Abstract
N-Methyl-d-aspartate (NMDA) receptors gate a slow and calcium-rich component of the postsynaptic glutamate response. Like all ionotropic glutamate receptors, NMDA subunits contain a highly conserved motif (SYTANLAAF) in the transmembrane (TM) 3 domain that is critically involved in channel gating. Mutation of an alanine in this domain (A7; underlined above) results in constitutively open receptors that show reduced sensitivity to several allosteric modulators. In this study, we examined the effects of ethanol, a substance that inhibits NMDA currents via an unknown mechanism, on tonically active NMDA receptors expressed in human embryonic kidney 293 cells. Ethanol (100 mM) inhibited currents from GluN1(A7R)/GluN2A and GluN1(A7R)/GluN2B receptors by approximately 50%, whereas those from GluN1/GluN2B(A7R) receptors were reduced by less than 10%. In cysteine-substituted GluN1 and GluN2 A7 mutants, estimated ethanol IC50 values for agonist-gated currents were 101, 117, 103, and 69 mM for GluN1(A7C)/GluN2A, GluN1(A7C)/GluN2B, GluN1/GluN2A(A7C), and GluN1/GluN2B(A7C) receptors, respectively. After exposure to the thiol-modifying reagent 2-(trimethylammonium)ethyl methanethiosulfonate (MTSET), A7C mutants showed robust agonist-independent currents and reduced sensitivity to ethanol (IC50 values of 371, 256, 715, and 958 mM, respectively, as above). In contrast, cysteine modification of the ligand-binding domain resulted in constitutively open receptors that showed robust ethanol inhibition. Ethanol inhibition of MTSET-treated GluN1(A7C) receptors was further reduced by TM3/TM4 mutations previously shown to reduce ethanol sensitivity of agonist-gated receptors. Overall, these results show that ethanol affects NMDA receptor function at a site distal from agonist binding and appears to exert greater effects via perturbation of GluN2 subunits.
Introduction
Ethanol is one of the most widely used substances, and its use and abuse have significant economic and social impact throughout the world. Although ethanol from one to two drinks produces feelings of warmth and well being, signs of behavioral impairment including slurred speech, changes in gait and posture, and altered mood appear as concentrations reach intoxicating levels (U.S. drink drive limit: 0.08% blood ethanol concentration ∼17 mM). Sedation and coma occur at higher blood concentrations (0.3–0.4%; ∼60–80 mM), and death due mainly to respiratory difficulty can occur in severely intoxicated individuals (Koob and Le Moal, 2006). Although the correlation between blood ethanol concentration and behavioral impairment is well characterized, the sites and mechanisms of ethanol that underlie these effects are less clear. Work over the last 25 years shows that ethanol interacts with a variety of ion channel subtypes that regulate neuronal activity. Among these are channels gated by the neurotransmitter glutamate, the major excitatory transmitter in the mammalian brain. Glutamate activates three major types of ion channels termed GluA1–4 (α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid), GluK1–5 (kainate), and GluN1/GluN2 (NMDA). All three channel subtypes show some sensitivity to ethanol, and NMDA receptors are among those most consistently affected by concentrations of ethanol associated with human consumption (Dopico and Lovinger, 2009). Given the key role of NMDA receptors in synaptic transmission, neuronal plasticity, and learning and memory, understanding how ethanol alters NMDA receptor function is an important goal.
Lovinger et al. (1989) were among the first to show that NMDA currents evoked in brain neurons are inhibited by ethanol. Subsequent studies using radiolabeled neurotransmitter release, calcium uptake, and ratiometric calcium indicators confirmed this finding and established that neuronal NMDA receptors across a wide range of brain regions are inhibited by ethanol (Woodward, 2000). After the cloning of the family of NMDA receptor subunits, several studies showed that recombinant NMDA receptors are also inhibited by ethanol (Woodward, 2000). Although there is now almost universal agreement that ethanol inhibits NMDA receptor function, the mechanism of action for this effect remains unclear. The actions of ethanol on NMDA currents are voltage-independent and occur with no change in single channel conductance, suggesting that ethanol does not act as a pore blocker (Lima-Landman and Albuquerque, 1989; Wright et al., 1996). Ethanol also does not behave as a competitive antagonist at either the glycine or glutamate site on the receptor (Peoples and Weight, 1992; Mirshahi and Woodward, 1995), although glycine may affect the degree of inhibition at low ethanol concentrations (Rabe and Tabakoff, 1990; Woodward and Gonzales, 1990). In addition, although intracellular C-terminal domains and phosphorylation sites can, in some instances, modulate ethanol sensitivity of NMDA responses (Woodward, 2000; Maldve et al., 2002; Xu et al., 2010), receptors lacking the intracellular C terminus retain sensitivity to ethanol (Peoples and Stewart, 2000). In addition, examination of other well characterized modulatory sites on the receptor has not revealed any direct interaction of ethanol with these sites (Woodward, 2000).
The activation of the NMDA receptor is complex, given that the receptor requires two different agonists, glutamate and glycine binding, to two different subunits, GluN2 and GluN1, for full activity (Traynelis et al., 2010). Agonist-induced closure of extracellular bilobed ligand-binding domains (LBDs) in both subunits is thought to exert strain on transmembrane (TM) domains that make up the channel gate, thus leading to pore opening (Furukawa and Gouaux, 2003). To date, all studies of ethanol action on NMDA receptors have used agonist binding to induce channel opening, and these studies are hampered by the large number of steps that link agonist binding to channel opening. In this study, we use a series of mutant receptors that are constitutively open in the absence of agonists. These constructs generate tonically active receptors through perturbation of either a highly conserved motif (SYTANLAAF) in TM3 that may form the activation gate of the receptor (Jones et al., 2002; Yuan et al., 2005; Chang and Kuo, 2008) or a motif in which the LBD is locked via disulfide bonds into its active closed-cleft form (Blanke and VanDongen, 2008a; Kussius and Popescu, 2010). The results of these studies indicate that the ethanol sensitivity of these mutants varies, depending on the location of the mutation and the subunit on which it is expressed.
Materials and Methods
Molecular Biology, Cell Culture, and Transfection.
