Abstract
Long interspersed element-1 (LINE-1 or L1) retrotransposons encode two proteins (ORF1p and ORF2p) that contain activities required for conventional retrotransposition by a mechanism termed target-site primed reverse transcription. Previous experiments in XRCC4 or DNA protein kinase catalytic subunit-deficient CHO cell lines, which are defective for the nonhomologous end-joining DNA repair pathway, revealed an alternative endonuclease-independent (ENi) pathway for L1 retrotransposition. Interestingly, some ENi retrotransposition events in DNA protein kinase catalytic subunit-deficient cells are targeted to dysfunctional telomeres. Here we used an in vitro assay to detect L1 reverse transcriptase activity to demonstrate that wild-type or endonuclease-defective L1 ribonucleoprotein particles can use oligonucleotide adapters that mimic telomeric ends as primers to initiate the reverse transcription of L1 mRNA. Importantly, these ribonucleoprotein particles also contain a nuclease activity that can process the oligonucleotide adapters before the initiation of reverse transcription. Finally, we demonstrate that ORF1p is not strictly required for ENi retrotransposition at dysfunctional telomeres. Thus, these data further highlight similarities between the mechanism of ENi L1 retrotransposition and telomerase.
Long interspersed element-1 sequences (LINE-1s or L1s) are abundant non-long terminal repeat (non-LTR) retrotransposons that constitute approximately one-sixth (i.e., ≈17%) of human nuclear DNA (1). An estimated 80–100 full-length, retrotransposition competent L1s (RC-L1s) are present in a typical diploid human genome, and a small number, termed “hot L1s” exhibit high retrotransposition efficiencies in cultured human cells (2). Human RC-L1s are ≈6 kb (3, 4) and encode two proteins (ORF1p and ORF2p) required for retrotransposition (5). ORF1p is a 40-kDa protein (6) with RNA binding (7–12) and nucleic acid chaperone activities (13–15). ORF2p is a 150-kDa protein (16, 17) with endonuclease (EN) (18, 19) and reverse transcriptase (RT) (20, 21) activities.
The mobilization of RC-L1s can be mutagenic (22); germ line and, less often, somatic L1 insertions have led to ≈65 sporadic cases of disease in humans (reviewed in ref. 23). ORF1p and/or ORF2p also can act in trans to mobilize short interspersed elements [e.g., Alu (24) and SINE-VNTR-Alu (25) elements], certain noncoding RNAs [e.g., U6 snRNA and small nucleolar RNAs (24, 26–29)], and some cellular mRNAs, which leads to the formation of processed pseudogenes (30–32). In total, these trans retrotransposition events account for at least an additional 10% of human DNA (1). Moreover, several recent studies have further demonstrated that L1-mediated retrotransposition events are responsible for a significant proportion of interindividual genetic variation in the human population (33–39) and may cause intraindividual variation in the mammalian nervous system (40, 41).
L1 retrotransposition likely occurs by a mechanism termed target-site primed reverse transcription (TPRT) (42). Full-length human L1 mRNA is transcribed from an internal RNA polymerase II promoter located within its 5′ UTR (43, 44) and is exported to the cytoplasm. Upon translation, ORF1p and ORF2p exhibit cis-preference and bind their encoding mRNA (30, 32) to form a ribonucleoprotein particle (RNP) (7, 16, 45, 46). Although ORF1p is relatively abundant in cytoplasmic RNPs (7, 45, 46), the unconventional translation mechanism of human L1 ORF2 likely gives rise to fewer molecules of ORF2p per L1 mRNA (16, 47, 48). The L1 RNP translocates into the nucleus by a process that can occur in the absence of nuclear envelope breakdown (49). During TPRT, L1 EN makes a single-strand nick in genomic DNA at a loosely defined consensus site (5′-TTTT/A-3′), exposing a free 3′-OH group that the L1 RT can use to prime first-strand L1 cDNA synthesis (18, 50, 51). Short regions of complementarity between single-stranded DNA at the target site and the L1 mRNA poly (A) tail may facilitate TPRT (52). The processes of second-strand target site DNA cleavage and second-strand L1 cDNA synthesis remain enigmatic, although insight into these processes has been gained from the study of the R2 retrotransposon from Bombyx mori (53). The end result of TPRT is the integration of an L1 at a new genomic location, which in most cases is flanked by variable length target-site duplications.
