Abstract
Heterozygous mutations in the gene encoding isocitrate dehydrogenase-1 (IDH1) occur in certain human brain tumors, but their mechanistic role in tumor development is unknown. We have shown that tumor-derived IDH1 mutations impair the enzyme’s affinity for its substrate and dominantly inhibit wild-type IDH1 activity through the formation of catalytically inactive heterodimers. Forced expression of mutant IDH1 in cultured cells reduces formation of the enzyme product,α-ketoglutarate (α-KG), and increases the levels of hypoxia-inducible factor subunit HIF-1α, a transcription factor that facilitates tumor growth when oxygen is low and whose stability is regulated by α-KG. The rise in HIF-1α levels was reversible by an α-KG derivative. HIF-1α levels were higher in human gliomas harboring an IDH1 mutation than in tumors without a mutation. Thus, IDH1 appears to function as a tumor suppressor that, when mutationally inactivated, contributes to tumorigenesis in part through induction of the HIF-1 pathway.
Gliomas are the most common type of human brain tumors and can be classified based on clinical and pathological criteria into four grades. The grade IV glioma, commonly known as glioblastoma multiforme (GBM), has one of the worst prognoses among all types of human tumors and can develop either de novo (primary GBM) or through progression from low-grade tumors (secondary GBM). Although pathologically indistinguishable, primary and secondary GBM exhibit distinct patterns of cancer gene alterations (1). A recent cancer genome sequencing project revealed that the gene encoding IDH1 is somatically mutated predominantly in secondary GBM (2). Three subsequent studies of targeted IDH1 gene sequencing confirmed this finding, together identifying IDH1 mutations in more than 70% of secondary GBM or low-grade gliomas but infrequently in primary GBM (about 5%) (3–5). Notably, all of the IDH1 mutations identified to date produce a single amino acid substitution at Arg132 (R132) and no obvious inactivating (frame-shift or protein-truncation) mutations were found. This observation, together with the fact that the tumors do not show loss-of-heterozygosity (LOH), has led to speculation that R132 mutations lead to oncogenic activation of the enzyme.
IDH enzymes catalyze the oxidative decar-boxylation of isocitrate (ICT) to produce α-KG. The human genome has five IDH genes coding for three distinct IDH enzymes whose activities are dependent on either nicotinamide adenine dinucleotide phosphate (NADP+-dependent IDH1 and IDH2) or nicotinamide adenine dinucleotide (NAD+-dependent IDH3). Both IDH2 and IDH3 enzymes are localized in the mitochondria and participate in the citric acid (TCA) cycle for energy production, whereas IDH1 is localized in the cytoplasm and peroxisomes (6). The R132 residue is conserved in all NADP+-dependent IDHs.
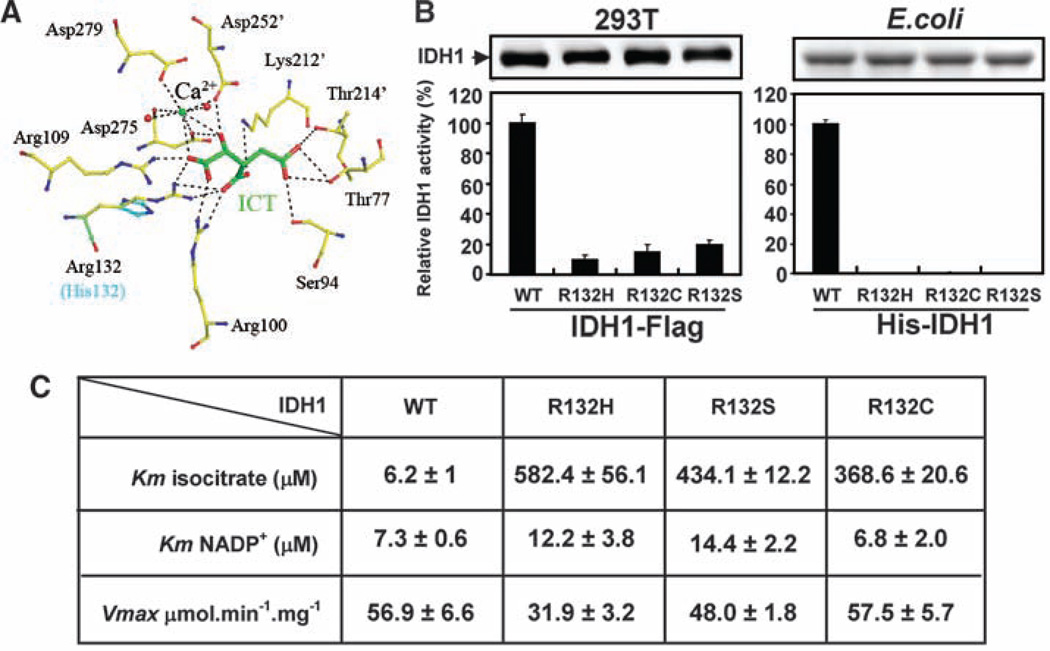
