SUMMARY
The NF-E2-related factor 2 (Nrf2) is a key transcriptional regulator of antioxidant defense and detoxification. To directly monitor stabilization of Nrf2, we fused its Neh2 domain, responsible for the interaction with its nucleocytoplasmic regulator, Keap1, to firefly luciferase (Neh2-luciferase). We show that Neh2 domain is sufficient for recognition, ubiquitination, and proteasomal degradation of Neh2-luciferase fusion protein. The Neh2-luc reporter system allows direct monitoring of the adaptive response to redox stress and classification of drugs based on the time course of reporter activation. The reporter was used to screen the Spectrum library of 2000 biologically active compounds to identify activators of Nrf2. The most robust and yet nontoxic Nrf2 activators found—nordihydroguaiaretic acid, fisetin, and gedunin—induced astrocyte-dependent neuroprotection from oxidative stress via an Nrf2-dependent mechanism.
INTRODUCTION
Oxidative stress is a major contributor to aging, insulin resistance, and neurodegeneration. An emergent strategy for restoring redox homeostasis involves activation of the transcription factor, Nrf2 (nuclear factor erythroid 2-related factor 2), a member of the cap’n’collar family of basic leucine zipper transcription factors that regulates a coordinated adaptive gene program (Moi et al., 1994). Indeed, activators of the Nrf2 response are beneficial for the treatment and prevention of chronic degenerative diseases, whereas inhibitors of Nrf2 may help to fight cancer (Calabrese et al., 2008; Hayes and McMahon, 2009; Lau et al., 2008). A major challenge in the development of effective Nrf2 activators is to identify those that lead specifically to Nrf2 stabilization and consequent promoter activation without imposing general oxidative/electrophilic stress.
Nrf2 is sequestered under homeostatic conditions by binding to its inhibitory protein, Keap1 (Kelch-like ECH-associated protein-1) (Motohashi and Yamamoto, 2004; Itoh et al., 1999). Keap1 serves as a bridge between Nrf2 and the Cul3-Rbx1 E3 ubiquitin ligase, leading to Nrf2 ubiquitination and thereby targeting Nrf2 for degradation by the 26S proteasome (Kobayashi et al., 2004; Cullinan et al., 2004; Zhang et al., 2004). Upon exposure to oxidative stress, xenobiotics, or electrophilic compounds, the Nrf2 protein is released from its complex with Keap1 and translocates to the nucleus. There, it forms heterodimers with other transcription regulators, such as small Maf proteins, and induces the expression of antioxidant genes controlled by the antioxidant response element (ARE) (Kaspar et al., 2009).
Nrf2 is composed of Neh1–Neh6 domains, among which Neh2 is the putative negative regulatory domain that interacts with Keap1, Neh4 and Neh5 are transactivation domains, and Neh1 is the binding domain for ARE (Tong et al., 2006b). The functional domains of Keap1 are the Broad complex, Tramtrack and Bric-a-Brac (BTB), the intervening region (IVR), the double glycine repeats domain (DGR), and the C-terminal region (CTR) (Tong et al., 2006b). Two motifs in the Neh2 domain, i.e., ETGE and DLG, are recognized by the Keap1 homodimer in a hinge-latch mode (Tong et al., 2006a, 2006b, 2007). Keap1 mediates poly-ubiquitination of the positioned lysines within the central α helix of the Neh2 domain under homeostatic conditions. Under oxidative/electrophilic stress, reactive cysteines within Keap1 are modified, and thus, Keap1 undergoes conformational changes that lead to the detachment of the weak-binding DLG, resulting in Nrf2 stabilization. However, debate remains as to whether Nrf2 is completely released from its complex with Keap1 (Zhang, 2006) or not. Nrf2 activators identified so far are represented by potent alkylating agents (Dinkova-Kostova et al., 2004) and redox active compounds like diphenols, aminophenols, and phenylene diamines, the precise mechanism of action of which is controversial. Recent data show an enhanced effect of these compounds in the presence of exogenously added copper (Wang et al., 2010).
Current techniques for monitoring Nrf2 activation include the ARE-luciferase (Moehlenkamp and Johnson, 1999), Nrf2 responsive element-luciferase (Westerink et al., 2010), or ARE-human placental alkaline phosphatase reporter systems (Son et al., 2010). Recently, a GFP fusion protein with the Nrf2 ZIP domain was utilized to study Nrf2 nuclear translocation (Theodore et al., 2008), whereas GFP fusion with the C. elegans Nrf2 analog was used to analyze Nrf2 activation by proteasomal dysfunction (Kahn et al., 2008). The ARE-GFP reporter assay was used to screenthe library of 2000 biologically active compounds (Spectrum library), and 45 hits were identified (Shaw et al., 2010), with andrographolidebeing the most potent. The use of ARE-luciferase reporter for high throughput screening (HTS) purposes has been recently published (Hur et al., 2010). The screen of 1.5 million compounds resulted in discovery of novel alkylating agents targeting Cys 151 in Keap1 as well as a dozen other cellular proteins, including phosphatase 2a, and HDAC1 and HDAC2 (Hur et al., 2010). Here, we present a new reporter construct, the Neh2 domain fused to a luciferase gene (Neh2-luc), as a powerful tool for the HTS and real-time monitoring of Nrf2 activation. We also demonstrate the utility of the Neh2-luc model to identify and classify compounds capable of inducing Nrf2-specific astrocyte-dependent neuroprotection from oxidative stress.
RESULTS
Construction and Validation of Neh2-luc Reporter
The PCMV-driven Neh2-luc reporter supports the constitutive, intracellular synthesis of a fusion protein composed of the human Neh2 domain (1–97 aa) and firefly luciferase. Because the Neh2 domain is known to be sufficient for recognition by the ubiquitin-ligase complex and subsequent ubiquitination of the fusion protein, the recombinant luciferase-labeled protein should undergo proteasomal degradation. The steady-state concentration of the fusion protein should correspond to the equilibrium between its synthesis and degradation (Figure 1A). The background luminescence signal calibrated with recombinant luciferase allows us to estimate the steady-state concentration of the Neh2-luciferase fusion protein: the background is ~15–20 rlu, which corresponds to 0.25–0.33 pg luciferase protein and is more than two orders of magnitude lower than that observed for the cell line expressing wild-type (WT) luciferase under control of the same promoter. The low steady-state luciferase activity (recalculated as 0.6–0.8 nM fusion protein for 30,000 cell/well density and 233 μ3 single-cell volume) suggests that despite forced expression of the Neh2-luciferase fusion protein, it is successfully recognized by the endogenous Keap1-Cul3 complex and almost fully degraded. The findings support prior observations that the Neh2 domain is critical for Keap1 binding and sufficient for recognition and degradation of Neh2-containing fusion protein (Zhang et al., 2004).
Figure 1. Development of Neh2-luc Reporter.
(A) Schematic presentation of reporter functioning.
(B) Time course of the Neh2-luc reporter response to TBHQ compared to that for the commonly used ARE-luc reporter.
(C) Time course of Neh2-luc and HIF ODD-luc reporter responses to lactacystin showing the lag period shortening with rising concentrations of the proteasomal inhibitor and, thus, confirming the switch of the rate-limiting step from specific recognition to proteasomal degradation. All values are presented as mean ± SEM. Increased expression of Nrf2-regulated genes in the Neh2-luc reporter line as a result of rescue of endogenous Nrf2 in the presence of the overexpressed Neh2-luciferase fusion, and the reporter response to canonical Nrf2 activators—PGJ2, TBHQ, and sulforaphane—in comparison to the absence of any response for HIF ODD-luc reporter (to confirm the specificity of each reporter) are shown in Figure S1.
The overexpressed Neh2-luciferase fusion protein successfully competes with endogenous Nrf2 for Keap1 binding and, thus, rescues endogenous Nrf2 from degradation: the reporter cell line shows a 4- to 6-fold increase in mRNA for Nrf2-regulated genes such as HO-1 and GSLM (see Figure S1A available online). The reporter exemplifies the action of an “ideal Nrf2 activator” that stabilizes endogenous Nrf2 by competing for Keap1 binding, and not by modifying Keap1 chemically. Of note, stabilization of endogenous Nrf2 and the upregulated expression of protective genes may explain the increased stability of the reporter cell line as compared to the original nontransfected cell line.