The wild-type GluN1 and GluN2 NMDA receptor cDNAs used in these experiments were kindly provided by Drs. S. Nakanishi (Kyoto University, Kyoto, Japan) and P. Seeburg (Max-Planck Institute for Medical Research, Heidelberg, Germany) and were subcloned into mammalian expression vectors as required. Dr. G. Popescu (SUNY, Buffalo, NY) kindly provided the GluN2A(K499C:N687C) cDNA. Site-directed mutagenesis was performed using a QuikChange Mutagenesis Kit (Invitrogen, Carlsbad, CA), and mutants were confirmed by DNA sequencing. Human embryonic kidney 293 cells were obtained from American Type Culture Collection (Manassas, VA) and were maintained in feeder flasks containing serum-supplemented Dulbecco's modified Eagle's medium in a humidified incubator supplied with 5% CO2 (Xu et al., 2010). For recordings, cells were plated onto polyornithine-coated 35-mm dishes and transfected with plasmids encoding various NMDA receptor subunits (typically 1 μg each) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendation. In each set of transfections, a cDNA encoding enhanced green fluorescent protein was included to allow for detection of transfected cells. Plasmids were used at a ratio of 1:1:1 unless otherwise indicated. After transfection, the NMDA antagonist 2-amino-5-phosphonovaleric acid (AP5) (200 μM) was added to the medium to prevent glutamate-mediated excitotoxicity. In cells transfected with spontaneously active mutants, AP5 was replaced by 10 mM MgCl2 to block receptor activity during incubation. Medium containing AP5 or elevated magnesium was removed by extensive washing just before recording.
Electrophysiology.
Dishes containing transfected cells were mounted on the stage of an Olympus IX50 inverted microscope and perfused with extracellular recording solution at 1 to 2 ml/min. The recording solution contained 135 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 5 mM HEPES, and 10 mM glucose (pH adjusted to 7.4 and osmolarity adjusted to 310–325 mOsM with sucrose). In experiments with constitutively open receptors, two recording solutions were used and contained either 10 mM MgCl2 or 10 mM CaCl2. Patch pipettes (2–5 MΩ) were pulled from borosilicate glass (1.5 × 0.86 mm) and filled with internal solution containing 140 mM KCl, 6 mM MgCl2, 1 mM CaCl2, 5 mM EGTA, 10 mM HEPES, 2 mM tetraethylammonium chloride, and 4 mM Na-ATP (pH adjusted to 7.2 with KOH). Transfected cells were identified by enhanced green fluorescent protein fluorescence, and whole-cell voltage clamp recordings were performed at room temperature using an Axon 200B amplifier (Molecular Devices, Sunnyvale, CA). Cells were held at −60 mV to monitor seal breakthrough and maintained at this potential unless otherwise noted. Series resistance was monitored over the course of the experiment, and cells with unstable holding currents or significant changes in series resistance were not used for analysis.
NMDA receptor currents were evoked using a Warner FastStep multibarrel perfusion system (Warner Instruments, Hamden, CT) to switch between normal extracellular solution and solutions containing agonist or agonist plus ethanol (30–500 mM). The order of solutions was interleaved with control applications of agonists to monitor current rundown. Data were filtered at 1 to 2 kHz and acquired at 5 kHz using an InstruTECH ITC-16 digital interface (InstruTECH Corp., Port Washington, NY) controlled by IGOR Pro software (WaveMetrics, Lake Oswego, OR) running the Pulse Control Acquisition module. Data were analyzed offline using AxoGraph X software (Axograph, Sydney, NSW, Australia), and amplitudes were measured during the last 0.5 s of agonist application when currents had reached steady-state levels. Ethanol inhibition was calculated as a percentage of the average control current obtained before and after application of ethanol. Ethanol was purchased from Aaper Alcohol and Chemical Company (Shelbyville, KY), and all other chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted.
Molecular Modeling.
A model of the heteromeric GluN1/GluN2A receptor was generated using the crystal structure of the homomeric GluA2 (α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid)-subtype glutamate receptor (Protein DataBank 3KG2) as a template (Sobolevsky et al., 2009). We first aligned the GluN1 and GluN2A sequences with the respective sequences in 3KG2. Then we concatenated the GluN1 and GluN2A sequences to form four subunits in the order N1/N2A/N1/N2A. We used the Sequence Alignment module of Modeler in Discovery Studio 2.5 (Accelrys, San Diego, CA) to produce an initial alignment and then manually aligned the sequences as described in Supplemental Fig. 2 of Sobolevsky et al. (2009). This manual alignment, based on the X-ray structure, assured that the residues important to the present studies were correctly displayed. We submitted the alignment of N1/N2A/N1/N2A with 3KG2 to the Modeler module to build known and conserved di-cysteine cross-links found in the GluA2 crystal structure. We then ran two “loop refinement” protocols on each of the initial models and chose the “best” on the basis of the force field energy calculated by CHARMM. To check on the validity of the best model, we separated the first (NR1) and second (NR2) subunits and submitted them to the ProQM server, a server trained on quality of membrane proteins (Ray et al., 2010). As a further improvement, the model was then optimized with CHARMM to a gradient of 0.0001 kcal/mol-Å in the presence of harmonic restraints on the backbone atoms of 10 kcal/Å2. Then we relaxed the structure with 100,000 1-fs steps of molecular dynamics at 300 K with the same restraints on the backbone atoms. Finally, we reoptimized the model as described above. The glutamate-bound crystal structure of the GluN2A LBD (Protein Data Bank 2A5S) was used to model the locked ligand-binding domain mutants. Protein structures were visualized using the DeepView Swiss-PdbViewer (Guex and Peitsch, 1997), and figures were rendered using MegaPOV software (http://megapov.inetart.net).
Data Analysis.
Data are expressed as means ± S.E.M. and were analyzed by analysis of variance or t test (as indicated) using Prism 4.0 software (GraphPad Software Inc., San Diego, CA). Prism software was also used to generate concentration-response curves for ethanol inhibition using unweighted least-squares nonlinear regression of log concentration values versus percent inhibition.