To explore the functional impact of the tumor-associated mutations at R132, we performed modeling studies based on the previously reported human cytosolic IDH1 crystal structure (7). Among all residues involved in binding with ICT, the side chain of R132 uniquely forms three hydrogen bonds with both the α- and β-carboxyl groups of the substrate ICT, whereas other residues involved in ICT binding form no more than two hydrogen bonds (Fig. 1A). Substitution of R132 with any one of the six amino acids observed in gliomas (His, Ser, Gly, Cys, Val, and Leu) would impair interactions of the enzyme with ICT both sterically and electrostatically. Representative modeling of H132, which corresponds to the most prevalent IDH1 mutation in human gliomas, is shown in Fig. 1A. We determined the in vitro enzymatic activities of three tumor-derived IDH1 mutants, R132H, R132C and R132S, expressed in transformed human embryonic kidney (HEK) 293T cells and found that all three mutants have a greater than 80% reduction in activity as compared with the wild-type (WT) IDH1 (Fig. 1B). Analysis of recombinant IDH1 mutant proteins purified from Escherichia coli likewise displayed little activity in vitro compared with wild-type controls (Fig. 1B). Kinetic analyses of recombinant IDH1 proteins revealed that all three mutant IDH1s had a dramatically reduced affinity for ICT: The Michaelis constants (Kms) of IDH1R132C, IDH1R132S, and IDH1R132H for ICT were increased by factors of 60, 70, and 94, respectively (Fig. 1C). In contrast, the Km for NADP+ and the maximum velocity (Vmax) were not appreciably altered (Fig. 1C). Similarly, mutation of an arginine residue in pig mitochondrial IDH2 equivalent to R132 in human IDH1 caused a dramatic increase in Km for isocitrate (by a factor of 165), with minimal effect on Vmax (8). Because the normal cellular concentration of ICT is 20 to 30 µM (9), which is lower than the Km of the IDH1 mutants, the mutant enzymes are likely to have limited activity under physiological conditions. Together, these structural and biochemical analyses indicate that the tumor-associated IDH1 mutations inactivate the enzyme.
Fig. 1.
Tumor-derived IDH1 mutants have reduced catalytic activity because of impaired isocitrate binding. (A) Structural modeling predicts that mutation of R132 in IDH1 would weaken hydrogen bonding of the enzyme to ICT. Shown is a view of the catalytic active site of human IDH1 bound with NADP+ (omitted for clarity), ICT (green), and Ca2+. The residues interacting with ICT from the adjacent subunit are labeled with an apostrophe. Hydrogen-bonding interactions are indicated with dashed lines. Simulated H132 mutation (cyan) is superimposed on R132. (B) Tumor-derived IDH1 mutants have reduced catalytic activity in vitro. Left, FLAG-tagged wild-type and mutant IDH1 were expressed in HEK293T cells, purified by immunoprecipitation and eluted by FLAG peptide; right, HIS-tagged wild-type and mutant IDH1 were expressed in E. coli and purified by nickel resin. Specific IDH1 activities for all proteins were measured in the presence of NADP+ (10 µM) and ICT (30 µM), with the presence of 2 mM Mn2+. Shown are mean values of triplicate experiments ±SD. (C) Kinetic parameters of wild-type and mutant IDH1. Shown are mean values of duplicate experiments ±SD.
