists through several generations of homozygous rde-1 or rde-4 mutants. Only in the F4 generation of rde-1(−/−) and rde-4(−/−) worms did a small percentage of the animals start to express the virus, as measured by GFP fluorescence (Table 1; Suppl. Fig.1). Among the progeny of F4 GFP/Virus(+) worms we found an increasing number of worms that failed to silence the virus and were thus GFP/Virus(+) (Table 1; Suppl. Fig.1). This inherited silencing effect can eventually “wear off”, as evidenced by continued isolation and propagation of GFP/Virus(+) worms which then produced progenies that were 100% GFP/Virus(+) upon heat-shock induction (Suppl. Fig.1).
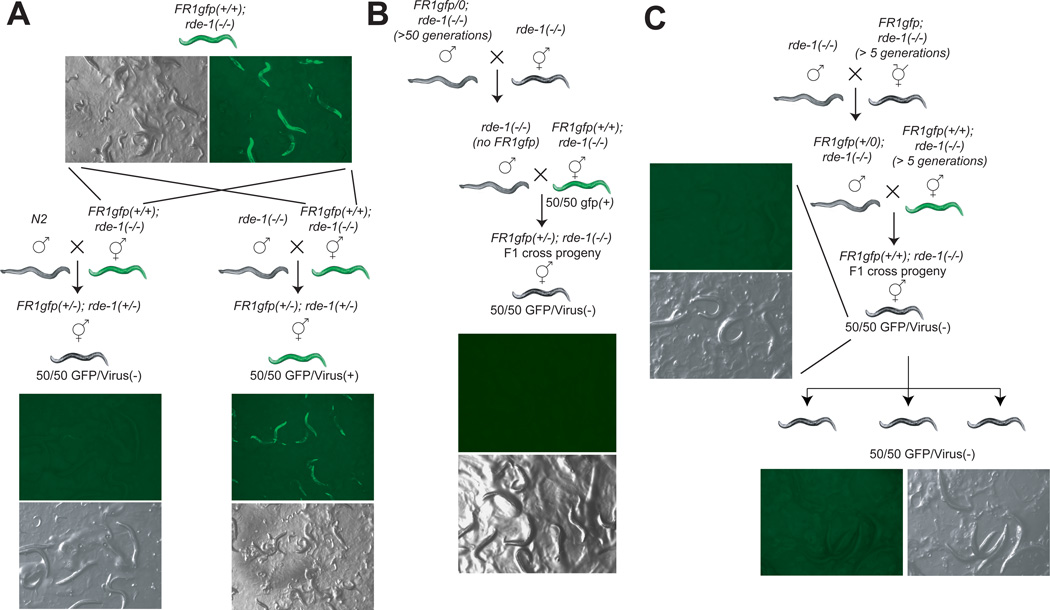
Crossing GFP/Virus(+), homozygous rde-1(−/−) mutant worms, in which the inherited antiviral silencing effect has worn off, with RNAi-competent wild-type males produced cross progeny in which the viral genome is again silenced (Figure 2A). This demonstrates that the restitution of the RNAi machinery, achieved through provision of a functional copy of rde-1, reestablished the antiviral response. When rde-1(−/−) males, rather than wild-type males, were crossed to the same, GFP/Virus(+) >F5 generation rde-1 homozygous mutant hermaphrodites, the heat-shocked cross progeny remained GFP/Virus(+) since it still lacked the ability to mount an RNAi response (Figure 2A). This control also demonstrated that unpairing of the transgenic DNA during meiosis is insufficient by itself to trigger silencing, as has been observed in Neurospora ("meiotic silencing by unpaired DNA")(Shiu et al., 2001).
Fig.2. Genetic analysis of the transgenerational inheritance of an antiviral response.
Animals of all the genotypes schematically shown in this figure were tested for whether they express GFP after heat shocking adult animals (see Experimental Procedures), as a measure to assess viral silencing; dark animals do not express GFP after heat shock, green animals do. Numbers are shown for the most relevant genotypes. The X-linked FR1gfp transgene is present only when specifically indicated.
(A) Reconstitution of the RNAi machinery re-silences viral production.