Canonical activators of Nrf2 such as 15-deoxy-prostaglandin J2 (15d-PGJ2) (Itoh et al., 2004), sulforaphane (Myzak and Dashwood, 2006), and tert-butylhydroquinone (TBHQ) (Moehlenkamp and Johnson, 1999) disrupt the interaction in the Neh2luc-Keap1-Cul3 complex, leading to a measurable increase in luciferase activity (Figure S1B). This effect is not observed for the reporter cell line bearing another construct, HIF ODD-luciferase, where HIF-1α oxygen degradable domain is fused to luciferase (Smirnova et al., 2010), thus indicating the specific character of Neh2-luc reporter response (Figure S1B).
If compared to commonly used ARE-luc reporter, the newly developed one has an obvious advantage to monitor immediate changes upon the addition of Nrf2 activators: the response of ARE-luc reporter to TBHQ is 3 hr delayed (Figure 1B).
The response of both Neh2-luc and HIF ODD-luc reporters to a proteasomal inhibitor is similar (Figure 1C): there is a concentration-dependent delay (lag period) in reporter response. The shortening lag periods observed with rising concentrations of the proteasomal inhibitor provide evidence for the switch of the rate-limiting step from the disruption of the Neh2-Keap1-Cul3 complex to the proteasomal degradation step. The comparison of Neh2-luc and HIF ODD-luc reporter performance with respect to Nrf2 activators (Figure S1B) and proteasomal inhibitors (Figure 1C) proves the specific character of each reporter.
The Neh2-luc reporter system is a novel tool to monitor the direct effect of a particular compound on the first step controlling Nrf2 stability, i.e., Nrf2-Keap1 and/or Keap1-Cul3 interaction. Validation studies further performed using traditional approaches (Figure 2) demonstrate that Keap1 regulates the stability of the Neh2 reporter in the same manner as for endogenous Nrf2: forced expression of Keap1 in the Neh2-luc reporter cell line (Figure S2A) led to a 3.5-fold decrease in the background luminescence (Figure 2A). In contrast, Keap1 reduction by siRNA resulted in a steady-state increase in Neh2-luc reporter activity (Figure 2B) and an induction of transcription of Nrf2-regulated genes in both Neh2-luc and WT-luc-expressing cell lines (Figure S2B). Keap1 depletion had no effect on the levels or activity of a native firefly luciferase expressed under the same CMV promoter, confirming the key role of the Neh2 domain in the Keap1-dependent regulation of the Neh2-luciferase fusion protein (Figure 2C). The results of Keap1 overexpression (Figure 2A) or siRNA-mediated reduction in Keap1 levels (Figure 2B) establish that the stability of the Neh2-luc reporter directly depends on the expression level of Keap1.
Figure 2. Neh2-Luc Reporter Response to Upregulation and Downregulation of Keap1 Levels.
(A) Keap1 overexpression results in a decreased luminescence in Neh2-luc cells transduced by Keap1 adenovirus. The efficiency of transfection of Neh2-luc cell line with FLAG-labeled Keap1-overexpressing adenovirus was 45%–70% as judged by immunostaining with anti-FLAG antibodies (see Figure S2A).
(B) siRNA Keap1 knockdown results in an increased level of luminescence only in Neh2-luc cell line, but not in WT-luc line.
(C) siRNA Keap1 knockdown has no effect on luminescent signal in WT-luc line. All values are presented as mean ± SEM. The siRNA Keap1 knockdown was confirmed by RT-PCR: it decreased levels of Keap1 mRNA and increased levels of mRNA of Nrf2-regulated genes in both Neh2-luc and WT-luc cell lines (see Figure S2B).
In contrast to the previously utilized ARE-based promoter-reporter constructs, the Neh2-luc reporter provides real-time monitoring of Nrf2 stabilization and can be successfully used for HTS purposes (see below) as well as in vivo bioluminescent imaging.
Pilot HTS of Spectrum Library
The reporter cell line was stable for more than 1 year, providing constant readings for all control Nrf2 activators. It has been shown to be suitable for HTS purposes: the results of a pilot screen of the Spectrum library using the Neh2-luc reporter cell line with 10 μM TBHQ as a positive control are presented below. TBHQ has been used in vivo for prophylaxis against ischemic stroke (Shih et al., 2005). TBHQ was chosen among other canonical activators tested because the concentration titration curve had no peaks and showed a saturation plateau (Figure S1B), and thus, was ideal for signal normalization. Induction of luciferase activity is reported throughout as percentage of activation by 10 μM TBHQ.
The screen revealed 224 hits exhibiting Neh2-luc reporter activity equal or higher than 25% of TBHQ; among those, 100 showed activation of at least 75% of that induced by TBHQ. Thus, 5% of biologically active compounds and drugs presented in the Spectrum library are at least 75% as potent as TBHQ in activation of Nrf2. The prevalence of hits may reflect the important role that Nrf2 plays in xenobiotic detoxification of a large number of chemical entities.
As a further test of specificity of the identified Nrf2 activators, we compared our 200 putative Nrf2 activators to almost 30 hits from HTS of the same library found using a HIF1 ODD-luc reporter as described in Smirnova et al. (2010). Upon hydroxylation at proline 564 in normoxia, the ODD-luciferase recruits the E3 Ubiquitin Ligase, von Hippel-Lindau protein, targeting the ODD-luciferase for proteasomal degradation (Smirnova et al., 2010). The observation that the Nrf2 (Neh2-luc) or HIF1 (ODD-luc) screens of the identical 2000 compound library give hits that do not overlap is the strongest evidence for specific chemical control of the stability of both reporters. The findings suggest that the rate-limiting step in reporter activation is determined by Neh2 (of Nrf2) or ODD (of HIF1α), and not by proteasomal degradation. In other words the reporters select unique activators of Nrf2 and HIF1, respectively, and not common inhibitors of proteasomal degradation, as one may have criticized the approach initially.
We found well-known drugs and hormones as potent activators of the Neh2-luc reporter, e.g., minocycline (Kuang et al., 2009), sulindac, auranofin (Kataoka et al., 2001), teniposide, and podophyllotoxin derivatives, all of which showed 200% activation over the canonical TBHQ-induced Neh2-luc response. Purpurogallin carboxylates (Figure 3, Ic), prevalent components of black tea, were extremely potent in activating the reporter up to 500% of TBHQ levels. It is of interest to note that drinking black tea three times a day was recently reported to delay Parkinson’s disease symptom onset by more than 7 years (Kandinov et al., 2009). We found representatives of all structural classes (Figure 3) that were described previously as inducers of the Nrf2-regulated gene nicotinamide quinone oxidoreductase 1 (NQO1) (Dinkova-Kostova et al., 2004). This fact provides additional evidence for reliability of the Neh2-luc reporter, which is capable of identifying all hits reported previously using ARE-luc reporter or those inducing Nrf2-dependent genes.
Figure 3. Structural Formulas of HTS Hits.
Activation effects are shown in percentage (%) for 16 μM of the representative hits. See also Figure S3 for other novel scaffolds found.
The hits included: phenolic antioxidants; diphenols (Figure 3, I); aminophenols or their derivatives, e.g., acetaminophen exhibiting more than 50% activation; phenylene diamines; substituted coumarines, especially those containing adjacent hydroxy groups (Figure 3, II); other cyclic lactones and enones; Michael reaction acceptors such as fumaric, maleic, acrylic, crotonic, ferulic, and caffeic acid derivatives, with bis-salicyl fumarate (Figure 3, IIIa) being the most potent hit in this group (>300% activation); chalcones providing activation up to 400% (Figure 3, IIIc); sappanones and sappanols; flavanones; and flavones (Figure 3, IV), such as 3,7,3′4′-tetrahydroxyflavone, fisetin, and 3,5,7,3′,4′-pentahydroxyflavone, quercetin (showing >300% activation), and isoflavones such as koparin (>200%) and genistein (>100%).
Structure-activity relationship studies for flavones indicate the necessary presence of 3-hydroxy group because 3′,4-dimethoxy-3-hydroxyflavone and kaempferol (3,5,7,4′-tetrahydroxyflavone) are 2.5-fold less effective than quercetin and fisetin. Luteolin (5,7,3′,4′-tetrahydroxyflavone) has an effect similar to kaempferol, and thus, is much less active than fisetin and quercetin, although they all have two adjacent hydroxy groups on a freely rotating phenyl ring. Additionally, double Michael reaction acceptors such as curcumins showing more than 200% activation (Figure 3A, V), dithiolethiones, dimercaptans, and isothiocyanates (Figure 3, VI) came up as hits. Sulforaphane (Figure 3, VIc) is the prototypic activator of Nrf2 (Figure S1A). Heavy metals, such as cadmium and cisplatin, were also hits showing modest activation of 30%–50%.