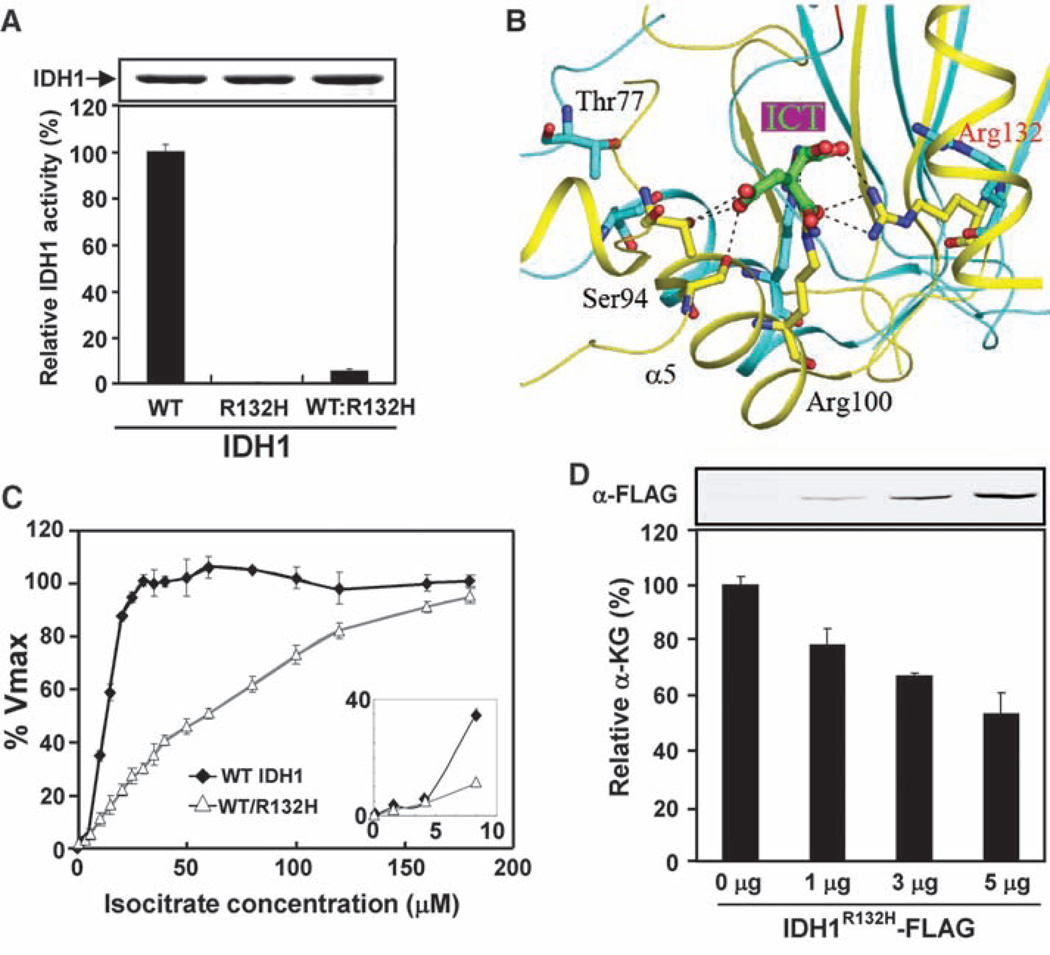
Given that IDH1 normally functions as a homodimer, we hypothesized that the mutant IDH1 molecules in tumor cells form heterodimers with wild-type molecules and, in so doing, dominantly inhibit the activity of wild-type IDH1. To test this hypothesis, we coexpressed His-tagged wild-type and FLAG-tagged R132H mutant IDH1 in E. coli and isolated the heterodimer by sequential affinity purification using first nickel resin and then FLAG beads. Formation of either WT:WT homodimer or WT:R132H heterodimer was confirmed by gel filtration (fig. S1). As expected, the WT:WT homodimers were fully active and the R132H:R132H homodimers were nearly completely inactive (Fig. 2A). Notably, the WT:R132H heterodimer exhibited only 4%of the activity shown by the wild-type enzyme when assayed with limited ICT concentration (Fig. 2A). Normally, IDH1 can adopt at least three distinct conformations during catalysis: a quasi-open conformation when it is in a complex with NADP+, a quasi-closed conformation when it is in a complex with ICT, and a closed conformation when it is in a complex with both NADP+ and ICT (fig. S2A) (7). The two IDH1 subunits act in a cooperative manner and undergo conformational changes in a concerted way. Our modeling study suggests that the impairment in enzyme binding with ICT conferred by the R132 mutation in one subunit might also impair the binding of ICT to the second wild-type subunit. As a result, both subunits would be locked in an unliganded or quasi-open (NADP+-bound) conformation, thereby inhibiting catalytic activity (Fig. 2B for the close-up of the catalytic active site) (fig. S2, B and C). Consistent with this model, we found that the wild-type IDH1 exhibits a sigmoidal curve of cooperative binding to ICT, whereas the WT:R132H heterodimer displayed a hyperbolic curve with a much higher Km (Fig. 2C), indicating that the heterodimer not only loses affinity but also cooperativity toward ICT.