(B) The antiviral RNA silencing can pass through the sperm and is independent of the presence of the viral template. Because the FR1gfp array is on the X chromosome, F1 males originating from the cross of male FR1gfp/0 (indicating hemizygosity); rde-1(−/−) with rde-1(−/−) hermaphrodites will not contain the array.
(C) Long-term silenced worms contain a non-chromosomally encoded dominant spreading signal. Crossed animals carry the same genotype in regard to the transgene and rde locus, yet one strain is long-term silenced, while the other has lost its ability to silence. The long-term silenced strain is able to silence the non-silent strain in trans. Viral silencing was always assessed by heat-shock treatment to induce the gfp-tagged viral transcript. If the FR1gfp transgene was not present in a strain, it is not shown in the genotype.
It was previously shown that cells infected with FHV produce highly abundant RNA transcripts, as abundant as ribosomal RNAs (Ball et al., 1994; Johnson and Ball, 1997). Our heat-shock induction of viral transcripts should therefore not produce nonphysiological levels or RNAs. We nevertheless tested whether much lower level induction of viral transcript can induce an inherited response. To this end we made use of the inherent, slight leakiness of the heat shock promoter at 15°C and simply maintained the strain containing FR1gfp at 15°C. We find that the rde-1(−/−) F2 progeny of the cross between wildtype males and rde-1(−/−) animals is able to silence viral propagation even if viral replication in the F1 rde-1(+/−) cross-progeny were kept at 15°C (50/50 animals are GFP/Virus(−)). These results indicate that even very low levels of viral product are sufficient to trigger an antiviral response.
In addition to examining the impact of the small RNA biogenesis genes, rde-1 and rde-4 on viral silencing, we also checked other components of the RNAi pathway (mut-2, mut-7, mut-14, mut-16, rde-2, ergo-1, CSR-1, and C04F12.1). We find that the activity of these genes is not required for virus silencing as their elimination did not result in viral/GFP expression (>50 animals tested for each gene), likely due to the documented redundancy of the RNAi silencing machinery in the worm, which contains 27 known Argonautes (Yigit et al., 2006).
Transgenerational transmission of the antiviral response is template-independent
We next tested whether the antiviral RNA agent can be passed to ensuing generations independently of the viral template. To this end, we used the genetic strategy shown in Figure 2B. We crossed rde-1 mutant males that carry the X-linked FR1gfp transgene and display silencing because of the inherited RNA agent (and therefore are GFP/Virus(−)) to rde-1(−/−) hermaphrodites that never encountered the viral transgene. Cross-progeny males from this cross do not carry the X-linked FR1gfp due to its X linkage. These cross-progeny rde-1(−/−); FR1gfp(−/0) males were crossed with GFP/Virus(+) positive, F5 generation rde-1(−/−); FR1gfp(+/+) hermaphrodites. We find that the rde-1(−/−) progeny of this cross had their viral GFP signal eliminated (Figure 2B). This experiment demonstrates that the antiviral agent can be transmitted in the absence of its template. As a side note, the experiment also shows that the antiviral agent can be transmitted through sperm, consistent with previous experiments that tested the transmission of silencing effects against exogenously provided dsRNA (Alcazar et al., 2008; Grishok et al., 2000).
Long-term silencing is transmitted in a non-Mendelian manner and requires the RNA-dependent RNA polymerase rrf-1
One notable feature that became apparent throughout handling FR1gfp; rde-1(−/−) and FR1gfp; rde-4(−/−) animals over the course of many months was that in many animals the GFP/Virus(+) signal never reappeared after the initial construction of those strains. This indicates that in addition to the “fading” (~3 generations) mode of silencing that we described above, a second and more stable mode of inherited silencing can also take place. Such long-term mode of inherited silencing was also previously observed in response to exogenously added dsRNA (Alcazar et al., 2008; Vastenhouw et al., 2006). Based on genetic loss of function analysis, it has been proposed that this long-term silencing effect requires specific chromatin modifying factors (Vastenhouw et al., 2006). Similarly, it has previously been reported that transgenes can become silenced over many generations (Kelly et al., 1997)(a scenario unlikely to apply here, because the rol-6 injection marker that is present on the FR1gfp-containing array is still expressed in these animals). A number of chromatin factors have been identified whose knock-down affects either exogenous RNAi-mediated, long-term silencing (hda-4, K03D10.3, isw-1, mrg-1) or transgene silencing (mes-2, mes-3, mes-4, mes-6, mys-1, m03c11, zfp-1, rba-1, cin-4, gfl-1) or both (hda-4, K03D10.3, isw-1, mrg-1) (Kim et al., 2005; Vastenhouw et al., 2006; Wang et al., 2005). We tested all these factors in the same manner as they were tested in earlier studies and found that the silencing of heat-shock induced FR1gfp expression is not affected upon knockdown of any of these factors (>50 animals tested for each gene).