Of the 45 hits from the ARE-GFP screen of the same library (Shaw et al., 2010), 37 of those were among our hits. The conditions of HTS were very different, in particular the incubation time (24 hr ARE-GFP versus 3 hr Neh2-luc), so some of the hits we missed were likely to induce extremely delayed effects. The lesser number of hits in ARE-GFP screen could reflect both prolonged incubation and lesser sensitivity of the assay: the cell number per well was at least seven times higher and ebselen as a positive control induced only a 3-fold increase in the reporter signal (Shaw et al., 2010) compared to more than 10-fold activation by TBHQ in the case of Neh2-luc reporter (Figure 1B).
Previously Unknown Classes of Nrf2 Activators
The previously unknown classes of hits included:
All members of gedunin/khivorin family (18 compounds) were among the hits (see Figure 3, group IX). The finding of numerous gedunins as hits was unexpected. Moreover, some of the tricyclic hits (Figure 3, group VII) resembled the structure of gedunin very closely. The stereo effects in play are obvious from comparison of tanshinone (Figure 3, VIIIa) and dihydrotanshinone (VIIIb), the major components of danshen, one of the most important traditional Chinese medicines widespread in Asian countries: both compounds have a clear quinone motif, but the change from planar to 3D scaffold leads to a significant increase in the reporter activation. Although one may ascribe the effects of group VII and VIII compounds (Figure 3) exclusively to the presence of neighboring hydroxy groups/quinone moiety, the activation by dihydroabietamide (VIIc) cannot be explained by alkylation or redox-cycling mechanism. The structure-activity relationship within the gedunin/khivorin group (Figure 3, IX) clearly points to the structural effects in play: the most remarkable is the comparison between α- and β-dihydrogedunols that differ only by the orientation of a hydroxy group (activation effects are 40% and 220%, respectively).
Planar Zn2+ chelators such as 8-hydroxyquinoline and chloroacetoxyquinoline (60% activation). The presence of Zn2+-atom in Keap1 was documented for the recombinant protein produced in E. coli, and an estimate for Zn2+ binding constant was on the order of pM (Dinkova-Kostova et al., 2005). We recently identified a number of novel branched oxyquinolines as inhibitors of the HIF prolyl hydroxylases (Smirnova et al., 2010). None of these compounds (which are also zinc chelators with Ki below 200 nM) showed any Neh2-luciferase activation, pointing to specific structural requirements for oxyquinoline zinc chelators as Nrf2 activators. 3-Hydroxyflavone was found as a modest Nrf2 activator and is known to bind zinc better than 5-hydroxyflavone or 3′4′-dihydroxyflavone (Lapouge et al., 2006).
Adenosine, azathioprine, bromonitroindazole were modest hits in our screen: they resemble the recently published structures of Nrf2 inducers supposedly targeting the IVR of Keap1 (Wu et al., 2010) (see Figure S3). In that paper the authors performed virtual screening of chemical databases for putative Nrf2 inducers showing best scores for docking into the 3D model of the Keap1-intervening domain with subsequent verification by ARE-luciferase-based assay (Wu et al., 2010). They found substituted purines with a freely rotating tetrahydrothiophene ring in the seventh position (BM10 and BM31 in Figure S3), with lower potency than sulforaphane (Wu et al., 2010). Of note, the tetrahydrothiophene ring is extremely sensitive to oxidation, and it is not clear to what extent the mechanism of action of these compounds can be ascribed to specific interaction with Keap1.
Time Course of Reporter Activation as a Tool for Hit Classification
As mentioned, the Neh2-luc reporter provides the possibility of real-time monitoring for changes in the stability of Nrf2 in the form of the luciferase-labeled Neh2 domain for the first time. By following the kinetics of reporter activation, one may expect to discriminate the mechanism of action of various Nrf2 activators, i.e., direct activators will exert immediate effects, whereas those acting indirectly will show lag periods of different durations.
The mechanism of Nrf2 activation has been postulated to occur due to the chemical modification of key thiols in Keap1. Accordingly, all alkylating agents tested were hits. The exact mechanism of action of redox-cycling compounds like ortho-or para-dihydroxy-phenols is not known, although they are supposed to undergo oxidation resulting in formation of potential alkylating compounds.
Among well-known classes of hits, particularly those of catechol type, with two adjacent hydroxy groups, e.g., fisetin, quercetin, but not luteolin (class IV, Figure 3), and nordihydroguaiaretic acid (NDGA, class I, Figure 3), demonstrated the best parameters of activation, i.e., the lowest half-activation concentration, the highest amplitude, and the lowest toxicity in the concentration range, providing maximum activation of the reporter. Moreover, in contrast to other hits of the screen, and especially in comparison with the established Nrf2 activators showing a gradual response on a concentration titration curve (Figure S1B), NDGA and fisetin exhibit a very steep concentration response curve (Figures 4A and 4B).
Figure 4. Confirmation of Neh2-luc Fusion Accumulation Using Anti-Luciferase Antibodies.
Concentration dependence of luciferase signal for “switch”-type hits (A and B) and confirmation of fusion protein accumulation by western blot (C). F, 3 hr treatment with 5 μM fisetin; N, 5 μM NDGA; Q, 4 μM quercetin. The control cell lines WT-luc and HIF ODD-luc (Smirnova et al.,2010) did not accumulate luciferase fusion under the same exposure conditions (see Figure S4).
We decided to undertake a separate study to use the kinetics of reporter activation to compare the mechanism of action of our best hits using the Neh2-luc reporter system. In addition to providing a novel categorization of Nrf2 activators, our central interest was to further characterize our best hits, which exhibited a very steep concentration response over a very narrow range of concentrations (Figure 4). An increase in Neh-2 luciferase activity was shown to correspond to the accumulation of the fusion protein monitored by immunoblotting with selective anti-luciferase antibodies after treatment with our most potent hits (Figure 4C). Under basal conditions, no fusion protein was detectable, consistent with a model in which Keap1 binding to the Neh2-luciferase triggers its efficient proteasomal degradation (Figure 4C; Figure S4).
For the comparative studies we selected a number of hits, suspected to work via different mechanisms: TBHQ, ortho-phenylene diamine (oPD), o-catechol, NDGA, quercetin, and fisetin as representatives of redox-cycling compounds; sulforaphane, and pyrithione as alkylating compounds; Cd2+, as a heavy metal of unknown mechanism of action; and geldanamycin, specific inhibitor of Hsp90 working via blockade of ATP-binding site (Obermann et al., 1998), trichostatin A (TSA), a general inhibitor of HDACs resulting in destabilization of Hsp90, and gedunin, which is supposed to disrupt the association of Cdc37 and Hsp90 (Brandt et al., 2008).
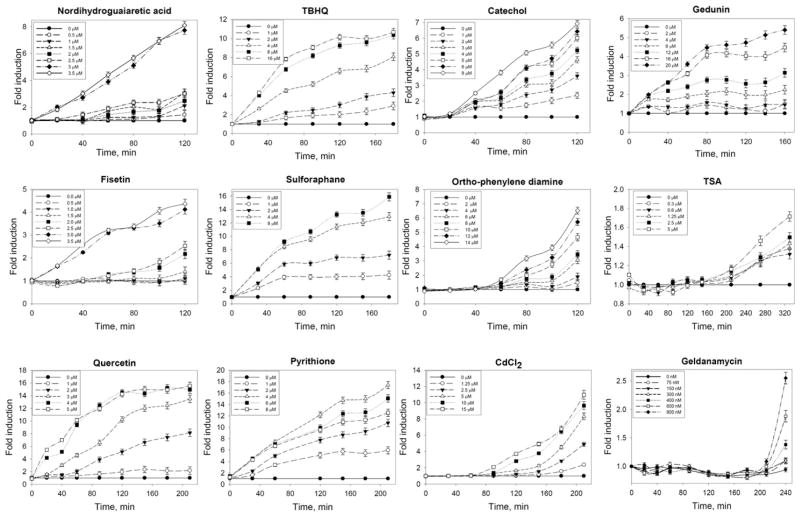
In accord with the time course of reporter activation (Figure 5), we have classified hits into five groups: (1) immediate activation but gradual stabilization over time, like sulforaphane, pyrithione, TBHQ, quercetin, gedunin; (2) gradual stabilization with a barely detectable (20 min) lag period (catechol); (3) gradual stabilization with a short lag period of 40–50 min (oPD); (4) stabilization after a prolonged lag period, 1–3 hr (Cd2+, TSA, geldanamycin); and (5) activation via a switch or receptor, i.e., showing sharp conversion from almost no effect to full activation over a narrow concentration range (NDGA and fisetin—the best hits in the screen).