Results
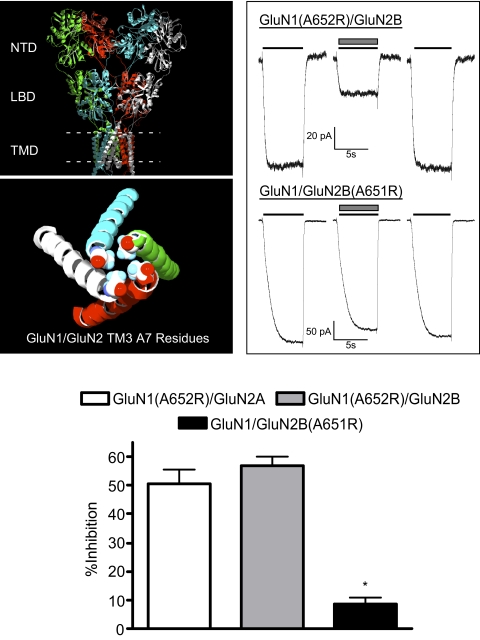
Figure 1 shows a molecular model of the tetrameric GluN1/GluN2A receptor and the relationship between the TM3 domains that contain the highly conserved SYTANLAAF motif. In this model, GluN1 subunits are in the A/C conformation with GluN2 subunits in the B/D positions. This arrangement is consistent with experimental evidence showing that NMDA receptors assemble in a N1/N2/N1/N2 formation (Sobolevsky et al., 2009; Rambhadran et al., 2010; Salussolia et al., 2011). In addition, the manual structural alignment, following the “automated sequence alignment” had high homology in the regions of interest of this study, in particular F639 in GluN1 and A825 in GluN2A. As a result, we have confidence in their relative interactions and proximity in the TM domains. In initial experiments, we generated and tested mutants in which arginine replaced alanine at the A7 position of the SYTANLAAF motif in both GluN1 and GluN2 subunits. As reported previously, these mutants generate agonist-independent currents that retain sensitivity to magnesium (Yuan et al., 2005; Chang and Kuo, 2008). We used this property to activate receptor currents by stepping from a recording solution containing 10 mM MgCl2 to one in which magnesium was replaced by 10 mM CaCl2. As shown in Fig. 1B, A7R mutants yielded significant inward currents upon exposure to calcium-containing solutions. The mean amplitude of these currents was 118.2 pA (±35.7; n = 6) for GluN1(A652R)/GluN2A, 90.7 pA (±22.30; n = 8) for GluN1(A652R)/GluN2B, and 313.6 pA (±68.9; n = 13) for GluN1/GluN2B(A651R). Nontransfected cells or those expressing wild-type receptors showed no responses when exposed to these solutions (data not shown). In addition, although the GluN2A(A650R) mutant generated currents in the presence of glutamate and glycine (data not shown), reproducible currents in the absence of agonists were not observed, and this mutant was not tested further. Ethanol (100 mM), applied acutely, inhibited currents from both GluN1(A652R)/GluN2A- and GluN1(A652R)/GluN2B-expressing cells by approximately 50%. This inhibition was similar to that obtained for wild-type receptors activated by glutamate and glycine (Fig. 1, dashed lines). In contrast, this same concentration of ethanol produced less than 10% inhibition of GluN1/GluN2B(A651R) receptors. In all cases, the effects of ethanol were reversed after washout.
Fig. 1.
Ethanol inhibition of A7 arginine-substituted NMDA receptors. A, top panel shows a model of the GluN1/GluN2A receptor with extracellular amino terminal domain (ATD), LBD, and transmembrane domain (TMD) (dashed line represents approximate location of the plasma membrane). Bottom panel shows a top-down view of TM3 helices (GluN1, white/green; GluN2A, blue/red) with alanines at position 7 of the conserved SYTANLAAF domain shown as space-filling molecules. B, traces show representative currents from single cells expressing A7R mutants [top, GluN1(A652R)/GluN2B; bottom, GluN1/GluN2B(A651R)]. In each set of traces, currents were activated by switching from a solution containing magnesium to one containing calcium (both at 10 mM; solid line) in the absence and presence of 100 mM ethanol (▩). C, summary of effects of 100 mM ethanol on spontaneous currents from arginine-substituted A7 mutants. Dashed lines show mean ethanol inhibition of GluN1/GluN2A (41.7 ± 4.9%; blue line) and GluN1/GluN2B (49.4 ± 4.7%; red line) for comparison. Data are means ± S.E.M. from 8 to 14 cells for each receptor combination. *, value significantly different from that for GluN1(A652R)/GluN2B; unpaired t test (p < 0.0001).
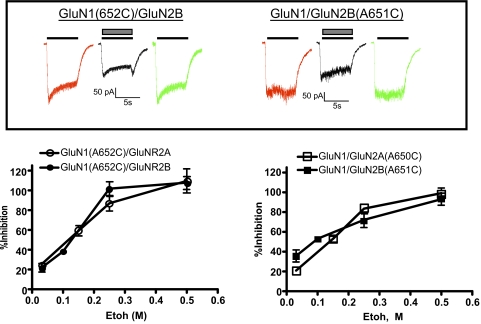
Unlike arginine-substituted receptors, A7 cysteine mutants are not tonically active under control conditions, but like wild-type receptors, currents can be evoked by application of agonists (e.g., glutamate/glycine). These same receptors become agonist-independent after exposure to bulky thiol-reactive compounds applied in the presence of agonists (Jones et al., 2002; Yuan et al., 2005). We thus generated A7C mutants in both GluN1 and GluN2 subunits and determined the ethanol sensitivity of agonist-dependent and -independent currents. To establish the baseline ethanol sensitivity of the A7C mutants before (2-(trimethylammonium)ethyl methanethiosulfonate (MTSET) treatment, we first determined the degree of ethanol inhibition of A7C mutants during activation by glutamate and glycine (both at 10 μM). Figure 2A shows representative currents obtained from GluN1(A652C) and GluN2B(A651C) mutants during exposure to agonists. Coapplication of ethanol significantly inhibited these agonist-dependent currents and responses returned to normal after washout. Dose-response curves for ethanol inhibition were then generated for cells expressing each combination of the GluN1(A7C) and GluN2(A7C) mutant. As shown in Fig. 2B, ethanol inhibited agonist-dependent currents similarly from cells expressing GluN1(A652C) and either wild-type GluN2A or GluN2B. This inhibition was concentration-dependent over the range tested (30–500 mM), and currents were almost completely inhibited at ethanol concentrations greater than 250 mM. Analysis of the dose-response curves by nonlinear regression revealed estimated IC50 values of 101 mM (95% CI 69–147.7) for GluN1(A652C)/GluN2A and 117.1 mM (95% CI 96.5–142.3) for GluN1(A652C)/GluN2B. As shown in Fig. 2C, ethanol also dose dependently inhibited agonist-evoked currents in cells expressing either GluN1/GluN2A(A650C) or GluN1/GluN2B(A651C) mutants. Estimated ethanol IC50 values for these receptors were 102.9 mM (95% CI 78.9–134.1) and 69.3 mM (95% CI 48.4–99.2), respectively.
Fig. 2.