To further test the transgene silencing issue, but also to examine whether long term-silenced worms carry a trait that segregates in a Mendelian manner (as would be expected from a silenced transgenic array or from mutation of some other, secondary locus), we conducted a genetic experiment schematically shown in Figure 2C. We crossed long term-silenced (i.e. GFP/Virus(−)), FR1gfp-containing animals that were more than F5 generation homozygous for rde-1(−/−) with >F5 generation rde-1(−/−); FR1gfp animals that have lost their silencing ability after a few generations and are therefore GFP/Virus(+) upon viral induction. Since the RNAi machinery is not reinstated in the cross progeny (all animals used are rde-1(−/−)), a hypothetical genomically encoded locus that suppresses GFP/Virus production should segregate in a Mendelian manner in the ensuing generations; similarly, if the GFP/Virus(−) worms simply had their transgene silenced, three quarters of the progeny should be GFP/Virus(+). If, in contrast, the silencing agents are diffusible trans-acting factors (as would be expected from viRNAs), then all F2 progeny should be silenced. We observed that all the F2 progeny had the virally produced GFP signal eliminated (Figure 2C). This experiment indicates that inherited silencing in rde-1(−/−); FR1gfp worms, in which the virus is still silenced after >F5 generation, is achieved by a spreading and DNA-independent element.
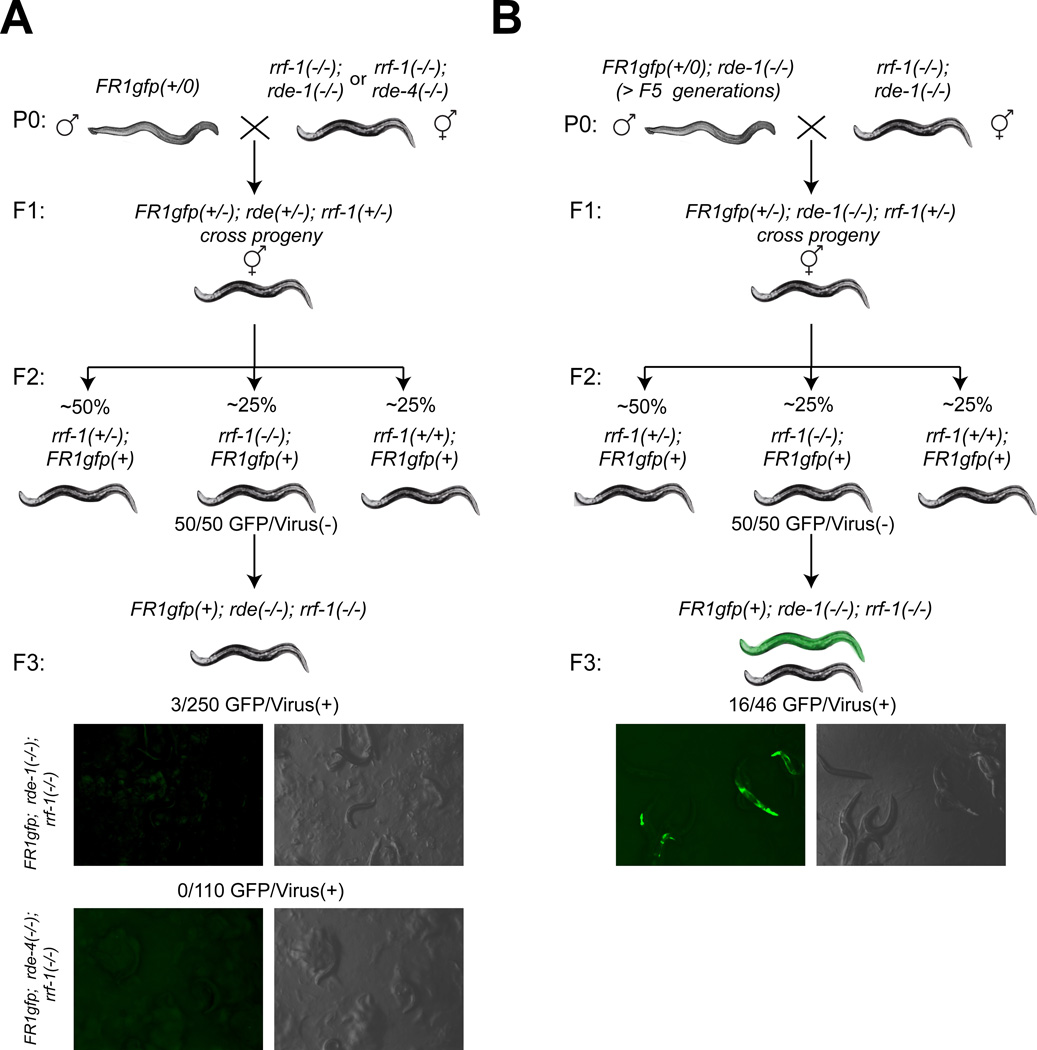
It is conceivable that long term silencing would be maintained by constant amplification of the original RNAi response (Alcazar et al., 2008; Groenenboom et al., 2005). Even though the endogenous RdRP rrf-1 has been shown in the past to amplify viRNAs (Parameswaran et al., 2010; Schott et al., 2005), we find that rrf-1 is not required for the initial transgenerational viral silencing that is observed in rde-1 or rde-4 homozygous mutant animals that were derived from RNAi-competent parents (Figure 3A). Intriguingly, however, rrf-1 dependence is observed in animals that have been long-term silenced in the complete absence of rde-1 (Figure 3B). We draw this conclusion from the reinstated ability of the virus to be expressed (i.e. a GFP/Virus(+) phenotype) in the rrf-1(−/−) homozygous F3 progeny of a cross between stably-silenced GFP/Virus(−) >F5 generation rde-1(−/−); FR1gfp worms and rde-1(−/−); rrf-1(−/−) double mutants (Figure 3B).
Fig.3. The RdRP amplification by rrf-1 is required only for long-term silencing.