Fig. 2.
The R132H mutation dominantly inhibits IDH1 activity and reduces cellular levels of α-KG. (A) The WT:R132H heterodimer of IDH1 has low specific activity. The specific activities of WT:WT, R132H:R132H, and WT:R132H dimers were measured under conditions of NADP+ (10 µM), ICT (30 µM), and 2 mM MnCl2. Activities were normalized by protein levels, and wild-type activity was arbitrarily set as 100%. Shown are mean values of triplicate experiments ±SD. (B) A close-up view showing the conformational differences between the IDH1-NADP+ and IDH1-NADP+-ICT complexes at the active site. The enzyme adopts a quasi-open conformation in the IDH1-NADP+ complex (cyan) and a closed conformation in the IDH1-NADP+-ICT complex (yellow). The bound ICT and the side chains of several residues in IDH1 involved in ICT binding are shown. (C) The WT:R132H heterodimer loses cooperative binding to ICT. The activities of the WT:WT and WT:R132H enzymes were assayed with increasing concentrations of ICT in the presence of 100 µM NADP+ and 2 mM Mn2+. Shown are mean values of duplicate assays ±SD. The inset is an expanded view showing the IDH1 activities at lower isocitrate concentrations. (D) Cellular α-KG levels decrease with increasing IDH1R132H expression. The upper panel is a Western blot showing expression levels of the transfected IDH1R132H mutant in U-87MG cells. The α-KG level in cells transfected with empty vector was set as 100%, and this value was used to calculate the relative α-KG level in cells transfected with different amounts of IDH1R132H mutant. Shown are mean values of triplicate assays ±SD.