Ethanol (Etoh) inhibition of agonist-activated cysteine-substituted A7 mutants. A, traces show representative currents from single cells expressing A7C mutants [left, GluN1(A652C)/GluN2B; right, GluN1/GluN2B(A651C)]. Currents were activated by switching from normal recording solution to one containing glutamate and glycine alone (10 μM each; solid line, red and green traces) or with 100 mM ethanol (▩, black trace). B, dose-response curve for ethanol inhibition of agonist-evoked currents in cysteine-substituted A7 mutants. Data represent the mean ± S.E.M. from five to seven cells for each receptor combination. Estimated ethanol IC50 values were 101 mM GluN1(A652C)/GluN2A, 117.1 mM GluN1(A652C)/GluN2B, 102.9 mM GluN1/GluN2A(A650C), and 69.3 mM GluN1/GluN2B(A651C). See text for more details.
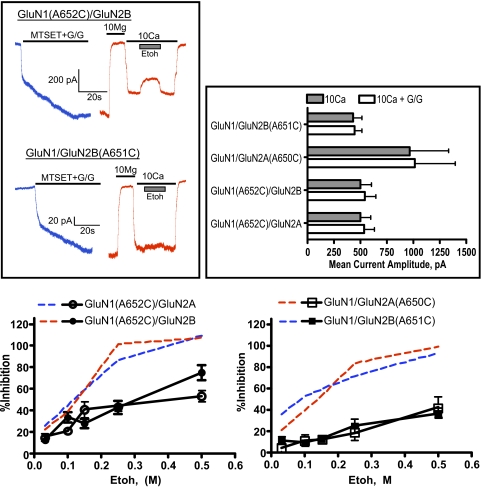
To investigate the ethanol sensitivity of constitutively open A7C mutants, cells were first patched and then exposed to a solution containing 0.4 mM MTSET and glutamate and glycine (both at 10 μM). As shown in Fig. 3A, coapplication of MTSET and agonists results in large sustained currents that persist after washout of the agonist/MTSET solution. MTSET-treated A7C mutants can be blocked by magnesium, and solutions containing either 10 mM MgCl2 or CaCl2 were used to control receptor activity. To verify that A7C receptors were fully activated after MTSET/agonist treatment, cells were exposed to solutions containing just the elevated CaCl2 or elevated CaCl2 plus glutamate and glycine (30 μM each). As shown in Fig. 3B, for each subunit combination tested, current amplitudes in MTSET-treated cells were similar whether agonists were present or not. The effect of ethanol on MTSET/agonist-treated A7C mutants was determined by switching between solutions containing magnesium or calcium plus varying concentrations of ethanol. As shown by the traces in Fig. 3A and the dose-response curves in Fig. 3C, the magnitude of ethanol inhibition of MTSET-treated receptors (solid lines) was reduced compared with that of non-MTSET-treated A7C mutants (dashed lines), and this effect was subunit dependent. For MTSET-treated GluN1(A652C)/GluN2A and GluN1(A652C)/GluN2B receptors, estimated ethanol IC50 values were 370.8 mM (95% CI 249.7–550.6) and 256.2 mM (95% CI 197.8–331.9), respectively. These values represent a 3.7- and 2.2-fold increase, respectively over those calculated from agonist-dependent currents. A more dramatic reduction in ethanol sensitivity was observed for MTSET-treated GluN2 A7C receptors. Estimated ethanol IC50 values for these receptors were 714.6 mM (95% CI 383.5–1332) for GluN1/GluN2A(A650C) and 958 mM (95% CI 494.2–1857) for GluN1/GluN2B(A651C). These values represent a rightward shift of approximately 7- and 14-fold, compared with values determined for agonist-evoked currents. Note that these IC50 values are only estimates because ethanol inhibition of MTSET-treated GluN2 mutants did not reach 50% even at 500 mM, the highest concentration that could be tested without compromising seal integrity and holding current. MTSET treatment itself had no significant effect on ethanol inhibition of wild-type receptors activated by glutamate and glycine (data not shown). As described above, there was a clear difference in the ethanol sensitivity of currents from MTSET-treated GluN1 and GluN2 A7C mutants. To determine whether these effects were additive, the ethanol inhibition of single and double A7C mutants was determined after treatment of cells with MTSET. Ethanol (100 mM) inhibited GluN1(A651C)/GluN2A receptors by 24.9% (±2.0; n = 8), whereas currents recorded from GluN1/GluN2A(650C) receptors showed an 11.6% (±4.9; n = 18) reduction. In the double GluN1(A651C)/GluN2A(A650C) mutant, ethanol (100 mM) inhibited spontaneous currents by 9.2% (±1.2; n = 6), a value similar to that obtained for the GluN1/GluN2A(A650C) mutant alone.
Fig. 3.
Ethanol (Etoh) inhibition of spontaneously active cysteine-substituted A7 mutants. A, traces show representative currents from single cells expressing A7C mutants [top, GluN1(A652C)/GluN2B; bottom, GluN1/GluN2B(A651C)]. In each set of traces, cells were first exposed to a solution containing MTSET (0.4 mM) plus glutamate/glycine (10 μM each; blue traces). After washout of the MTSET/agonist solution, currents were blocked by a solution containing 10 mM MgCl2 (red traces). Currents were then activated with a solution containing 10 mM CaCl2 (solid line) during which ethanol (100 mM) was applied (▩). B, current amplitudes (mean ± S.E.M.; n = 6–7 cells/group) from MTSET-treated cells in the absence (▩) or presence (□) of glutamate/glycine (30 μM each). C, dose-response curve for ethanol inhibition of spontaneously active cysteine-substituted A7 mutants. Data represent the mean ± S.E.M. from 4 to 21 cells for each receptor combination. Dashed lines are from Fig. 2 and represent ethanol inhibition before MTSET treatment and are shown for comparison. Estimated ethanol IC50 values for MTSET cells were: 370.8 mM GluN1(A652C)/GluN2A, 256.2 mM GluN1(A652C)/GluN2B, 714.6 mM GluN1/GluN2A(A650C), and 958 mM GluN1/GluN2B(A651C). See text for more details.