Animals of all the genotypes schematically shown in this figure were tested for whether they express GFP after heat shocking adult animals (see Experimental Procedures), as a measure to assess viral silencing; dark animals do not express GFP after heat shock, green animals do. Numbers are shown for the most relevant genotypes. FR1gfp(+) indicates that animals contain the FR1gfp array but it was not tested whether it is contained in the homozygous (FR1gfp(+/+)) or heterozygous (FR1gfp(+/−)) state. If the FR1gfp transgene was not present in a strain, it is not shown in the genotype.
(A) RNAi competent grandparents initiate an RNAi response that is sufficient for viral silencing for at least 2 generations in the absence of rrf-1.
(B) The virus escapes long-term silencing (GFP/Virus(+)) when rrf-1 is neutralized in otherwise GFP/Virus(−) >F5 generation rde-1(−/−) animals.
Physical detection of inherited viRNA molecules
We next tried to detect viRNAs transgenerationally transmitted from RNAi-competent parents through small RNA isolation and ensuing deep sequencing. Although both rde-1 and rde-4 mutants are required for initiation of a de novo (but not inherited) RNAi response, we chose to sequence viRNAs from rde-4 mutant animals rather than rde-1 mutant animals because of the different roles that the two proteins play in the RNAi pathway. rde-4 mutants are depleted of siRNAs because the RDE-4 protein acts upstream in the dsRNA processing pathway by binding long dsRNA and guiding it toward dicing (Aoki et al., 2007). rde-1, on the other hand, is not defective in primary small RNAs synthesis (or primary viRNA synthesis)(Wu et al., 2010) because its protein activity is required only for the separation of the double strand RNA duplex but not for the accumulation of short dsRNAs (Aoki et al., 2007; Parrish and Fire, 2001; Steiner et al., 2009). We chose to use a cloning protocol that enriches for rare rde-4-dependent primary small RNAs (Parameswaran et al., 2010), because rde-4 (as well as rde-1) animals are not defective in secondary siRNA production (Blanchard et al., 2011), and thus can theoretically continue to amplify secondary siRNAs de novo if even a single “trigger” siRNA is inherited (Groenenboom et al., 2005). Detected primary siRNAs, however, are guaranteed to be derived from the original RNAi-competent parents, because rde-4 mutant animals can not produce them (Alcazar et al., 2008; Groenenboom et al., 2005).