We previously characterized an unconventional endonuclease-independent (ENi) L1 retrotransposition pathway in CHO cells that are defective for both the nonhomologous end-joining (NHEJ) DNA repair pathway and p53 function (54, 55). The resultant ENi retrotransposition events generally do not integrate at an L1 EN consensus site and lack structural hallmarks associated with conventional L1 retrotransposition (55). ENi retrotransposition events also are often 3′ truncated, sometimes contain cellular cDNAs at the L1/genomic DNA junction sequences, and can be associated with genomic deletions (55). Remarkably, some ENi retrotransposition events in DNA protein kinase catalytic subunit (DNA-PKcs)-deficient V3 CHO cells occur at dysfunctional telomeres (56). Here, we further examined the roles of L1 EN and ORF1p in ENi retrotransposition and have uncovered intriguing parallels between ENi retrotransposition and telomerase function.
Results
ENi Retrotransposition Occurs with Additional EN Active Site Mutants.
The L1-ORF2p amino terminus has homology to apurinic/apyrimidinic endonucleases (APE) (18, 19, 57). We previously demonstrated that L1s containing mutations in the EN active site (D205A or H230A) (58) were capable of ENi retrotransposition (55). Here, we extended those analyses and assayed an additional active site mutant (D145A) (58), as well as a mutant in a conserved EN residue (N14A) (18) for ENi retrotransposition (Fig. 1A and Fig. S1A). Consistent with previous data, mutations in the ORF2p EN domain severely compromised L1 retrotransposition in NHEJ-competent CHO cells (AA8) and human HeLa cells (Fig. 1B and Fig. S1 B and C) (18, 55, 56, 59). In contrast, the N14A and D145A EN mutants retrotransposed in the DNA-PKcs–defective V3 CHO cell line at comparable levels to D205A and H230A EN mutants (Fig. 1B and Fig. S1C) (55).
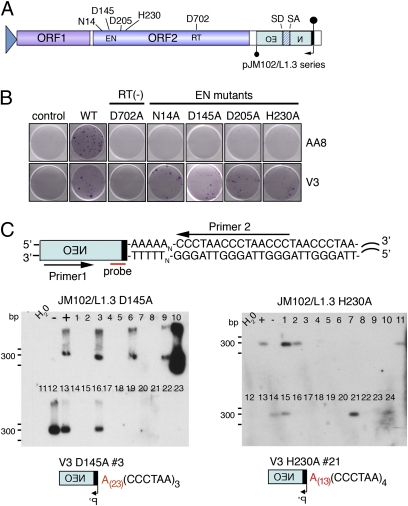
Fig. 1.
L1-EN mutants can retrotranspose to dysfunctional telomeres. (A) An RC-L1 is shown with conserved residues in the EN (N14, D145, D205, and H230) and RT (D702) domains. The blue triangle represents a CMV promoter in the pJM102/L1.3 constructs. A neomycin phosphotransferase reporter cassette interrupts the 3′ UTR. The cassette contains its own promoter (upside-down arrow) and is interrupted by an intron with splice donor (SD) and splice acceptor (SA) sites. “Lollipops” mark polyadenylation signals for both transcriptional orientations. (B) L1 constructs with mutations in the EN or RT domain are unable to retrotranspose in AA8 CHO cells (Upper) but can retrotranspose in V3 CHO cells (Lower). Mock transfected cells = control. (C) Schematic of the PCR/Southern blot assay. Relative positions of the primer specific to the retrotransposed L1 (Primer 1), a telomere primer (Primer 2), and the probe are shown. Panels of the PCR/Southern blot assay show that some clonal V3 cell lines harbor an ENi retrotransposition event at a telomere (pJM102/L1.3 wild type and pJM102/L1.3 N14A are shown in Fig. S2). A schematic of a representative PCR product positive for retrotransposition at telomeres from a clonal cell line is shown.