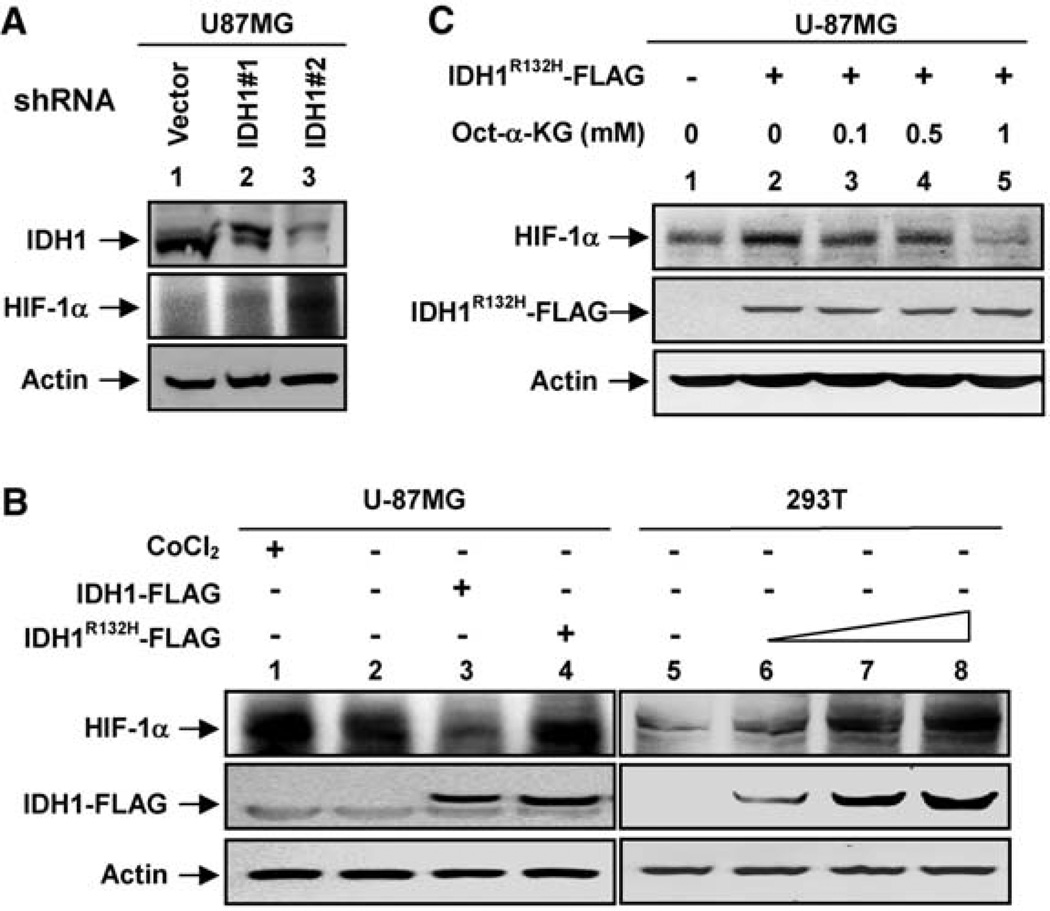
We next investigated whether loss of IDH1 activity would alter cellular levels of α-KG, the product of IDH1 catalysis. We used RNA interference to down-regulate endogenous IDH1 and determined the α-KG levels in U-87MG human glioblastoma cells. Two independent short hairpin RNAs (shRNAs) decreased IDH1 mRNA by more than 75% and reduced cellular α-KG levels by up to 50% (fig. S3). Expression of the IDH1R132H mutant at a level similar to the endogenous protein (fig. S4A) in the cytoplasm of U-87MG cells caused a dose-dependent reduction of α-KG levels (Fig. 2D) (fig. S4, A and B). Together, these data indicate that tumor-derived mutant IDH1 dominantly inhibits the wild-type IDH1 by forming a catalytically inactive heterodimer, resulting in a decrease of cellular α-KG.