Thus, we prepared and sequenced small RNA libraries from four different types of animals: (a) FR1gfp-transgenic worms that can mount an RNAi response and should contain viRNAs (positive control); (b) rde-4(−/−) mutants (negative control as no viRNAs are expected to be produced); (c) FR1gfp; rde-4(−/−) worms that are two generations away from their rde-4(+/−) grandparents, i.e. worms that can themselves not produce viRNAs, but may have inherited viRNA from their grandparents; and (d) the F3 progeny of wild-type animals that contained the FR1gfp transgene (and therefore produced viRNAs), but have lost this transgene through outcrossing with non-transgenic wild-type worms. This tests whether the silencing reagent can exist without its template.
In agreement with the functional assays we detected different viRNAs complementary to several regions of the viral genome in the positive control (FR1gfp), no viRNAs in the negative control (rde-4(−/−)), and a number of viRNAs in the worms that could not generate their own viRNAs and thus inherited these viRNAs from their grandparents (F3 generation FR1gfp; rde-4(−/−))(Table 2; Figure 4). Moreover, we detect viRNAs in the worms in which the FR1gfp transgene had been crossed out, confirming that the viRNA transmit in a template-independent manner. The inherited viRNA matched the two most abundant types of viRNAs detected in the positive control (Figure 4) and these viRNAs were all of the reverse orientation (negative strand, which typically exists in much lower quantities than the positive strand (Felix et al., 2011)); both observations make it highly unlikely that the detected viRNAs merely represent unspecific break-down products of the viral RNA. In regard to the low number of inherited viRNA reads it needs to be considered that our protocol enriches specifically for rare primary viRNA species. Moreover, these viRNA species are derived from a response mounted in RNAi-competent grandparents, and are therefore possibly diluted over the course of several generations. Taken together, our results support the genetic experiments that argued for the existence of trans-acting factors that are transmitted in a non-Mendelian manner to ensuing generations.
Table 2.
Molecular identification of transgenerationally inherited viRNAs
| Genotype | # 20–30bp sequences * |
# viRNAs against FHV ** |
viRNA reads per total # small RNA reads (× 106) |
|---|---|---|---|
| FR1gfp | 3,641,082 | 115 | 31.6 |
|
F3 generation FR1gfp; rde-4(−/−) |
1,131,284 | 10 | 8.8 |
|
F3 generation after FR1gfp outcross |
567,549 | 2 | 3.5 |
| rde-4 (−/−) | 966,052 | none | none |
Numbers correspond to reads after adaptors and poly A tails have been removed.
See Experimental Procedure for more information and comments about these viRNA reads and Figure 4 for the mapping of the viRNAs on the viral genome.
Fig.4. viRNA match to specific epitopes in the viral genome.
The number and strandedness of 20–30nt viRNAs are shown with respect to their alignment to the FHV genome. The thickness of lines, which indicate the location of individual reads, is proportional to number of reads. Note that the transmitted viRNA reads match to the main two epitopes of the virus. Clustering of epitopes to the 3' end of FHV have been noted before (Parameswaran et al., 2010). See Experimental Procedures and Table 2 for more details on the reads.
DISCUSSION
We have described here a series of genetic experiments that provide robust support for the existence of non-Mendelian, multigenerational inheritance of extrachromosomal information. This information is transmitted in the form of small RNAs, viRNAs, which are induced by an episode of viral replication and which are propagated through the germline in a non-template dependent manner. Our results therefore support the Lamarckian concept of the inheritance of an acquired trait.