Figure 5. Classification of HTS Best Hits Based on Kinetics of Reporter Activation.
Switch-type activators (NDGA and fisetin); immediate alkylators (TBHQ, quercetin, sulforaphane, pyrithione); redox-cycling compounds undergoing prior oxidation and showing lag period (catechol, o-phenylene diamine); heavy metals (cadmium) working via inhibition of thiol-disulfide exchange and corresponding enzymes; Hsp90 inhibitors/destabilizers showing prolonged lag period (geldanamycin, TSA); and gedunin. Protein concentration 5.1 ± 0.2 μg per well. All values are presented as mean ± SEM. Keap1-labeling experiments in the presence of selected hits show that only sulforaphane behaves as a potent alkyating agent (see Figure S5).
The similar behavior of TBHQ, sulforaphane, auranofin, pyrithione, and gedunin permits their classification into one group of “alkylators.” Catechol is likely to undergo quick transformation and then also works as an “alkylator.” Apparently, oPD and catechol behave differently: oPD has a clearly defined short lag period, which may reflect the additional modification step of the inducer, such as enzymatic oxidation with copper-dependent enzymes (Wang et al., 2010); oPD is possibly the one working through redox cycling.
The effect of Cd2+ is more than 1 hr delayed, so it either has problems with getting into the cell or, more likely, has an indirect effect on the system via inactivation of thiol-disulfide exchange by inhibiting thioredoxin reductase/thioredoxin system. It is of interest to note that increased concentrations of Cd2+ shorten the lag period, whereas in the case of oPD, the lag period duration barely depends on the inducer concentration.
The activation effect observed with geldanamycin, a selective Hsp90 inhibitor, was rather modest (2- to 3-fold in the range of 0.5–1.5 μM), with toxicity dominating at increased concentrations. A characteristic feature of geldanamycin-induced Neh2-luciferase stabilization was an extremely prolonged (up to 3 hr) lag period, similar to that observed for the global histonedeacetylase inhibitor, TSA (Figure 5). Of TSAs numerous effects, it is known to lead to acetylation of Hsp90 and inhibition of its chaperone activity. The long lag period of geldanamycin and TSA-induced activation suggest Nrf2 activation as a downstream effect of Hsp90 inhibition. Although gedunin has also been described as an Hsp90 inhibitor, the absence of a lag period in gedunin-induced activation of Neh2-luciferase (Figure 5) likely reflects direct disruption of Neh2-Keap1 association. The titration behavior is similar to the effect of alkylators of Cys151 in Keap1, except the magnitude of the effect was much lower, and the activation plateau is clearly observed at low, nonsaturated concentrations of gedunin.
As an independent approach to test the mechanism of action of selected hits in comparison with the well-known controls, we performed Keap1-labeling experiments (Figures S5A and S5B) in the presence of sulforaphane (positive control, alkylating agent), TBHQ (positive control, redox-cycling compound), fisetin (hit), quercetin (hit), gedunin (hit), geldanamycin (negative control, working via Hsp90), and ciclopirox (negative control, not a hit). All hits induce upregulation of Nrf2 target genes (Figure S5C). As one may expect, only sulforaphane being a potent alkylating agent shows a decent competition for the overexpressed Keap1, whereas TBHQ, fisetin, quercetin, and gedunin (redox-cycling compounds) demonstrate very modest competition (Figure S5B), indicative of either reversible modification of Keap1 cysteines or preference for particular cysteine residues in Keap1. The labeling approach does not allow one to discriminate between the mechanism of action of the hits, whereas the Neh2-luc reporter assay clearly shows that all hits exert immediate effects, although the time course patterns are different in shape and magnitude.
Neuroprotective Effects of the Best Hits
To confirm that the best Neh2-luc activators identified from our screen induce a neuroprotective response, we examined the biological effects of these activators on astrocyte-dependent neuroprotection using an astrocyte-neuron coculture model of oxidative stress. Specifically, Nrf2 activation in astrocytes induces non-cell-autonomous neuroprotection via the transcriptional regulation of genes involved generally in the antioxidant response, including those involved in the biosynthesis, use, and export of the major antioxidant glutathione (GSH) (Shih et al., 2003). Glutamate or homocysteic acid (HCA, glutamate analog) treatment of immature neurons leads to substantial glutathione depletion in neurons and astrocytes and subsequent oxidative stress-induced death of immature neurons; because astrocytes possess ten times as much glutathione as neurons, HCA-treated astrocytes remain viable (Haskew-Layton et al., 2010). Thus, primary cultured astrocytes were pretreated with NDGA, fisetin, or gedunin for 24 hr followed by the addition of adjacent neurons in the presence of the GSH-depleting compound, HCA. Pretreatment of the astrocytes with NDGA, fisetin, or gedunin induced significant neuroprotection (Figures 6A–6C). As expected, all hits induced overexpression of Nrf2 target genes (Figures 6D and 6E) and a corresponding increase in HO-1 protein levels (Figure 6F), the major Nrf2-regulated gene. The treatment of astrocytes with NDGA and gedunin clearly shows a boost in GSH, actually higher than classic Nrf2 activators, whereas fisetin does not show the same level of enhancement and is comparable to what we find with the classical Nrf2 activator TBHQ (Figure S6A). Fisetin is thought to have multiple targets such as LOX, estrogen receptor, and kinases, and therefore, its protective effect may be cumulative, and off-target effects may negatively influence GSH levels. The absence of a significant effect of fisetin on GSH levels does not point to a Nrf2-independent mechanism because the Nrf2 target gene HO-1 in astrocytes has also been found to be neuroprotective (Vargas et al., 2005).
Figure 6. Nrf2 Mediates the Astrocyte-Dependent Protective Effect of the Neh2-Luc Reporter Activators.
(A–C) Cultured primary astrocytes were treated for approximately 24 hr with NDGA (A), fisetin (B), or gedunin (C). Immediately following complete wash off of the treatments, primary immature neurons were plated in the presence or absence of HCA. Forty-eight hours later, neuronal viability was determined.
(D and E) Astrocytes were treated for 24 hr with 5 μM sulforaphane, 20 μM tBHQ, 10 μM NDGA, 24 μM gedunin, or 20 μM fisetin followed by RNA isolation. mRNA for NADPH quinone oxidoreductase 1 (NQO1) (D) or heme oxygenase-1 (HO-1) (E) was quantified with real-time PCR.
(F) Astrocytes were treated for 24 hr with 5 μM sulforaphane, 20 μM fisetin, 24 μM gedunin, 5 μM NDGA (NDGA (5)), or 10 μM NDGA (NDGA (10)). Immunoblots show HO-1 and β-actin, used as a loading control, immunoreactivity. The last lane is recombinant HO-1.
(G) Astrocytes were transfected with transfection reagent alone (Tsx-Ctl), a scrambled siRNA sequence (siScrml), or siRNA targeted against Nrf2 (siNrf2-1, siNrf2-2, siNrf2-3) and treated with 5 μM sulforaphane (SF) for 24 hr (total siRNA treatment 48 hr). Immunoblots show HO-1 and β-actin immunoreactivity. The last lane is recombinant HO-1 protein.
(H–J) Astrocytes were treated with siRNAs for 24 hr (Ctl, nontreated; Tsx, transfection reagent alone; siScr, scrambled siRNA, or siRNA targeted against Nrf2, siN1, siN2, or siN3), followed by treatment with NDGA (H), fisetin (I), or gedunin (J) for 24 hr (total siRNA = 48 hr). Immediately following complete wash off of the treatments, primary immature neurons were plated with or without HCA. Statistical significance was determined via one-way ANOVA, followed by post hoc Dunnett’s test (A–C and H–J) or Student’s t test with Bonferroni correction (D and E). (A–C) *p < 0.05, comparisons are made within 5 mM HCA treatment groups and are versus 5 mM HCA alone. (D and E) **p < 0.01 all groups versus control. (H–J) *p < 0.05, versus Ctl NDGA plus HCA, Ctl fisetin plus HCA, or Ctl gedunin plus HCA. See Figure S6 for control experiments.