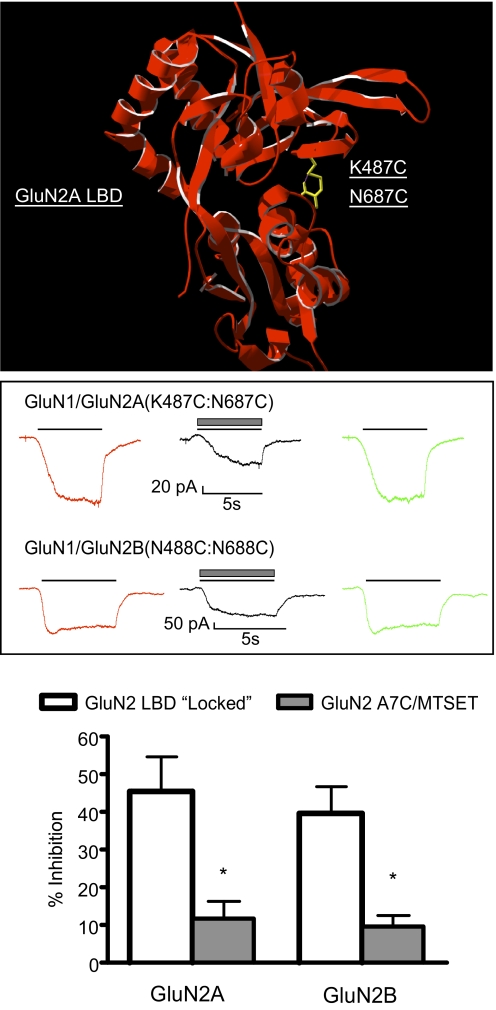
The results described above show that ethanol had less effect on agonist-independent currents in cells expressing A7-modified GluN2 subunits than on those elicited by agonists. Whether this holds true for other constitutively open NMDA receptors is not known. To test this possibility, we used mutants with cysteines introduced at key residues in the extracellular LBD as illustrated in Fig. 4A, which shows the structure of the GluN2A LBD with residues 487 (lysine) and 687 (asparagine) replaced with cysteines. During spontaneous transitions between open and closed cleft conformations, these cysteine residues form disulfide bonds and covalently lock the LBD in a state that mimics glutamate binding (Blanke and VanDongen, 2008a; Kussius and Popescu, 2010). In the absence of any added agonists, both GluN1/GluN2A(K487C:N687C) and GluN1/GluN2B(K488C:N688C) mutants generated reproducible currents (mean ± S.E.M.; 134.8 ± 21.1 and 178.8 ± 22.0 pA, respectively) when switching from the 10 mM MgCl2 solution to one containing 10 mM CaCl2 (Fig. 4B). Ethanol (100 mM) inhibited these responses by approximately 45 to 50% in both subunit combinations (Fig. 4C), a value similar to that in subunits expressing the GluN1(A7R) mutant (Fig. 1). A similar degree of inhibition of these receptors was observed when recordings were performed in the presence of 10 μM glycine, the agonist for the unmodified GluN1 subunit [percent inhibition by 100 mM ethanol, 47.4 ± 7.9; n = 9; GluN1/GluN2A(K487C:N687C)]. For comparison, Fig. 4C also shows data for MTSET-treated GluN2(A7C) receptors and demonstrates the large difference in ethanol inhibition between the two classes of constitutively open receptors.
Fig. 4.
Ethanol inhibition of NMDA receptors with locked ligand-binding domains. A, model of the LBD of the GluN2A subunit showing cysteines substituted for lysine at position 487 (K487C) and asparagine at position 687 (N687C). The proximity of these cysteine-substituted resides locks the LBD in the closed (activated) conformation. B, traces show representative currents from single cells expressing locked LBD mutants [top, GluN1/GluN2A(K487C:N687C); bottom, GluN1/GluN2B(N488C:N688C)]. Currents were activated by switching from a recording solution containing 10 mM MgCl2 to one containing 10 mM CaCl2 (solid line, red and green traces) alone or with 100 mM ethanol (▩, black trace). C, summary of effects of 100 mM ethanol on spontaneous currents from locked LBD mutants (□). Ethanol inhibition of MTSET-treated A7C mutants is shown for comparison (▩; taken from Fig. 3). Data are the mean ± S.E.M. from 7 to 18 cells for each receptor combination. *, value significantly different from that for the corresponding locked LBD GluN2 mutant; unpaired t test (p < 0.01).
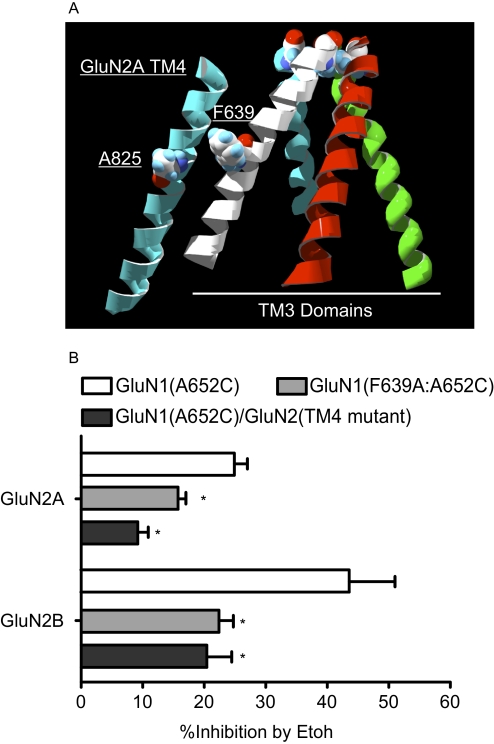
Results from previous studies in this laboratory and others show that the ethanol inhibition of agonist-dependent NMDA receptors is reduced by a phenylalanine to alanine substitution in the TM3 domain of GluN1 (F639A) or an alanine to tryptophan mutation in the TM4 domain of GluN2A (A825W) (Ronald et al., 2001; Honse et al., 2004; Smothers and Woodward, 2006). In the model of the TM domains shown in Fig. 5A, these residues are within 10 Å of one another and may be involved along with other residues in defining an alcohol-sensitive site on the receptor. Because MTSET-treated GluN1(A7C) mutants retained some sensitivity to ethanol (Fig. 2 and IC50 values cited above), we tested whether adding the TM3/TM4 mutations would reduce ethanol inhibition of these receptors. As summarized in Fig. 5B, GluN1(A652C)/GluN2A and GluN1(A652C)/GluN2B receptors were inhibited by 100 mM ethanol. This action was reduced when the GluN1 F639A mutation was added to generate the double mutant GluN1(F639A:A652C). Likewise, expressing the GluN1(A652C) subunit with a TM4 GluN2 mutant (A825W for GluN2A; G826W for GluN2B) also significantly reduced the inhibition of spontaneous currents by 100 mM ethanol.
Fig. 5.