Our results suggest a physiologically relevant role for these inherited molecules that may lie in transmitting an antiviral agent to protect future generations from viral propagation. We infer this notion from our use of a transgenic system that expresses a viral genome that is from the same family as a natural virus recently discovered in a wild isolate in France (Felix et al., 2011). This transgenic system permitted us to measure viral expression and silencing through a gfp tag rather than relying on the relatively subtle phenotypes induced by the natural virus. Moreover, the use of a well-defined minimal transgenic system in which the viral genome can be unambiguously removed through genetic crosses eliminates potentially confounding issues of viral contamination and also enabled us to show that an antiviral response is transgenerationally segregated in the absence of a template. Lastly, since the FR1gfp transgene does not harm the worm (unlike the natural virus), it can be used to study inherited immunity without fear of accidentally selecting for worms that acquired viral resistance through secondary mutations.
Our findings suggest a physiological context for the previously reported transmission of gene silencing effects elicited by exogenously added dsRNA (Alcazar et al., 2008; Fire et al., 1998; Grishok et al., 2000; Vastenhouw et al., 2006). As these previously described inherited effects show a non-understood specificity for only some target genes, it was unpredictable a priori whether an antiviral RNAi response, triggered by perhaps only a few dsRNA molecules, would also be transgenerationally transmitted and be effective in ensuing generations. Moreover, the mechanistic basis for previously reported transmission of gene silencing effects has been controversial (Alcazar et al., 2008; Fire et al., 1998; Grishok et al., 2000; Vastenhouw et al., 2006). Aside from suggesting biological context, our results also indicate that the inheritance of the viral silencing effect is mediated by inherited small RNA molecules acquired after viral replication.
It is conceivable that other biological functions may be controlled by transgenerationally transmitted, extrachromosomal agents as well. For example, one may speculate that the recently described inheritance of an olfactory memory (Remy, 2010) could also be the result of inherited small RNA molecules. Intriguingly, our deep sequencing of rde-4(−/−) animals identified not only transgenerationally transmitted viRNA molecules but hint also to the possibility that several classes of rde-4-dependent endo-siRNAs may be inherited (O.R., G.M. and O.H., unpubl. data), suggesting that the transmission of extrachromosomal information may be a common phenomenon. Such a mode of inheritance may provide adaptive advantages to an animal.
EXPERIMENTAL PROCEDURES
Genetics
Worms were cultured on NGM plates seeded with OP50 bacteria. FR1gfp transgenic worms were a kind gift from S.W Ding (Lu et al., 2009). Animals carrying the FR1gfp transgene were followed by monitoring their rolling behavior caused by the rol-6(d) transgene marker. In order to follow worms that were homozygous for RNAi-deficiency mutations ((rde-1(ne300) or rde-4(ne299)) after crossing to FR1gfp animals, we used the WM36 rde-1(ne300) unc-42(e270) and WM35 rde-4(ne299) unc-69(e587) strains, in which the rde mutations are linked to unc marker mutations. RNAi-deficient FR1gfp worms, OH10354 and OH1035, were created by singling Unc Rol worms for >10 generations (OH10354 genotype: FR1gfp; rde-1(ne300) unc-42(e270) and OH1035 genotype: FR1gfp; rde-4 (ne299) unc-69(e587)). Worms with the OH10435 genotype: FR1gfp; rde-1(ne300) unc-42(e270); rrf-1(ok589) and the OH10436 genotype: FR1gfp; rde-4 (ne299) unc-69(e587); rrf-1(ok589) were created using standard genetic crosses.
To assess the dependency of inherited antiviral protection on chromatin remodeling factors we used the MT15795 isw-1(n3294) and SS186 mes-2(bn11) strains and also preformed RNAi-feeding to knock down these and a number of other chromatin modulators (isw-1, M03c11, mrg-1, rba-1, cin-4, mes-2, mes-3, mes-4, mes-6, mys-1, hda-4, gfl-1, zfp-1). The RNAi assays were performed using a previously described bacterial feeding protocol (Kamath et al., 2003). Briefly, NGM agar plates containing 6 mM IPTG and 100µg/ml ampicillin were seeded with bacteria expressing dsRNA. 10 FR1gfp L4 hermaphrodites were placed onto these plates and grown at 20°C. The P0 worms and the F1 progeny of these worms were heat shocked as young adults and scored for GFP expression 48 hours after.