To confirm that the astrocyte-dependent neuroprotective effects were specific to the activation of Nrf2, astrocytes were pretreated with siRNAs targeted against Nrf2. Three separate Nrf2 siRNA sequences lead to reduced Nrf2 mRNA and protein levels (Figures S6B and S6C) and reduced protein levels of Nrf2-regulated HO-1 (Figure 6G). Sulforaphane, a canonical Nrf2 activator, known to enhance astrocyte-dependent Nrf2-mediated neuroprotection, was used as a positive control. Consistent with prior results, Nrf2 knockdown with the Nrf2 siRNAs completely abrogated the sulforaphane-induced astrocyte-specific neuroprotection (Figure S6D). Additionally, the protective effects of NDGA, fisetin, or gedunin were also abrogated with Nrf2 knockdown (Figures 6H–6J). We do not believe that this reversal reflects the manifestation of toxic properties of the compounds because Nrf2 knockdown in the absence of oxidative stress did not lead to death in fisetin, NDGA, or gedunin-treated cocultures.
As electrophiles, many of the canonical Nrf2 activators are potential neurotoxins. Even a low level of electrophilic stress would not be ideal for many neurological conditions where oxidative stress is a contributor to disease pathology. Thus, the identification of nonelectrophilic activators of Nrf2 is a high priority. Importantly, in contrast to the neurotoxic effects of the canonical Nrf2 activators such as TBHQ, the hits from our screen (NDGA, fisetin, or gedunin) did not induce toxicity in isolated neurons using a sensitive assay of neuronal vulnerability (Figures S6E–S6I). It is worth noting that both NDGA and gedunin identified in this work as effective Nrf2 activators are key components of herbal medicines used for centuries by Native Americans (chaparral) and Indians (neem tree), respectively. These results demonstrate that the Neh2-luc reporter system can be used to identify potent and safe neuroprotective activators of the Nrf2 adaptive response.
DISCUSSION
Previous reporters of Nrf2 activation have utilized the ARE fused to coding regions of firefly luciferase or human alkaline phosphatase in vitro or in vivo. The ARE-GFP construct was used to screen the Spectrum library, and 45 hits were identified (Shaw et al., 2010). The ARE-based reporters allow monitoring effects of antioxidant response induced by Nrf2 stabilization only after 24 hr or longer. We have constructed a reporter system that allows immediate monitoring of drug-induced Nrf2 stabilization in the form of Neh2-luciferase fusion protein. The reporter appears to be a physiological surrogate for Nrf2 based on several observations: (1) Keap1 overexpression inhibits the reporter activity, whereas Keap1 depletion stabilizes the reporter (Figure 2); (2) canonical activators of Nrf2, which have been shown to act by alkylating Keap1, lead to expected increases in the Neh2-luciferase activity and protein (Figure S1B; Figure 4C); (3) representatives of all previously known classes of Nrf2 activators as well as the majority of ARE-GFP screen hits (Shaw et al., 2010) were identified in the Spectrum library using the Neh2-luc reporter, further validating the assay (Figure 3); and (4) activators of Nrf2 discovered in this screen protect neurons from oxidative death via an Nrf2-dependent mechanism in astrocytes (Figure 6).
The power of the Neh2-luc reporter allowed us to discriminate between direct and indirect effects on reporter stabilization induced by compounds tested in HTS, and, to our knowledge, for the first time identify gedunin as a direct activator of Nrf2. Recent studies suggest that gedunins are potent Hsp90 inhibitors (Brandt et al., 2008). Celastrol, a quinone methide triterpenoid, is a known Hsp90 inhibitor (Zhang et al., 2008, 2009) as well, and its derivative, dihydrocelastrol, was also found as a modest hit in the screen. Based only on structural similarities between gedunin and celastrol, one could speculate that gedunin utilizes a similar mechanism of action via disrupting the interaction between Hsp90 and Cdc37, the cochaperone providing a bridge between Hsp90 and client tyrosine kinases (Zhang et al., 2008, 2009), which being detached from the Hsp90 complex undergo fast inactivation (usually within 40–45 min). Of note, triterpenoids have been described as Nrf2 activators using ARE-reporter mice and NQO1 induction levels (Yates et al., 2007), and induce neuroprotection in a transgenic model of Huntingtons disease (Stack et al., 2010). Withanolides, closer analogs of gedunins, have been long known as inducers of NQO1 (Dinkova-Kostova et al., 2004), and are also known to disrupt Hsp90-Cdc37 interaction (Yu et al., 2010).
If gedunin works via the same mechanism as the aforementioned compounds, we should observe the delayed effect of Hsp90 downregulation with all three compounds, e.g., gedunin, geldanamycin, and TSA. However, the latter two show 3 hr lag period in reporter activation, in contrast to the immediate effect induced by gedunin (Figure 5). We may speculate that the direct effect of gedunin originates from its competition with Nrf2 for Keap1 based on the comparatively modest activation amplitude and observed plateau in the time course of reporter activation (Figure 5). This is in contrast to alkylating agents that drive the system to the maximum activation linearly (see quercetin and catechol in Figure 5). The plateau is a characteristic of re-equilibration of the system with reversible binding, or in other words, gedunins may bind Keap1 reversibly. It is tempting to speculate that gedunins compete with Nrf2 for Keap1 binding: the possibility to design mild peptide-type inhibitors displacing Nrf2 from Keap1 like p62 does in vivo (Komatsu et al., 2010) has been discussed in the paper with the resolved crystal structure of Neh2-Keap1 DGR (Tong et al., 2007). This speculation is supported by computer modeling: gedunins fit perfectly into the same Keap1-binding pocket as Nrf2 (Figure 7A), closely following the bending of the 83FEGTE79 portion of the Nrf2 peptide (Figure 7B).
Figure 7. Schematic Representation of Different Mechanisms of Nrf2 Level Regulation and Plausible Mechanism of Gedunin Action.
(A) Docking mode of gedunin in comparison with the binding mode of Neh2 portion into Keap1.
(B) Overlap between Neh2 peptide and gedunin, from perpendicular views.
(C) Hypothetic modes of Nrf2 level regulation (see text for details).
An important unanswered question is the mechanism of the “switch” effect demonstrated for our best hits, fisetin and NDGA. The time course of NDGA and fisetin clearly shows that they exert an immediate effect upon addition to the reporter cell line; therefore, they act “as is” without prior chemical modification. Both NDGA and fisetin have adjacent hydroxy groups on a freely rotating phenyl ring. This might suggest that these adjacent hydroxy groups lead to reduction of a critical disulfide bond. However, there is some doubt that fisetin and NDGA work via this mechanism because the flavones are strong reducing agents capable of immediate reduction of dithionitrobenzoate, a model disulfide, whereas NDGA is not. In addition, luteolin, a flavone with potent reducing properties, with 3,4-dihydroxy-phenyl group present in fisetin, but hydroxyl group in position 5, not 3, is a very poor Nrf2 activator. Moreover, catechol, being a very potent reducing agent, does show a 20 min lag period, which may reflect initial “priming,” most likely oxidation that results in formation of its form capable of alkylating Keap1. The fact that luteolin and catechol do not behave the same way argues against this potential mechanism and points to the special structural requirements for a “switch” mechanism of Nrf2 activation.
A common and intriguing feature of our most promising hits, fisetin and NDGA, is their steep concentration response, reminiscent of a ligand binding to a receptor. Of note, a common feature of these hits is that they all have been reported to act as inhibitors of protein tyrosine kinases, and NDGA in particular was reported to target IGF1-R kinase. We also identified genistein (100% reporter activation), which is well known for targeting this class of enzymes. Phosphorylation of Tyr141 in Keap1 is catalyzed by an unknown protein tyrosine kinase and is critical for Keap1 stability (Jain et al., 2008). Protein tyrosine kinases are also known to be stabilized by Hsp90, inhibitors of which also came out in our screen as hits.