Effect of TM3 and TM4 domain mutations on ethanol (Etoh) inhibition of spontaneously active A7C NMDA receptors. A, model of TM domains of the GluN1/GluN2A receptor showing TM3 helices (GluN1, white/green; GluN2A, blue/red) and TM4 helix (blue) from the GluN2A subunit. Other TM domains have been removed for clarity. Space-filling side groups are shown for the A7 position in the TM3 helices (top), phenylalanine in TM3 (F639) of GluN1, and alanine in TM4 (A825) of GluN2A. B, summary of effects of TM3 and TM4 mutations on ethanol inhibition of A7C mutants. Data are percent inhibition (mean ± S.E.M.; n = 6–13 for each group) by 100 mM ethanol from cells expressing GluN1(A652C)/GluN2 receptors (□), GluN1(F639A:A652C)/GluN2 receptors (▩), or GluN1(A652C)/GluN2(TM4 mutant; ■). For GluN2A, the TM4 mutation is A825W, whereas for GluN2B it is G826W. *, value significantly different from the value for GluN1(A652C)/GluN2; analysis of variance with Dunnett's multiple comparison test (p < 0.05).
Discussion
The results of this study show that ethanol inhibition of constitutively open NMDA receptors varies in a subunit-dependent and domain-specific fashion. Thus, receptors expressing A7R/C GluN2 subunits showed a marked reduction in ethanol sensitivity compared with that of wild-type receptors or those modified only on GluN1 subunits. The reduction in ethanol inhibition in cysteine-modified GluN2 receptors was not a direct result of the amino acid substitution at A7 because these mutants showed normal ethanol inhibition before treatment with MTSET. MTSET treatment has also been shown to reduce xenon inhibition of A7C GluN2 mutants expressed in Neuro2A cells (Weigt et al., 2008). In the present study, the reduced ethanol sensitivity of constitutively open NMDA receptors was specific for the A7 mutants because cysteine-modified LBD mutants were also active in the absence of agonists but showed robust inhibition by ethanol. Finally, mutations in GluN1 TM3 and GluN2 TM4 domains previously shown to reduce ethanol inhibition of agonist-evoked currents also blunted ethanol inhibition of A7C-modified NMDA receptors.
As noted above, mutants with a locked LBD showed appreciable inhibition by ethanol. This is perhaps not surprising because previous studies suggested that these receptors behave similarly to those activated by agonists. For example, Kussius and Popescu (2010) reported that the number of kinetic states measured in single-channel recordings was the same for both agonist-activated wild-type receptors and those with cross-linked LBDs. Likewise, receptors with cross-linked LBDs displayed a degree of proton inhibition similar to that of wild-type receptors (Blanke and VanDongen, 2008a). These findings suggest that artificially inducing cleft closure via disulfide bonds closely mimics agonist binding and confirms previous reports that ethanol inhibition of NMDA receptors does not involve a direct interaction with the agonist binding site. In contrast to the LBD mutants, ethanol inhibition was altered in A7-modified receptors, and the magnitude of this effect was influenced by which subunit was modified. Although constitutively open receptors could be generated by expression of either A7-substituted GluN1 or GluN2 subunits, GluN2 mutants showed a more dramatic reduction in ethanol inhibition. This finding could reflect differences in the site of action for ethanol on the two subunits or dissimilarities in the structural or functional role of GluN1 and GluN2 subunits in controlling receptor function.
Previous studies suggested that subunit makeup is one factor that regulates the overall ethanol sensitivity of NMDA receptors and subunit-dependent changes in ethanol inhibition have been reported for GluN2 subunits although not all reports show the same rank order of sensitivity (Lovinger et al., 1989; Kuner et al., 1993; Masood et al., 1994; Mirshahi and Woodward, 1995; Otton et al., 2009). Ethanol inhibition is also influenced by splice variants of GluN1 (Jin and Woodward, 2006), whereas GluN3 appears to modify ethanol sensitivity under certain conditions (Jin et al., 2008). In addition, ethanol inhibition can be reduced by introducing mutations into either GluN1 (Ronald et al., 2001) or GluN2 (Ren et al., 2003; Honse et al., 2004) subunits, suggesting that neither subunit is fully responsible for conferring ethanol sensitivity to the receptor. With respect to constitutively open NMDA receptors, Chang and Kuo (2008) substituted a variety of amino acids at the A7 position in either GluN1 or GluN2B and showed that even single tryptophan mutants (carrying the largest side chain) displayed only 10 to 30% tonic activity in the absence of agonists. In contrast, mimicking the thiol-modified cysteine by substituting arginine at position A7 in GluN2B resulted in receptors with a constitutive open index of 96%. The homologous substitution in GluN1 produced receptors with a slightly lower open index of 71%, whereas A7C mutants of both GluN1 and GluN2 appear to be fully active after thiol modification (Yuan et al., 2005; Chang and Kuo, 2008). These findings suggest that the difference in ethanol sensitivity between A7-modified GluN1 and GluN2 receptors is probably not due to subunit-dependent alterations in the degree of receptor activation. Results from previous cysteine accessibility studies suggested that the vertical orientation of the TM3 domain of the GluN1 subunit may be offset in comparison with the GluN2C subunit (Sobolevsky et al., 2007) and thus impart different effects on channel function. Whether such an arrangement occurs in GluN1/GluN2A receptors is not known, although the GluA2 crystal structure and GluN1/GluN2A model generated in this study suggest that these domains occupy similar positions at least in the closed state (Sobolevsky et al., 2009). However, it is clear that there are some subunit-dependent differences in the function of these residues, because in the present study A7R GluN2B receptors were active in the absence of agonists, whereas A7R GluN2A mutants did not show appreciable spontaneous currents. Because GluN2B-containing receptors have a higher affinity for glycine than GluN2A-containing receptors, they may be more easily activated by residual amounts of glycine that are present in the recording solution. Nonetheless, ethanol had a greater effect on A7-modified GluN1 receptors than on those containing altered GluN2B subunits. Although the mechanism underlying this difference remains unclear, it may reflect the distinct roles the two subunits play in controlling receptor activity. GluN1 subunits are modulatory because the binding of glycine, although critical for receptor activity, is not sufficient to induce significant channel activity in the absence of glutamate. In addition, although glycine transporters appear to maintain concentrations of glycine below saturating levels in the synapse, in brain slices, significant NMDA receptor currents are evoked in the absence of added glycine, suggesting that a significant fraction of GluN1 subunits are occupied by glycine even during periods of quiescence (Chen et al., 2003). In contrast, GluN2 subunits bind the neurotransmitter glutamate, whose release into the synaptic cleft requires activity-dependent exocytosis. The subunit-dependent alteration in ethanol inhibition of A7R/C mutants may reflect these fundamental differences in subunit function and would be consistent with the dominant role that GluN2 subunits play in determining receptor kinetics and pharmacological properties of NMDA receptors (Kussius and Popescu, 2010).