Previous experiments demonstrated that the L1.3 D205A EN mutant could integrate at dysfunctional telomeres in the V3 CHO cell line (56). To test whether integration at dysfunctional telomeres is a general property of L1 EN mutants, we examined whether N14A, D145A, and H230A could integrate at dysfunctional telomeres. Genomic DNAs from clonal cell lines containing ENi retrotransposition events were isolated and subjected to a PCR/Southern blot assay to detect L1 integration events adjacent to telomeric repeat sequences (Fig. 1C and Fig. S2) (56). Approximately 12–30% of N14A, D145A, and H230A ENi retrotransposition events could integrate adjacent to telomeric repeats (Fig. 1C and Fig. S2). Characterization of the resultant PCR products that yielded a positive signal in the Southern blot assay revealed L1-specific sequences with variable length poly (A) tails followed by a series of complementary telomeric repeat sequences (5′-CCCTAA-3′) (Fig. 1C and Table S1). Notably, independent clones derived from the N14A mutant contained similar, short intervening sequences between the telomeric repeats and the L1 poly (A) sequence (Fig. S2B). The identity of this intervening sequence requires further investigation, but its inclusion may reflect functional differences between mutations of the conserved N14 EN residue and the catalytic EN residues (D145, D205, and H230) (57).
RT Activity of EN Mutants.
During conventional L1 retrotransposition, L1 EN activity liberates a 3′-OH from genomic DNA to prime first-strand L1 cDNA synthesis (18, 42, 60). Recently, our laboratory has developed an assay, termed LINE-1 element amplification protocol (LEAP), to examine aspects of L1 RT in vitro (Fig. 2A) (52). We assayed wild-type L1.3 and the L1.3 EN mutants N14A, D145A, D205A, and H230A for LEAP activity using a poly-dT (T12) LEAP adapter. Consistent with previous studies, these EN mutants have LEAP activity (Fig. 2B) (16, 52) despite being severely compromised for retrotransposition in NHEJ-competent cells (Fig. 1B and Fig. S1). Sequencing confirmed that the cDNA products contained the 3′ end of the transfected L1, variable sized poly (A) tail, and the LEAP adapter (Fig. 2B). As expected, the wild-type L1.3 RNP (JJ101) exhibited robust LEAP activity on the T12 adapter, whereas the L1.3/D702A (JJ105) RT mutant lacked LEAP activity (Fig. 2B). Control Moloney murine leukemia virus (MMLV) RT-PCR reactions using RNA isolated from the L1 RNPs confirmed comparable levels of L1 and GAPDH mRNA in each RNP preparation (Fig. 2B). The slightly lower amount of mRNA in the N14A RNP sample may account for weaker LEAP activity. However, characterization of LEAP products confirmed L1-specific cDNA synthesis in these reactions. These data confirm that detecting L1 RT activity in the LEAP assay does not depend on L1 EN activity (16, 52).
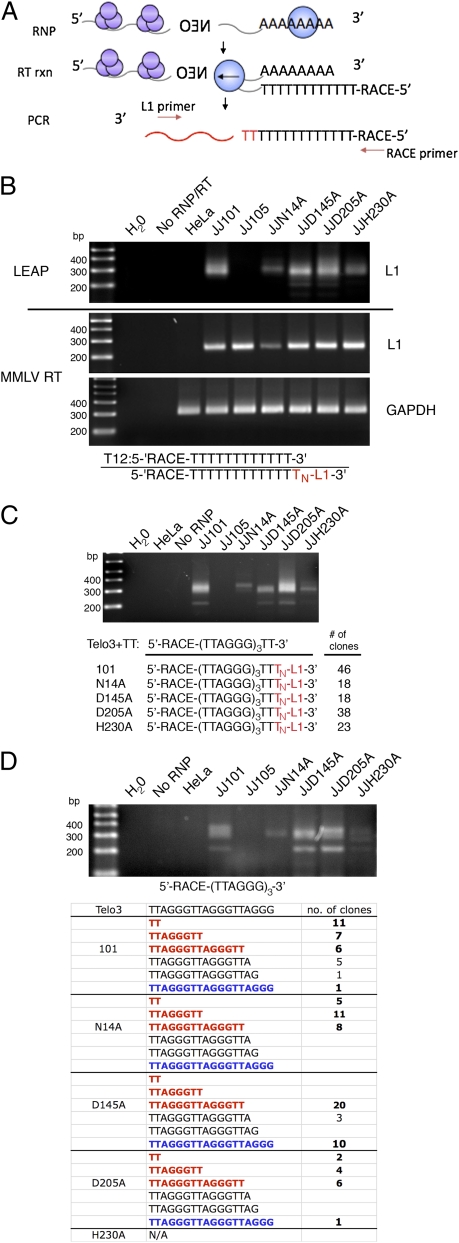
Fig. 2.