Because α-KG is required by prolylhydroxylases (PHD), enzymes that hydroxylate and promote the degradation of hypoxia-inducible factor 1α (HIF-1α), we hypothesized that decreased IDH1 activity might stabilize HIF-1α and increase its steady-state levels. We found that HIF-1α protein levels in U-87MG cells were elevated in response to shRNA-mediated knockdown of IDH1 (Fig. 3A). Conversely, overexpression of wild-type IDH1 reduced HIF-1α protein levels in HeLa (fig. S5A) and U-87MG cells (Fig. 3B). Notably, overexpression of IDH1R132H mutant increased HIF-1α protein levels in U-87MG and HEK293T cells (Fig. 3B). These results suggest that IDH1 regulates HIF-1α levels by controlling the level of α-KG. We tested this hypothesis by treating cells with octyl-α-KG, a cell-permeable derivative of α-KG that upon entering the cells is converted into α-KG after hydrolysis of the ester group (10). We found that octyl-α-KG suppressed the HIF-1α induction caused by either IDH1 knockdown in HeLa cells (fig. S5B) or overexpression of IDH1R132H mutant in U-87MG cells (Fig. 3C).We therefore conclude that a reduction in IDH1 activity produces a reduction in α-KG levels that in turn can lead to stabilization of HIF-1α.
Fig. 3.
α-KG mediates the HIF-1α induction in cells with a decreased IDH1 activity (A) IDH1 knockdown elevates HIF-1α levels in U-87MG glioblastoma cells. IDH1 and HIF-1α protein levels were determined by Western blotting from stable U-87MG cells transduced with empty retrovirus or retrovirus expressing different shRNAs silencing IDH1. (B) Ectopic expression of the IDH1R132H mutant elevates HIF-1α levels in U-87MG and HEK293T cells. The IDH1R132H mutant was overexpressed in U-87MG or HEK293T cells, and protein levels were detected by Western blot. CoCl2-treated cells (a mimetic of hypoxia) and cells overexpressing wild-type IDH1 were also included as controls. (C) A cell-permeable α-KG derivative blocks HIF-1α induction in cells expressing IDH1R132H. The U-87MG cells were transfected with IDH1R132H, and different concentrations of octyl-α-KG ester were added to each transfected cell for 4 hours. HIF-1α protein levels were assayed by Western blot.
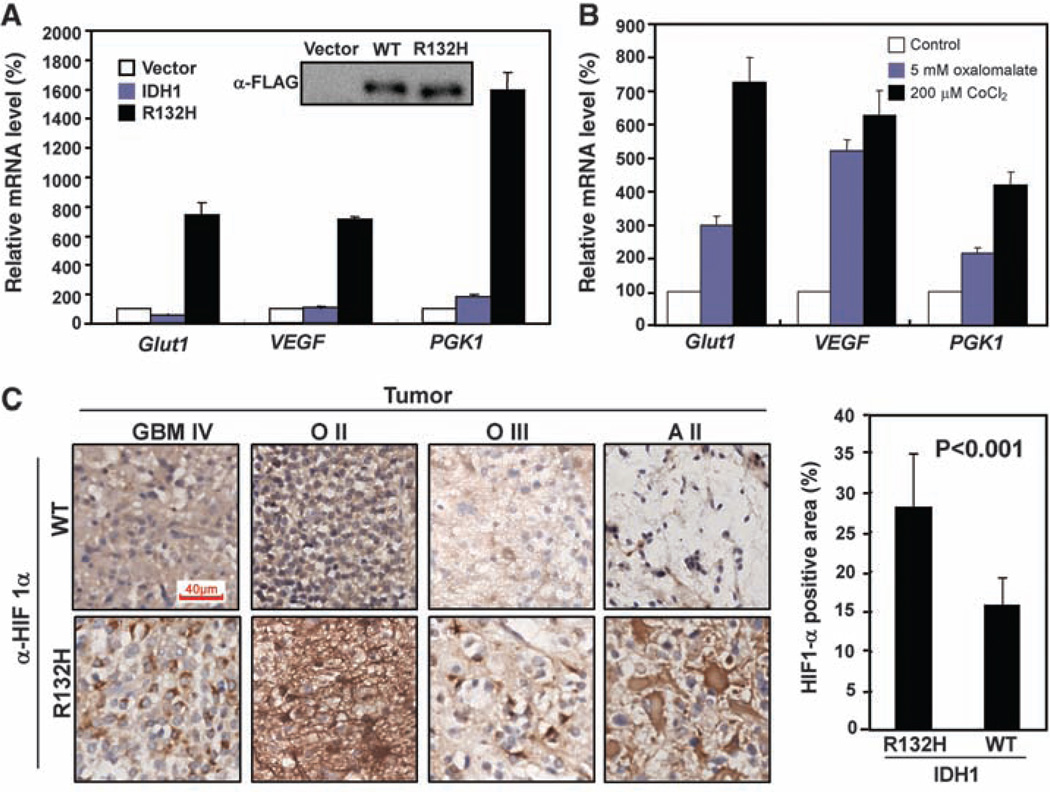