Measurement of viral silencing
All the animals in which viral expression was measured contained the FR1gfp array in the background. Our standard procedure of triggering viral propagation is to heat-shock young adults first for 1 hour at 37°C and then for an additional 3 hours at 33°C. 48 hours later the animals were scored in a binary manner as either producing ("GFP/Virus(+)") or not producing ("GFP/Virus(−)") any GFP signal. Production of GFP is usually observed in many tissue types. If animals with the FR1gfp transgene are kept at 15°C, RNAi-deficient animals will not produce a readily detectable GFP signal; however, we notice that even at 15°C there appears to be some small residual FR1gfp expression as those animals are able to produce viRNAs that can silence viral replication upon genetic crosses (see text). Such leakiness of heat-shock promoter expression has been noted in many instances in the literature (e.g. (Poole et al., 2011)).
Molecular and computational analysis of small RNAs
Small RNA libraries were constructed using a protocol that enriches for Dicer products which harbor a single phosphate at the 5’ end of the RNA (Zamore et al., 2000), such as primary siRNAs, and unlike RdRP products that harbor tri-phosphate ends (Parameswaran et al., 2010). This was the protocol of choice because we were interested in identifying viRNAs that are guaranteed to be derived from the original RNAi-competent parents (Alcazar et al., 2008), and rde-4 animals can not produce primary siRNAs (Grishok et al., 2000; Parrish and Fire, 2001). rde-4 animals are, however, not defective in secondary siRNA production (Blanchard et al., 2011), and can therefore continue to amplify secondary siRNAs de novo; such autonomously-produced siRNAs will not be distinguishable from inherited ones. The detection of rare primary siRNAs is important, as even a single “trigger” siRNA can induce a fullblown RNAi-response that is not proportional to the primary trigger (Groenenboom et al., 2005).
Worms were lysed using the TRIzol® reagent, and repetitive freezing, thawing and vortexing as previously described (Lee and Ambros, 2001). The mirVana kit (Ambion) was then used for isolation of <200nt RNAs following size selection of small RNAs of 18–30 bases by denaturing polyacrylamide gel fractionation. Small RNA library was constructed using the Illumina v1.5 Small RNA Cluster Generation Kit® and sequenced using the Illumina/Solexa GAIIX platform (Illumina, Inc., San Diego, CA). Sequences were preprocessed to remove flanking adaptor sequences and noise prior to analysis using the Hannon lab FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Reads were then size filtered using PRINSEQ (Schmieder and Edwards, 2011) so that only small RNAs in the range 20–30nt were considered for alignment. Alignments of viRNAs to FHV genome were performed using Geneious v5.4.3 (word length = 6) (Drummond AJ, 2011). Aligned reads were further examined for similarity to the C. elegans genome using BLASTN (Zhang et al., 2000) to rule out any small RNAs that might match both C. elegans and FHV genomes. No reads matching both FHV and C. elegans genomes were found.
The sequence analysis revealed a strong antisense polarity bias (113/115 viRNA reads in FR1gfp and all inherited reads), consistent with the notion that some of the much more abundant secondary viRNAs (that typically exhibit such a bias (Sijen et al., 2007)) are nevertheless captured despite the use of a protocol that enriches mostly for primary siRNAs (Parameswaran et al., 2010). This can possibly be due to nonenzymatic or post extraction loss of 5’ phosphates (Gent et al., 2010). However, a strong 3’ end bias that is typical to primary siRNAs (Sijen et al., 2007) and to (FHV-targeting) primary viRNAs in particular (Parameswaran et al., 2010) was observed in the detected viRNAs, suggesting that the cloning protocol successfully enriched for primary siRNAs. The fact that 98.26% of the viRNA transcripts were of the negative orientation, even though synthesis of sequences with this orientation is very rare during nodavirus replication (Felix et al., 2011) makes it highly unlikely that they represent non-specific degradation products.
Only reads containing 0–2 mismatches to FHV were considered for analysis. For consideration as a “viral epitope” (“viRNA hot spot”), an epitope had to contain at least one read with 0–1 mismatches and 2 mismatches containing reads were only counted when they corresponded to an epitope without mismatches. 65% of all reads contained 0 mismatches, 24% contained 1 mismatch, and 10% contained 2 mismatches.