Characterization of EN mutants in the LEAP assay. (A) Schematic of the LEAP assay (52). The L1 RNP contains L1 mRNA, ORF1p (in purple), and ORF2p (blue). ORF2p primes off of the LEAP adapter and reverse transcribes the L1 mRNA in the RT reaction (RT rxn). PCR using transfected L1-specific primers is used to amplify the L1 cDNA. (B) Top: LEAP activity of different L1-RNPs on the T12 adapter. Middle and Bottom: L1 RNA and GAPDH RNA, respectively, in each RNP sample amplified by MMLV-RT using the same T12 adapter. Subtle differences in LEAP activity among the EN mutants may be due to different stabilities of the ORF2p mutants (16, 52). Sequences of the LEAP adapter (Upper) and LEAP products (Lower) are shown below the gel panels. Letters in red represent sequences generated by L1 RT. (C) Representative experiment showing LEAP activity on the Telo3+TT LEAP adapter. Sequences of the LEAP products are shown in the table. Characterization of the lower band in the gel showed L1 cDNAs that initiated upstream of the L1-poly (A) tail. (D) Representative experiment showing LEAP activity on the Telo3 LEAP adapter. Although LEAP products are faint, we were able to clone products for sequence analysis (Materials and Methods). The processed adapter sequences in these LEAP products and the number of clones containing these sequences are shown in the table. Bold red sequences represent LEAP products with perfect L1 poly (A)/telomere repeat junctions. Bold blue sequence represents LEAP products that were made with an unprocessed LEAP adapter, still giving rise to a perfect L1 poly (A) tail/telomere repeat junction. Black sequences represent LEAP products that have partial telomere sequences at the L1 poly (A) tail/telomere junction.
We next used the LEAP assay to determine whether the L1 RT activity could extend a LEAP adapter sequence containing three telomeric repeats that end with two thymidines (Telo3+TT). Again, LEAP activity was detected from the wild-type (JJ101) and EN mutant (JJN14A, JJD145A, JJD205A, and JJH230A) derived RNP preparations but not from RT mutant (JJ105) RNPs (Fig. 2C). Sequencing of the LEAP products revealed the expected variable length L1 poly (A) tail juxtaposed to perfect telomere repeat sequences, as previously observed in V3 CHO cells (Figs. 1C and 2C) (56).
Processing Activity Is Associated with L1 Reverse Transcription.
The two thymidines in the Telo3+TT LEAP adapter might improve the complementarity of the 5′-TTAGGG-3′ repeat to the L1 mRNA poly (A) tail, facilitating the reverse transcription of L1 mRNA. Thus, we tested whether the same L1 poly (A) tail/telomere junction sequence could be formed using a LEAP adapter ending in a perfect telomere repeat (Telo3). LEAP activity on this adapter was not as robust as on the Telo3+TT adapter and yielded faint bands on the gel (Fig. 2D); however, product characterization revealed that the majority of events contained the expected L1 poly (A) tail/telomere junction sequence (Materials and Methods). Unexpectedly, the adapters seemed to be processed before their use as substrates in LEAP reactions by an activity associated with both the wild-type and the EN mutant-derived L1 RNPs (Fig. 2D). Although we detected processing of the adapter in the LEAP reactions, we cannot discriminate whether at least one thymidine is from the adapter or reverse transcribed from the L1 poly (A) tail (Fig. 2D). Unlike the in vivo telomeric ENi retrotransposition events, some LEAP products lacked L1 poly (A) tail/perfect telomere junction fragments (5′-ANCCCTAA-3′ vs. 5′-ANTAACCCTAA-3′; Figs. 1C and 2D).