We next determined whether inhibition of IDH1 enzyme activity leads to up-regulated expression of HIF-1α target genes. HIF-1α is a key component of HIF-1, a transcription factor that senses low cellular oxygen levels and that regulates the expression of genes implicated in glucose metabolism, angiogenesis, and other signaling pathways that are critical to tumor growth. Quantitative real-time fluorescence polymerase chain reaction (QPCR) of mRNAs corresponding to three well-established HIF-1α target genes, glucose transporter 1 (Glut1), vascular endothelial growth factor (VEGF), and phosphoglycerate kinase (PGK1) showed that IDH1 knockdown induced the expression of these HIF-1α target genes (fig. S6A). Moreover, expression of the IDH1R132H mutant, but not wild-type IDH1, strongly induced HIF-1α target gene expression (Fig. 4A). Oxalomalate, a competitive inhibitor of IDH1 (11) (fig. S6B), also induced expression of these HIF-1α target genes (Fig. 4B).
Fig. 4.
IDH1 activity affects the levels of HIF-1α and HIF-1α target genes in gliomas and cultured cells. (A) Overexpression of the IDH1R132H mutant in U-87MG cells stimulates expression of HIF-1α target genes (Glut1, VEGF, and PGK1) as assayed by QPCR. Shown are mean values of triplicate assays ±SD. (B) Inhibition of IDH1 by oxalomalate activates HIF-1α target genes. U-87MG cells were either untreated (control), treated with 5 mM oxalomalate, an IDH1 inhibitor, or treated with CoCl2, a hypoxia mimetic. HIF-1α target gene mRNAs were determined. Shown are mean values of triplicate assays ±SD. (C) Immunohistochemistry of HIF-1α was carried out in 12 human gliomas with wild-type IDH1 and 8 gliomas of similar grade harboring a mutated IDH1 allele. Shown are side-by-side comparisons of four gliomas representing different types or grades. Scale bar, 40 µM. Five fields (~173 µm2 each) were randomly selected from each sample for quantification of HIF-1α-positive staining area. Statistical analysis was performed using seven IDH1 wild-type and seven IDH1-mutated gliomas.
Finally, we determined whether IDH1 mutations correlate with elevated levels of HIF-1α in human gliomas. In a collection of 26 glioma samples, we identified 8 tumors that contained the R132H mutation in one allele of IDH1 (table S1) (fig. S7A). Using immunohistochemistry, we compared HIF-1α expression in gliomas with and without IDH1 mutations. We found that 8 tumors harboring the R132H mutation showed a statistically stronger HIF-1α signal than did 12 tumors that did not harbor this mutation (Fig. 4C). In IDH1 mutated tumors, 28.1 ±6.7% of the cells stained positive for HIF-1α, whereas in tumors with wild-type IDH1, only 15.8 ±3.5% of the cells stained positive (P < 0.001) (Fig. 4C). IDH1-mutated gliomas also exhibited an increase in VEGF levels compared with gliomas without IDH1 mutation of similar type and grade (fig. S7B).