We furthermore note that in addition to the perfectly matching inherited viRNAs we detected other unique viRNAs with mismatches in accordance to the template-derived FR1gfp RNA (these sequences were not present in the negative control). The ability to evade treatment by many drugs is granted to RNA viruses by poor proof reading transcription machinery (error every ~104 bp) (Lauring and Andino, 2010). Such viruses exist as swarms of similar variants that are continuously regenerated by their own polymerases, which amplify mutated versions of related sequences (Lauring and Andino, 2010). Even though we cannot exclude simple sequencing errors, it is attractive to speculate that some of the mismatched viRNA reads that were identified in our sequencing data have been produced off such mutated viral copies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Boyanov for small RNA preparation and deep sequencing, C. Mello for providing unc-linked rde strains, S.W. Ding for the FR1gfp transgene, Richard Friedman for advice on statistical methods and Piali Sengupta, Sarah Hall, Iva Greenwald and members of the Hobert lab for comments on the manuscript O.R. is funded by a Bikura postdoc fellowship and a Gruss Lipper postdoc fellowship and G.M. by a T32 predoctoral training grant. O.H. is an investigator of the HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alcazar RM, Lin R, Fire AZ. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics. 2008;180:1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. Embo J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball LA, Wohlrab B, Li Y. Nodavirus RNA replication: mechanism and harnessing to vaccinia virus recombinants. Arch Virol Suppl. 1994;9:407–416. doi: 10.1007/978-3-7091-9326-6_40. [DOI] [PubMed] [Google Scholar]
- Blanchard D, Parameswaran P, Lopez-Molina J, Gent J, Saynuk JF, Fire A. On the nature of in vivo requirements for rde-4 in RNAi and developmental pathways in C. elegans. RNA Biol. 2011;8 doi: 10.4161/rna.8.3.14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA. Epigenetic mechanisms and the transgenerational effects of maternal care. Front Neuroendocrinol. 2008;29:386–397. doi: 10.1016/j.yfrne.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, AB, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. Geneious v5.4.3. 2011 [Google Scholar]
- Felix MA, Ashe A, Piffaretti J, Wu G, Nuez I, Belicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD, et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans [see comments] Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell. 2010;37:679–689. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- Groenenboom MA, Maree AF, Hogeweg P. The RNA silencing pathway: the bits and pieces that matter. PLoS Comput Biol. 2005;1:155–165. doi: 10.1371/journal.pcbi.0010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KL, Ball LA. Replication of flock house virus RNAs from primary transcripts made in cells by RNA polymerase II. J Virol. 1997;71:3323–3327. doi: 10.1128/jvi.71.4.3323-3327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- Lauring AS, Andino R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010;6:e1001005. doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Yigit E, Li WX, Ding SW. An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS Pathog. 2009;5:e1000286. doi: 10.1371/journal.ppat.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W, et al. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 2010;6:e1000764. doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S, Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. Rna. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- Poole RJ, Bashllari E, Cochella L, Flowers EB, Hobert O. A genome-wide RNAi screen for factors involved in neuronal specification in Caenorhabditis elegans. PloS Genet. 2011 doi: 10.1371/journal.pgen.1002109. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy JJ. Stable inheritance of an acquired behavior in Caenorhabditis elegans. Curr Biol. 2010;20:R877–R878. doi: 10.1016/j.cub.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott DH, Cureton DK, Whelan SP, Hunter CP. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2005;102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–916. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- Steiner FA, Okihara KL, Hoogstrate SW, Sijen T, Ketting RF. RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:207–211. doi: 10.1038/nsmb.1541. [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Brunschwig K, Okihara KL, Muller F, Tijsterman M, Plasterk RH. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- Wang D, Kennedy S, Conte D, Jr, Kim JK, Gabel HW, Kamath RS, Mello CC, Ruvkun G. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436:593–597. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- Weismann A. Essays Upon Heredity. Oxford: Clarendon Press; 1889. [Google Scholar]
- Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M, Machaca K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- Wu Q, Luo Y, Lu R, Lau N, Lai EC, Li WX, Ding SW. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc Natl Acad Sci U S A. 2010;107:1606–1611. doi: 10.1073/pnas.0911353107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.