The finding that A7C-modified receptors are less sensitive to ethanol suggests that mutations that shift the receptor's activation profile toward greater channel opening also reduce the ability of ethanol to inhibit channel function. This finding is consistent with an allosteric mode of action for ethanol and mirrors findings from other studies demonstrating a coupling between TM domains and the potency of allosteric modulators of channel function. For example, substituting tyrosine or glutamine at GluN1 A7 enhanced glycine sensitivity of GluN2A-containing receptors and resulted in significant currents in the absence of agonists (Blanke and VanDongen, 2008b). These mutants as well as MTSEA-treated A7C receptors also show dramatically reduced sensitivity to protons and zinc that exert their effects via an interaction with the amino-terminal domain of the receptor (Yuan et al., 2005; Blanke and VanDongen, 2008b). The reduced effect of allosteric modulators on A7-modified receptors is thought to reflect an increase in open probability that occurs after modification of the A7 residue. In MTS-treated A7C receptors, open probability is essentially at its maximum and thus agents that slow channel opening such as protons and zinc have limited opportunity to act (Yuan et al., 2005). A similar effect could explain the reduced effect of ethanol on these channels because Wright et al. (1996) showed that although 200 mM ethanol reduced mean open time and frequency of bursts, it did not introduce new closed times. Thus, enhancing gating parameters via A7 mutations might be expected to generate receptors that are less sensitive to ethanol-induced changes in channel function. It should be noted that although the actions of various allosteric modulators including zinc, protons, and ifenprodil require the amino-terminal domain of the receptor, ethanol inhibition is unaltered in NMDA receptors lacking this domain (Smothers et al., 2009).
Changes in channel behavior by A7 mutations may also help explain the reduction in ethanol sensitivity observed when additional TM3/TM4 mutations were added to the A7C modified receptors. As mentioned previously, findings from this laboratory and that of Peoples identified specific amino acids in TM3 (F639) of the GluN1 subunit and TM4 (A825) of the GluN2A subunit that reduced the inhibition of recombinant NMDA receptors by ethanol (Ronald et al., 2001; Honse et al., 2004) and volatile anesthetics (Ogata et al., 2006). Although these amino acids may represent physical sites of action for these compounds, mutations of these residues may also induce changes in receptor gating or function that then affect or occlude ethanol action. Both mutants appear to alter normal receptor function because whole-cell currents from GluN1(F639A) mutants show enhanced sensitivity to glycine (Ronald et al., 2001; Smothers and Woodward, 2006) and currents from GluN2A(A825W) and GluN1(F639A) mutants display changes in single channel parameters consistent with enhanced gating (Honse et al., 2004; J. Woodward, unpublished observations). These effects are perhaps not surprising given the critical role of TM3 domains in gating and the close association of GluN1 TM3 with the TM4 domain of the neighboring GluN2 subunit (Sobolevsky et al., 2009). Changes in receptor gating induced by these TM3/TM4 mutations may add to those produced by the modified A7 residue, leading to channels that have very low sensitivity to ethanol.
In summary, the findings of this study have demonstrated that although ethanol inhibits spontaneously active NMDA receptors, the magnitude of this effect is critically dependent on how these channels are activated. These results also support previous findings suggesting that ethanol probably acts at a unique site on the receptor that is not shared with other allosteric modulators.
Acknowledgments
We thank Lalitha Kannan for excellent technical assistance and G. Popescu for supplying the GluN2A locked LBD mutant cDNA.
This work was supported by the National Institutes of Health National Institute on Alcohol Abuse and Alcoholism [Grants R37-AA009986, R01-AA013378].
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
- NMDA
- N-methyl-d-aspartate
- LBD
- ligand-binding domain
- TM
- transmembrane
- MTSET
- (2-(trimethylammonium)ethyl methanethiosulfonate)
- AP5
- 2-amino-5-phosphonovaleric acid
- CI
- confidence interval.
Authorship Contributions
Participated in research design: Xu, Smothers, Trudell, and Woodward.
Conducted experiments: Xu.
Contributed new reagents or analytic tools: Smothers and Trudell.
Performed data analysis: Xu, Trudell, and Woodward.
Wrote or contributed to the writing of the manuscript: Xu, Smothers, Trudell, and Woodward.
References
- Blanke ML, VanDongen AM. (2008a) Constitutive activation of the N-methyl-d-aspartate receptor via cleft-spanning disulfide bonds. J Biol Chem 283:21519–21529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanke ML, VanDongen AM. (2008b) The NR1 M3 domain mediates allosteric coupling in the N-methyl-d-aspartate receptor. Mol Pharmacol 74:454–465 [DOI] [PubMed] [Google Scholar]
- Chang HR, Kuo CC. (2008) The activation gate and gating mechanism of the NMDA receptor. J Neurosci 28:1546–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Muhlhauser M, Yang CR. (2003) Glycine tranporter-1 blockade potentiates NMDA-mediated responses in rat prefrontal cortical neurons in vitro and in vivo. J Neurophysiol 89:691–703 [DOI] [PubMed] [Google Scholar]
- Dopico AM, Lovinger DM. (2009) Acute alcohol action and desensitization of ligand-gated ion channels. Pharmacol Rev 61:98–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Gouaux E. (2003) Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J 22:2873–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 [DOI] [PubMed] [Google Scholar]
- Honse Y, Ren H, Lipsky RH, Peoples RW. (2004) Sites in the fourth membrane-associated domain regulate alcohol sensitivity of the NMDA receptor. Neuropharmacology 46:647–654 [DOI] [PubMed] [Google Scholar]