We next assayed LEAP activity on telomeric LEAP adapter sequences that represent a cohort of possible telomeric ends (Fig. S3). The majority of LEAP products contained a perfect L1 poly (A) tail/telomere junction and revealed evidence for processing of the adapter. By comparison, no processing was observed with the LEAP adapter ending in “5′-TT-3′” (Fig. 2C). These data suggest that L1 RNPs often process the telomere LEAP adapters before reverse transcription. We speculate that processing may facilitate optimal annealing of the adapter to the poly (A) tail of the L1 mRNA.
Processing of the telomere LEAP adapters suggests the presence of nucleolytic activity in crude L1 RNP preparations. Thus, we investigated whether this nuclease activity could process LEAP adapters that differed at their 3′ ends (Fig. 3A). The T12+VN adapter ends in two variable nucleotides (V = A, C, or G and N = any nucleotide) (52). Consistent with previous data, LEAP products generated with this adapter revealed that most products contain a combination of “VN” nucleotides embedded in the L1 poly (A) tail, which supports the hypothesis that L1 RT does not require terminal complementarity to initiate reverse transcription (Fig. 3 B and C) (52). However, some LEAP products from the T12+VN reactions did not contain the two variable nucleotides (Fig. 3C), again suggesting that adapter processing could occur before the initiation of L1 reverse transcription (52). As expected, control reactions revealed that MMLV-RT was only able to use nucleotides (VN = GT) that were complementary to the L1 mRNA/poly (A) junction as substrates to initiate reverse transcription (Fig. 3A) (52).
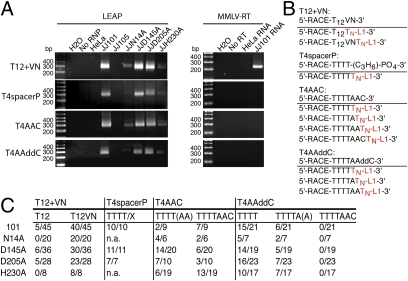
Fig. 3.
L1-RNPs are able to process adapters before reverse transcription. (A) LEAP activity on 3′ end modified adapters: nontemplated variable nucleotides (T12+VN, Top), phosphorylated carbohydrate-linker (T4spacerP, Upper Middle), deoxynucleotide (T4AAC, Lower Middle), and a dideoxynucleotide (T4AAddC, Bottom). Left: L1-RNP activity. Right: MMLV-RT activity using the same adapters with the RNA indicated. (B) Predicted sequences of LEAP products using the different adapters. Below the adapter sequence are the predicted LEAP products after adapter processing. Red letters represent predicted sequences generated by the L1 RT. (C) LEAP products from A were sequenced and contain the predicted features noted in B. The number of LEAP products sequenced for each adapter is shown. The clones are divided into processed and not-processed adapters.
To definitively test whether oligonucleotide adapters were being processed before their use as substrates in the LEAP reaction, we designed two adapters that have 3′ end modifications that would block DNA polymerization. The T4spacerP adapter has a three-carbon chain carbohydrate linker phosphorylated at its 3′ end. The T4AAddC adapter terminates in a dideoxycytidine. Removal of either the carbohydrate linker or the dideoxycytidine must occur before these adapters can be used as a primer in nucleotide polymerization reactions. RNPs derived from wild-type L1 (JJ101) and EN mutant L1s (JJD145A and JJD205A) displayed robust LEAP activity on both adapters (Fig. 3A). Sequencing of the products confirmed that the adapters were processed before their use as substrates in the LEAP reaction (Fig. 3C). Similarly, although the LEAP signal was weaker for JJN14A and JJH230A on the T4AAddC LEAP adapter, product characterization revealed processing of the LEAP adapter (Fig. 3C). Notably, we were unable to clone LEAP products from the JJN14A and JJH230A RNPs on the T4spacerP LEAP adapter, which could reflect technical difficulties or differential sensitivities of the EN mutants to the non-nucleic acid structure of the 3′ end of the adapter and requires further investigation. As expected, MMLV-RT was not able to use either adapter for reverse transcription (Fig. 3A). Thus, despite having mutations in the EN domain, the crude L1 RNP preparations contain a nuclease activity that can process the LEAP adapter. It remains unclear whether the nuclease activity is associated with the L1-encoded proteins or a cellular protein within the L1 RNP preparations (Discussion).