In summary, we have shown that IDH1 is likely to function as a tumor suppressor gene rather than as an oncogene. The glioma-associated mutations dominantly inhibit the activity of wild-type IDH1 through heterodimer formation (fig. S8). IDH1 gene mutations in gliomas exhibit two unique features: the lack of LOH and the lack of apparent inactivating mutations such as frame-shift or truncations. Our findings help to explain both features, as dominant inhibition would eliminate the selection pressure to mutate or lose the remaining wild-type allele (LOH) and frame-shift or premature termination would likely generate IDH1 fragments unable to dimerize with and inhibit the wild-type IDH1 protein.
The link between IDH1 and HIF-1α highlights an emerging theme in which mutationally altered metabolic enzymes are thought to contribute to tumor growth by stimulating the HIF-1α pathway and tumor angiogenesis. The genes encoding two TCA enzymes, fumarate hydratase (FH) and succinate dehydrogenase (SDH), have been found to sustain loss-of-function mutations in certain human tumors, which likewise correlate with an increase in HIF-1α levels (10, 11). In addition to affecting PHD, an alteration in α-KG might contribute to tumorigenesis by affecting other dioxygenases that use α-KG as a substrate. IDH1 also catalyzes the production of NADPH; thus, it is possible that a reduction in NADPH levels resulting from IDH1 mutation contributes to tumorigenesis through effects on cell metabolism and growth. Several dozen anticancer agents directly targeting HIF-1α are under development or being tested (12). Our finding that an α-KG derivative can reverse the induction of HIF-1α levels in cultured cells expressing mutant IDH1 suggests that drugs mimicking α-KG may merit exploration as a therapy for gliomas that harbor an IDH1 mutation.
Supplementary Material
Acknowledgments
We thank members of the Fudan Molecular and Cell Biology Laboratory for valuable input; Y. Liu, X. Liu, and H. Zhu for assistance with histology; Z. Bao, L. Yang, Q. Shi, and G. Zhao for clinical samples; and S. Jackson for reading the manuscript. This work is supported by the 985 program from the Chinese Ministry of Education, State Key Development Programs of China (2009CB918401, 2006CB806700), National 863 Program of China (2006AA02A308), China NSF grants (30600112 and 30871255) and Shanghai Key Basic Research Projects (06JC14086, 07PJ14011, and 08JC1400900), and NIH grants (to K.-L.G. and Y.X.). Y. Xiong, K.-L. Guan, and S. Zhao are applying for a patent related to the work on permeable alpha-ketogluterate.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/324/5924/261/DC1
Figs. S1 to S4
Table S1
References
References and Notes
- 1.Furnari FB, et al. Genes Dev. 2007;21:2683. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.Parsons DW, et al. Science. 2008;321:1807. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balss J, et al. Acta Neuropathol. 2008;116:597. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 4.Bleeker FE, et al. Hum. Mutat. 2009;30:7. [Google Scholar]
- 5.Yan H, et al. N. Engl. J. Med. 2009;360:765. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winkler BS, DeSantis N, Solomon F. Exp. Eye Res. 1986;43:829. doi: 10.1016/s0014-4835(86)80013-6. [DOI] [PubMed] [Google Scholar]
- 7.Xu X, et al. J. Biol. Chem. 2004;279:33946. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 8.Soundar S, Danek BL, Colman RF. J. Biol. Chem. 2000;275:5606. doi: 10.1074/jbc.275.8.5606. [DOI] [PubMed] [Google Scholar]
- 9.Albe KR, Butler MH, Wright BE. J. Theor. Biol. 1990;143:163. doi: 10.1016/s0022-5193(05)80266-8. [DOI] [PubMed] [Google Scholar]
- 10.MacKenzie ED, et al. Mol. Cell. Biol. 2007;27:3282. doi: 10.1128/MCB.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingebretsen OC. Biochim. Biophys. Acta. 1976;452:302. doi: 10.1016/0005-2744(76)90180-7. [DOI] [PubMed] [Google Scholar]
- 12.Semenza GL. Drug Discov. Today. 2007;12:853. doi: 10.1016/j.drudis.2007.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.