The analysis of kinetics of individual hits leads to the model scheme of Nrf2 regulation shown in Figure 7C. A key role is played by Keap1 Cys151, 273, 288, for which modification with alkylating agents causes a dramatic change in Keap1 conformation leading to Nrf2 stabilization. If Keap1 in vivo has a zinc atom in the structure, we may hypothesize that the small planar Zn2+ chelators identified in HTS may target and destabilize the thiol pair in Keap1 as well. The delayed effect of cadmium may reflect the inhibition of thioredoxin reductase/thioredoxin system, eventually compromising the redox status of key cysteines in Keap1. Regulation of Keap1 stability via Hsp90-Cdc37-tyrosine kinase interaction is upstream of immediate activation pathways. Hsp90 is a target for TSA and geldanamycin, whereas NDGA and fisetin inhibit tyrosine kinase activity. Gedunin, in addition to intercalation into the Hsp90-Cdc37 interface, exerts an immediate effect on Nrf2 stabilization, possibly by disrupting Nrf2-Keap1 interaction. With respect to fisetin and NDGA, we also cannot rule out a possibility of targeting an unknown site at the interface of Keap1 subunits (Figure 7C) resulting in an immediate change in Keap1 conformation and stabilization of Nrf2 because the scaffold of fisetin closely resembles those of the hits generated by the virtual screen in Wu et al. (2010) (Figure S3).
Canonical activators of Nrf2 such as TBHQ, isothiocyanates, and the recently identified AL-I (Hur et al., 2010) appear to act by modifying key cysteines in Keap1, the negative regulator of Nrf2 stability. A major potential problem with electrophile activators of Nrf2 is their ability to induce toxicity, particularly in cells vulnerable to redox stress such as neurons afflicted by ischemia or neurodegeneration. The challenge is to find Nrf2 activators that do not add to the overall oxidative load, and the reporter provides a valuable resource for future developments toward such medications. Here, we discover a number of Nrf2 activators that are nontoxic to neurons over the range of concentrations optimal for reporter activation (Figures S6E–S6I).
Activation of Nrf2 plays a key role in the antioxidant defense of the central nervous system and has been shown to be important for neuroprotection in several acute and chronic neuropathological conditions such as stroke, intracerebral hemorrhage, Parkinsons disease, Huntingtons disease, and amyotrophic lateral sclerosis. Yet, Nrf2 activators such as TBHQ, sulforaphane, or CDDO-triterpenoid are only now making their way into the clinic (Shih et al., 2005; Chen et al., 2009; Vargas et al., 2008). These findings highlight the biological and clinical importance of a real-time assay for screening and design of Nrf2 activators. The developed Neh2-luc reporter is perfectly suited for HTS purposes, for studying the mechanistic details of drug action, and by analogy with HIF ODD-luc system (Safran et al., 2006), we are confident that the reporter may be successfully used for in vivo imaging of Nrf2 activators in animals.
SIGNIFICANCE
Genetic antioxidant responses activated by electrophiles are currently monitored via the use of reporters such as firefly luciferase, human alkaline phosphatase, or GFP driven by a canonical antioxidant response element (ARE). Activators of this pathway lead to the stabilization of Nrf2 and induction of dozens of genes that have been shown to prevent cancer, neurodegeneration, proinflammatory states, and combat atherosclerosis. There is a lack of compelling bioassay to ensure real-time monitoring of anti-oxidant response. We present a reporter based on a principle different than the widely used ARE-luciferase. The developed reporter constitutively expresses the Neh2 domain of Nrf2 fused to firefly luciferase. The steady-state concentration of Nrf2 (as represented by Neh2 luciferase) established in cells can be manipulated by the addition of compounds affecting the individual steps controlling the Nrf2 stability. The Neh2-luc reporter allows monitoring the antioxidant response in real time, right after drug administration, and is suitable both for high throughput screening and elucidation of the mechanism of drug action. The power of the Neh2-luc reporter is illustrated by its application for screening of the Spectrum library followed by real-time monitoring of action of selected hits: in addition to the identification of Nrf2 activators, we make an insight into the mechanistic details of their action and offer a strategy to discriminate between the action of direct activators such as alkylating agents and those requiring additional transformation steps such as prior oxidation (catechols and diamines) or manipulation of upstream regulatory pathways (via Hsp90 inhibition). Gedunins and their structural analogs were identified as a novel pharmacological class of Nrf2 activators. We also provide biological evidence for Nrf2-dependent neuroprotective roles played by identified Nrf2 activators—fisetin, nordihydroguaiaretic acid, and gedunin—in an established model of oxidative stress in neuron-astrocyte coculture.
EXPERIMENTAL PROCEDURES
Materials
Cell lines, primary neuronal and astrocyte cultures, see Supplemental Experimental Procedures.
Reporter Plasmid Construction
DNA fragment encoding 1–97 aa residues of Neh2 domain of NRF2 was the product of PCR with a cDNA template obtained from total RNA isolated from SH-SY5Y cells by using NucleoSpin RNAII kit (MACHEREY-NAGEL) and used for cDNA synthesis by SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). Neh2 fragment flanked with HindIII and NarI sites was amplified using Advantage 2 Polymerase Mix (Clontech) and the following primers: HINDNRF: CCCAAGCTTGGATCCGAATTCGCCACCATGATGGACTTGGAGCTGCCGCCGCC; and NARNRF: TAGAATGGCGCCGGGCCTTTCTTTATGTTTTTGGCGTCTTCACTGGTTTCTGA. Then it was inserted into HIindIII and NarI sites of pGL3-control (Promega) to obtain pGL3NEH2LUC. The HindIII-XbaI DNA fragment of pGL3NEH2LUC-encoding fusion protein Neh2-luciferase was cloned into corresponding sites of pcDNA3 (Invitrogen) to obtain pcDNA3-Neh2LUC10. The HIF ODDLUC-encoding plasmid pcDNA3-ODDLUC8 was constructed as described previously (Smirnova et al., 2010). pcDNA3-LUC3 encoding plain luciferase was made by insertion of HindIII-XbaI fragment of pGL3-control into HindIII-XbaI sites of pcDNA3. The ARE-luciferase construct contained an ARE promoter consensus sequence as derived from the human NADPH quinone oxidoreductase gene (5′-CTCAGCCTTCCAAATCGCAGTCACAGTGACTCAGCAGAATC-3′), upstream of a luciferase reporter (Moehlenkamp and Johnson, 1999).
Methods
HTS Optimization and SAR Analysis
The assay was optimized for HTS format to provide Z values above 0.7. SH-SY5Y-Neh2-luc cells were plated into 384-well, white, flat-bottom plates at 7000 cell/well in 30 μl serum and incubated overnight at 37°C, 5% CO2. The next day, compounds were added to two final concentrations of 16 and 32 μM, plates were incubated for 3 hr at 37°C, and luciferase activity was measured using SteadyGlo reagent (Promega). Each plate had two internal standards, TBHQ (100%) and DMSO (0%). The reporter activation (%) was calculated as a ratio (L−LDMSO)/(LTBHQ−LDMSO). Hits were defined as those greater than 25%. HTS of 2000 compounds was performed at Rockefeller University HTS Resource Center. A total of 224 hits from the initial screen have been tested in duplicate, and 210 were confirmed. Classification into structural clusters has been done manually (see also Table S1). The line expressing WT luciferase under the same promoter was used to evaluate the effect of all compounds from the Spectrum library on luciferase activity. None was found to inhibit/enhance the luciferase activity under the experimental conditions, whereas 46 compounds were found to be toxic at 3 hr incubation and were excluded from consideration. The previously described HIF1 ODD-luc reporter line (Smirnova et al., 2010) was used as a control for specificity.
Extended SAR Analysis
Selected hits were tested in 96 format white, flat-bottom plates with varied concentrations of an inhibitor (0.05–25 μM). Cells were plated at the density of 25,000 cells per well using a WellMate multichannel dispenser from Matrix (Thermo Fisher Scientific) and grown overnight on DMEM/F12 plus GlutaMAX (100 μl per well). Then the inhibitor was added, and the plates were incubated for a fixed time interval; the medium was removed, cells were lysed in 20 μl (out of which 4 μl was taken for protein measurement), then BrightGlo reagent (Promega) was added to the wells and luciferase activity measured on a luminometer Lmax11384 (Molecular Devices). The reporter activation was normalized to the background luminescence divided by protein concentration. Kinetics of reporter activation were measured by adding varied fixed concentrations of an inhibitor at different time points followed by simultaneous cell lysis, protein determination, and luciferase activity measurement in the whole 96-well plate; this assay format minimizes experimental error originating from the well-known instability of luciferase reagent.