- Jin C, Smothers CT, Woodward JJ. (2008) Enhanced ethanol inhibition of recombinant N-methyl-d-aspartate receptors by magnesium: role of NR3A subunits. Alcohol Clin Exp Res 32:1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Woodward JJ. (2006) Effects of 8 different NR1 splice variants on the ethanol inhibition of recombinant NMDA receptors. Alcohol Clin Exp Res 30:673–679 [DOI] [PubMed] [Google Scholar]
- Jones KS, VanDongen HM, VanDongen AM. (2002) The NMDA receptor M3 segment is a conserved transduction element coupling ligand binding to channel opening. J Neurosci 22:2044–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob G, Le Moal M. (2006) Neurobiology of Addiction. Academic Press, London [Google Scholar]
- Kuner T, Schoepfer R, Korpi ER. (1993) Ethanol inhibits glutamate-induced currents in heteromeric NMDA receptor subtypes. NeuroReport 5:297–300 [DOI] [PubMed] [Google Scholar]
- Kussius CL, Popescu GK. (2010) NMDA receptors with locked glutamate-binding clefts open with high efficacy. J Neurosci 30:12474–12479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Landman MT, Albuquerque EX. (1989) Ethanol potentiates and blocks NMDA-activated single-channel currents in rat hippocampal pyramidal cells. FEBS Lett 247:61–67 [DOI] [PubMed] [Google Scholar]
- Lovinger DM, White G, Weight FF. (1989) Ethanol inhibits NMDA-activated ion current in hippocampal neurons. Science 243:1721–1724 [DOI] [PubMed] [Google Scholar]
- Maldve RE, Zhang TA, Ferrani-Kile K, Schreiber SS, Lippmann MJ, Snyder GL, Fienberg AA, Leslie SW, Gonzales RA, Morrisett RA. (2002) DARPP-32 and regulation of the ethanol sensitivity of NMDA receptors in the nucleus accumbens. Nat Neurosci 5:641–648 [DOI] [PubMed] [Google Scholar]
- Masood K, Wu C, Brauneis U, Weight FF. (1994) Differential ethanol sensitivity of recombinant N-methyl-d-aspartate receptor subunits. Mol Pharmacol 45:324–329 [PubMed] [Google Scholar]
- Mirshahi T, Woodward JJ. (1995) Ethanol sensitivity of heteromeric NMDA receptors: effects of subunit assembly, glycine and NMDAR1 Mg2+-insensitive mutants. Neuropharmacology 34:347–355 [DOI] [PubMed] [Google Scholar]
- Ogata J, Shiraishi M, Namba T, Smothers CT, Woodward JJ, Harris RA. (2006) Effects of anesthetics on mutant N-methyl-d-aspartate receptors expressed in Xenopus oocytes. J Pharmacol Exp Ther 318:434–443 [DOI] [PubMed] [Google Scholar]
- Otton HJ, Janssen A, O'Leary T, Chen PE, Wyllie DJ. (2009) Inhibition of rat recombinant GluN1/GluN2A and GluN1/GluN2B NMDA receptors by ethanol at concentrations based on the US/UK drink-drive limit. Eur J Pharmacol 614:14–21 [DOI] [PubMed] [Google Scholar]
- Peoples RW, Stewart RR. (2000) Alcohols inhibit N-methyl-d-aspartate receptors via a site exposed to the extracellular environment. Neuropharmacology 39:1681–1691 [DOI] [PubMed] [Google Scholar]
- Peoples RW, Weight FF. (1992) Ethanol inhibition of N-methyl-d-aspartate-activated ion current in rat hippocampal neurons is not competitive with glycine. Brain Res 571:342–344 [DOI] [PubMed] [Google Scholar]
- Rabe CS, Tabakoff B. (1990) Glycine site-directed agonists reverse the actions of ethanol at the N-methyl-d-aspartate receptor. Mol Pharmacol 38:753–757 [PubMed] [Google Scholar]
- Rambhadran A, Gonzalez J, Jayaraman V. (2010) Subunit arrangement in N-methyl-d-aspartate (NMDA) receptors. J Biol Chem 285:15296–15301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Lindahl E, Wallner B. (2010) Model quality assessment for membrane proteins. Bioinformatics 26:3067–3074 [DOI] [PubMed] [Google Scholar]
- Ren H, Honse Y, Peoples RW. (2003) A site of alcohol action in the fourth membrane-associated domain of the N-methyl-d-aspartate receptor. J Biol Chem 278:48815–48820 [DOI] [PubMed] [Google Scholar]
- Ronald KM, Mirshahi T, Woodward JJ. (2001) Ethanol inhibition of N-methyl-d-aspartate receptors is reduced by site-directed mutagenesis of a transmembrane domain phenylalanine residue. J Biol Chem 276:44729–44735 [DOI] [PubMed] [Google Scholar]
- Salussolia CL, Prodromou ML, Borker P, Wollmuth LP. (2011) Arrangement of subunits in functional NMDA receptors. J Neurosci 31:11295–11304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers C, Jin C, Woodward JJ. (2009) Effect of N-terminal domains of NR1 and NR2 subunits on ethanol inhibition of recombinant NMDA receptors. Alcohol Clin Exp Res 33:148A [Google Scholar]
- Smothers CT, Woodward JJ. (2006) Effects of amino acid substitutions in transmembrane domains of the NR1 subunit on the ethanol inhibition of recombinant N-methyl-d-aspartate receptors. Alcohol Clin Exp Res 30:523–530 [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Prodromou ML, Yelshansky MV, Wollmuth LP. (2007) Subunit-specific contribution of pore-forming domains to NMDA receptor channel structure and gating. J Gen Physiol 129:509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. (2009) X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462:745–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. (2010) Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 62:405–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigt HU, Adolph O, Georgieff M, Georgieff EM, Föhr KJ. (2008) Evidence that xenon does not produce open channel blockade of the NMDA receptor. J Neurophysiol 99:1983–1987 [DOI] [PubMed] [Google Scholar]
- Woodward JJ. (2000) Ethanol and NMDA receptor signaling. Crit Rev Neurobiol 14:69–89 [DOI] [PubMed] [Google Scholar]
- Woodward JJ, Gonzales RA. (1990) Ethanol inhibition of N-methyl-d-aspartate-stimulated endogenous dopamine release from rat striatal slices: reversal by glycine. J Neurochem 54:712–715 [DOI] [PubMed] [Google Scholar]
- Wright JM, Peoples RW, Weight FF. (1996) Single-channel and whole-cell analysis of ethanol inhibition of NMDA-activated currents in cultured mouse cortical and hippocampal neurons. Brain Res 738:249–256 [DOI] [PubMed] [Google Scholar]
- Xu M, Smothers CT, Woodward JJ. (2010) Effects of ethanol on phosphorylation site mutants of recombinant N-methyl-d-aspartate receptors. Alcohol 45:373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Erreger K, Dravid SM, Traynelis SF. (2005) Conserved structural and functional control of N-methyl-d-aspartate receptor gating by transmembrane domain M3. J Biol Chem 280:29708–29716 [DOI] [PubMed] [Google Scholar]