Computer Modeling
Docking experiments were performed using the CDOCKER algorithm, followed by force field minimization and binding energy calculations using the molecular mechanics algorithm CHARMm (as implemented in Discovery Studio 2.5, Accelrys, San Diego, CA, USA). The crystal structure of human Keap1 kelch domain with the bound 16-mer peptide of human Neh2 (2FLU.pdb) with hydrogen atoms added was used as the starting template structure.
siRNA Keap1 Knockdown
siRNA against human Keap1 and control nonspecific siRNA were purchased from Thermo Scientific Dharmacon. Neroblastoma SH-SY5Y cells carrying pcDNA3-Neh2LUC10 or pcDNA3-LUC3 were plated at 3 × 105 cells per well in 6-well plate. Next-day cells were transfected with ON-TARGETplus SMARTpool siRNA Keap1 and ON-TARGETplus Non-Targeting Pool using Lipofectamine 2000 (Invitrogen) according protocol. Transfected cells were probed in luciferase assays and quantitative real-time PCR analysis 24, 48, and 72 hr after transfection with siRNA.
Real-Time Polymerase Chain Reaction
Total RNA was isolated from SH-SY5Y cells by using NucleoSpin RNAII kit (MACHEREY-NAGEL) and used for cDNA synthesis by SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). Quantitative real-time PCR analyses of human KEAP1, GCLC, GCLM, HO-1, and NQO1 were performed by using the corresponding primers and probe set from Applied Biosystems on the ABI 7500 Fast Real-Time PCR TaqMan system (Applied Biosystems). GAPDH was used for normalization.
Western Blotting
Cell cultures were rinsed in PBS, then lysed and scraped in RIPA buffer (Boston BioProducts) with 1% Protease Inhibitor Cocktail (Sigma). Lysates were vortexed, incubated on ice for 15 min, sonicated, and stored at −80°C. Protein concentration was determined using BCA Protein Assay Kit (Pierce/Thermo Scientific, Rockford, IL, USA). Samples were diluted in water to equalize protein concentration, mixed with Laemmli SDS sample buffer (reducing, 4×), boiled at 100°C for 5 min, cooled on ice, and centrifuged at 13,000 × g for 1 min immediately before gel loading. Samples were resolved by SDS-PAGE using 10% gels run at 120 V for 2 hr and transferred onto nitrocellulose membranes at 100 V for 1 hr. Quantitative western blots were performed according to the Western Blot Analysis protocol supplied by LI-COR Biosciences (Doc# 988-09288). Primary antibodies used were mouse monoclonal antibody for luciferase sc-74548 diluted 1:1,000 (Santa Cruz Biotechnology), rabbit polyclonal antibody for β-actin A2066 diluted 1:10,000 (Sigma), and a rabbit polyclonal antibody for heme oxygenase-1 (Stressgen; 1:1,000). Secondary antibodies used were goat anti-Rabbit IR dye 680 and goat anti-mouse IRDye 800CW (LI-COR Biosciences).
Keap1 Labeling by Sulfoxythiocarbate-Alkyne in Cells
Keap1-labeling experiments were performed as described previously (Ahn et al., 2010) with following modifications. HEK293 cells transiently expressing FLAG-Keap1 were incubated with 200 μM competing compounds (sulforaphane, fisetin, quercetin, gedunin, TBHQ, ciclopirox, geldanamycin) in serum-free DMEM for 1 hr. After washing with PBS, cells were further incubated with 10 μM STCA for 30 min at 37°C. FLAG-Keap1 was immunoprecipitated from cell lysates, subjected to click reaction with biotin azide on beads, and eluted with SDS-loading buffer. Eluted samples were immunoblotted with Streptavidin-HRP (Pierce) and anti-FLAG antibodies (Sigma).
Adenoviral Transduction
Adenoviral vectors containing cDNA for Nrf2 or Keap1 were obtained from the laboratory of Timothy H. Murphy. Nrf2 was driven by a CMV promoter, and a separate CMV promoter also drove the expression of GFP. Keap1 was driven by a CMV promoter and contained a FLAG tag. Cells were treated with the adenoviral plasmids at a multiplicity of infection (MOI) equal to 25 for 4 hr in serum-free Opti-MEM media and used ~24–48 hr following transduction.
Neuronal Viability
Neuronal viability was quantified using a modified protocol (Carrier et al., 2006). Astrocyte-neuron cocultures were 4% paraformaldehyde fixed for 0.5 hr at 37°C, then incubated with antibodies against the neuronal-specific marker microtubule-associated protein 2 (polyclonal anti-MAP2, 1:500, in 4% normal goat serum and 0.3% Triton-X 100) overnight at 4°C. Then the cells were incubated with rabbit secondary antibodies conjugated with horseradish peroxidase (anti-rabbit-HRP, 1:1250, in 4% normal goat serum and 0.3% Triton-X 100) for 0.5 hr at RT. The fixed cells were incubated with a reaction buffer containing 150 μM Amplex Red and 800 μM H2O2 made up in basal media (135 mM NaCl, 3.8 mM KCl, 1.2 mM MgSO4, 1.3 CaCl2, 1.2 mM KH2PO4, 10 mM D-glucose, 10 mM HEPES [pH 7.4]) for approximately 0.5 hr at RT; the formation of resorufin was measured on a SpectraMax Plus 384 (Molecular Devices) at 560 nm at RT. To account for the nonspecific binding of MAP2 to astrocytes, values determined for astrocytes alone were subtracted from coculture values.
Supplementary Material
Acknowledgments
We thank the Rockefeller University High-Throughput Screening Resource Center and Ronald Realubit, BS, for help with HTS. We also thank Dr. F. Glickman for the information provided for the Table S1 on small molecule screening data. We are grateful to Dr. H. Aleyasin for the valuable comments on neuroprotection experiments. We also thank Dr. T. Murphy for the generous gift of the Keap1 and Nrf2 adenovirus expression construct and Dr. J. Johnson for the ARE-luciferase plasmid. The work was funded by the Winifred Masterson Burke Relief Foundation, the Adelson Foundation for Neurorehabilitation and Repair, NYS DOH Center of Research Excellence # CO19772, and Thomas Hartman Foundation for Parkinsons Research. N.A.S., R.E.H.-L., R.R.R., and I.G.G. conceived the project, designed research, and wrote the paper. N.A.S., R.E.H.-L., I.R., R.E.S., M.B., J.B.P., Y.-H.A., P.A.C., J.T.P., and I.G.G. performed research. D.M.H. performed computer modeling. N.A.S. and I.G.G. analyzed HTS and SAR data.
References
- Ahn YH, Hwang Y, Liu H, Wang XJ, Zhang Y, Stephenson KK, Boronina TN, Cole RN, Dinkova-Kostova AT, Talalay P, Cole PA. Electrophilic tuning of the chemoprotective natural product sulforaphane. Proc Natl Acad Sci USA. 2010;107:9590–9595. doi: 10.1073/pnas.1004104107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt GE, Schmidt MD, Prisinzano TE, Blagg BS. Gedunin, a novel hsp90 inhibitor: semisynthesis of derivatives and preliminary structure-activity relationships. J Med Chem. 2008;51:6495–6502. doi: 10.1021/jm8007486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese V, Cornelius C, Mancuso C, Pennisi G, Calafato S, Bellia F, Bates TE, Giuffrida Stella AM, Schapira T, Dinkova Kostova DT, Rizzarelli E. Cellular stress response: a novel target for chemoprevention and nutritional neuroprotection in aging, neurodegenerative disorders and longevity. Neurochem Res. 2008;33:2444–2471. doi: 10.1007/s11064-008-9775-9. [DOI] [PubMed] [Google Scholar]
- Carrier RL, Ma TC, Obrietan K, Hoyt KR. A sensitive and selective assay of neuronal degeneration in cell culture. J Neurosci Methods. 2006;154:239–244. doi: 10.1016/j.jneumeth.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Chen W, Sun Z, Wang XJ, Jiang T, Huang Z, Fang D, Zhang DD. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34:663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Fahey JW, Talalay P. Chemical structures of inducers of nicotinamide quinone oxidoreductase 1 (NQO1) Methods Enzymol. 2004;382:423–448. doi: 10.1016/S0076-6879(04)82023-8. [DOI] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Wakabayashi N. Keap1, the sensor for electrophiles and oxidants that regulates the phase 2 response, is a zinc metalloprotein. Biochemistry. 2005;44:6889–6899. doi: 10.1021/bi047434h. [DOI] [PubMed] [Google Scholar]
- Haskew-Layton RE, Payappilly JB, Smirnova NA, Ma TC, Chan KK, Murphy TH, Guo H, Lnagley B, Sultana R, Butterfield DA, et al. Controlled enzymatic production of astrocytic hydrogen peroxide protects neurons from oxidative stress via an Nrf2 independent pathway. Proc Natl Acad Sci USA. 2010;107:17385–17390. doi: 10.1073/pnas.1003996107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Hur W, Sun Z, Jiang T, Mason DE, Peters EC, Zhang DD, Luesch H, Schultz PG, Gray NS. A small-molecule inducer of the antioxidant response element. Chem Biol. 2010;17:537–547. doi: 10.1016/j.chembiol.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Mochizuki M, Ishii Y, Ishii T, Shibata T, Kawamoto Y, Kelly V, Sekizawa K, Uchida K, Yamamoto M. Transcription factor Nrf2 regulates inflammation by mediating the effect of 15-deoxy-Delta(12,14)-prostaglandin j(2) Mol Cell Biol. 2004;24:36–45. doi: 10.1128/MCB.24.1.36-45.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AK, Mahajan S, Jaiswal AK. Phosphorylation and dephosphorylation of tyrosine 141 regulate stability and degradation of INrf2: a novel mechanism in Nrf2 activation. J Biol Chem. 2008;283:17712–17720. doi: 10.1074/jbc.M709854200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem J. 2008;409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- Kandinov B, Giladi N, Korczyn AD. Smoking and tea consumption delay onset of Parkinsons disease. Parkinsonism Relat Disord. 2009;15:41–46. doi: 10.1016/j.parkreldis.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K, Handa H, Nishizawa M. Induction of cellular antioxidative stress genes through heterodimeric transcription factor Nrf2/small Maf by antirheumatic gold(I) compounds. J Biol Chem. 2001;276:34074–34081. doi: 10.1074/jbc.M105383200. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Kuang X, Scofield VL, Yan M, Stoica G, Liu N, Wong PK. Attenuation of oxidative stress, inflammation and apoptosis by minocycline prevents retrovirus-induced neurodegeneration in mice. Brain Res. 2009;1286:174–184. doi: 10.1016/j.brainres.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge C, Dangleterre L, Cornard JP. Spectroscopic and theoretical studies of the Zn(II) chelation with hydroxyflavones. J Phys Chem A. 2006;110:12494–12500. doi: 10.1021/jp064362q. [DOI] [PubMed] [Google Scholar]
- Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehlenkamp JD, Johnson JA. Activation of antioxidant/electrophile-responsive elements in IMR-32 human neuroblastoma cells. Arch Biochem Biophys. 1999;363:98–106. doi: 10.1006/abbi.1998.1046. [DOI] [PubMed] [Google Scholar]
- Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Myzak MC, Dashwood RH. Chemoprotection by sulforaphane: keep one eye beyond Keap1. Cancer Lett. 2006;233:208–218. doi: 10.1016/j.canlet.2005.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M, Kim WY, OConnell F, Flippin L, Gunzler V, Horner JW, Depinho RA, Kaelin WG., Jr Mouse model for noninvasive imaging of HIF prolyl hydroxylase activity: assessment of an oral agent that stimulates erythropoietin production. Proc Natl Acad Sci USA. 2006;103:105–110. doi: 10.1073/pnas.0509459103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Mead R, Higginbottom A, Barber S. Therapeutics for neurological disorders. #0918626.3. UK Patent Application. 2010 Priority Date 24.10.2008. Publ Date 05.05.2010.
- Shih AY, Johnson DA, Wong G, Kraft AD, Jiang L, Erb H, Johnson JA, Murphy TH. Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress. J Neurosci. 2003;23:3394–3406. doi: 10.1523/JNEUROSCI.23-08-03394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AY, Li P, Murphy TH. A small-molecule-inducible Nrf2-mediated antioxidant response provides effective prophylaxis against cerebral ischemia in vivo. J Neurosci. 2005;25:10321–10335. doi: 10.1523/JNEUROSCI.4014-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova NA, Rakhman I, Moroz N, Basso M, Payappilly J, Kazakov S, Hernandez-Guzman F, Gaisina IN, Kozikowski AP, Ratan RR, et al. Utilization of an in vivo reporter for high throughput identification of branched small molecule regulators of hypoxic adaptation. Chem Biol. 2010;17:380–391. doi: 10.1016/j.chembiol.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son TG, Camandola S, Arumugam TV, Cutler RG, Telljohann RS, Mughal MR, Moore TA, Luo W, Yu QS, Johnson DA, et al. Plumbagin, a novel Nrf2/ARE activator, protects against cerebral ischemia. J Neurochem. 2010;112:1316–1326. doi: 10.1111/j.1471-4159.2009.06552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack C, Ho D, Wille E, Calingasan NY, Williams C, Liby K, Sporn M, Dumont M, Beal MF. Triterpenoids CDDO-ethyl amide and CDDO-trifluoroethyl amide improve the behavioral phenotype and brain pathology in a transgenic mouse model of Huntingtons disease. Free Radic Biol Med. 2010;49:147–158. doi: 10.1016/j.freeradbiomed.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodore M, Kawai Y, Yang J, Kleshchenko Y, Reddy SP, Villalta F, Arinze IJ. Multiple nuclear localization signals function in the nuclear import of the transcription factor Nrf2. J Biol Chem. 2008;283:8984–8994. doi: 10.1074/jbc.M709040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006a;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem. 2006b;387:1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas MR, Pehar M, Cassina P, Martınez-Palma L, Thompson JA, Beckman JS, Barbeito L. Fibroblast growth factor-1 induces heme oxygenase-1 via nuclear factor erythroid 2-related factor 2 (Nrf2) in spinal cord astrocytes. J Biol Chem. 2005;280:25571–25579. doi: 10.1074/jbc.M501920200. [DOI] [PubMed] [Google Scholar]
- Vargas MR, Johnson DA, Sirkis DW, Messing A, Johnson JA. Nrf2 activation in astrocytes protects against neurodegeneration in mouse models of familial amyotrophic lateral sclerosis. J Neurosci. 2008;28:13574–13581. doi: 10.1523/JNEUROSCI.4099-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Hayes JD, Higgins LG, Wolf CR, Dinkova-Kostova AT. Activation of the NRF2 signaling pathway by copper-mediated redox cycling of para- and ortho-hydroquinones. Chem Biol. 2010;17:75–85. doi: 10.1016/j.chembiol.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Westerink WM, Stevenson JC, Horbach GJ, Schoonen WG. The development of RAD51C, Cystatin A, p53 and Nrf2 luciferase-reporter assays in metabolically competent HepG2 cells for the assessment of mechanism-based genotoxicity and of oxidative stress in the early research phase of drug development. Mutat Res. 2010;696:21–40. doi: 10.1016/j.mrgentox.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Wu JH, Miao W, Hu LG, Batist G. Identification and characterization of novel Nrf2 inducers designed to target the intervening region of Keap1. Chem Biol Drug Des. 2010;75:475–480. doi: 10.1111/j.1747-0285.2010.00955.x. [DOI] [PubMed] [Google Scholar]
- Yates MS, Tauchi M, Katsuoka F, Flanders KC, Liby KT, Honda T, Gribble GW, Johnson DA, Johnson JA, Burton NC, et al. Pharmacodynamic characterization of chemopreventive triterpenoids as exceptionally potent inducers of Nrf2-regulated genes. Mol Cancer Ther. 2007;6:154–162. doi: 10.1158/1535-7163.MCT-06-0516. [DOI] [PubMed] [Google Scholar]
- Yu Y, Hamza A, Zhang T, Gu M, Zou P, Newman B, Li Y, Gunatilaka AA, Zhan CG, Sun D. Withaferin A targets heat shock protein 90 in pancreatic cancer cells. Biochem Pharmacol. 2010;79:542–551. doi: 10.1016/j.bcp.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Hamza A, Cao X, Wang B, Yu S, Zhan CG, Sun D. A novel Hsp90 inhibitor to disrupt Hsp90/Cdc37 complex against pancreatic cancer cells. Mol Cancer Ther. 2008;7:162–170. doi: 10.1158/1535-7163.MCT-07-0484. [DOI] [PubMed] [Google Scholar]
- Zhang T, Li Y, Yu Y, Zou P, Jiang Y, Sun D. Characterization of celastrol to inhibit hsp90 and cdc37 interaction. J Biol Chem. 2009;284:35381–35389. doi: 10.1074/jbc.M109.051532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.