ORF1p Is Not Required for ENi Retrotransposition at Telomeres.
To test whether L1 ORF1p is required for L1 integration at dysfunctional telomeres, we transfected parental AA8 and DNA-PKcs–deficient V3 CHO cells lines with an expression construct containing only L1 ORF2 and the neomycin phosphotransferase retrotransposition indicator cassette (AJ101 series; Fig. 4A). Surprisingly, in this context, the wild-type and EN mutants could retrotranspose in the V3 cells, albeit at only 1.5–2.4% the efficiency of wild-type full-length L1.3 (Fig. S4A compared with Fig. 1B and Fig. S1C). As expected, the RT mutant could not retrotranspose in V3 cells, and the EN-deficient mutants could not retrotranspose in the parental AA8 CHO cell line or in HeLa cells (Fig. S4A and Fig. S1C) (5).
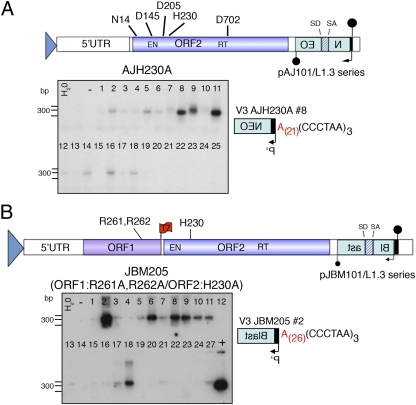
Fig. 4.
L1-ORF2 only is sufficient for retrotransposition in NHEJ-deficient cells. A schematic of the (A) pAJ101/L1.3 and (B) pJBM105 plasmid constructs. Hallmarks of the constructs are noted as in Fig. 1A except pJBM101/L1.3 constructs contain an mblastI reporter and a T7 tag on ORF1p (45). A PCR/Southern blot assay shows some clonal V3 cells harbor an ENi retrotransposition event at telomeres (A, pAJH230A/L1.3; and B, pJBM205/L1.3). A schematic of a representative PCR product positive for retrotransposition at telomeres from a clonal cell line is shown.
To confirm the above analyses, a full-length L1.3 construct with an ORF1p RR261-262AA double mutant (JBM105), which results in little to no detectable ORF1p in L1 RNPs (16, 45), and a construct harboring the ORF1p double mutant as well as the H230A ORF2p EN mutation (JBM205) (Fig. 4B) also were assayed for retrotransposition. Both JBM105 and JBM205 could retrotranspose at a low efficiency in the V3 cell line (3.4% and 5.4% of the level of L1.3, respectively) but could not retrotranspose in the parental AA8 CHO cell line (Fig. S4B and Fig. S1C). PCR/Southern blot analysis revealed that only constructs that harbored the H230A mutation (AJH230A and JBM205) gave rise to retrotransposition events at dysfunctional telomeres (Fig. 4 and Fig. S4 C and D). The retrotransposition events had perfect L1 poly (A) tail/telomere junctions (Fig. 4 and Table S1). Notably, although these constructs retrotransposed at a lower efficiency than those containing a wild-type ORF1, a similar proportion (≈20–30%) of drug-resistant clones contained ENi retrotransposition events at dysfunctional telomeres. These data suggest that ORF1p is not strictly required for directing ENi L1 retrotransposition to dysfunctional telomeres.
Discussion
We previously found that ENi L1 retrotransposition may occur at endogenous DNA lesions or at dysfunctional telomeres in NHEJ-defective CHO cells, which also are reported to be p53-defective (54–56). The ENi retrotransposition events differ from canonical retrotransposition events and often lack L1 structural hallmarks. Here, we extended those analyses and further highlight similarities between the mechanism of ENi retrotransposition and telomerase activity.
An increasing body of evidence suggests that retrotransposons provide an alternative mechanism to maintain telomere structure in a variety of organisms. For example, Drosophila lacks a formal telomerase and relies on three retrotransposons (HetA, TART, and TAHRE) to maintain its telomeres (61–65). Interestingly, TART and TAHRE have an apparent APE domain, suggesting that, unlike ENi L1 retrotransposition, this putative endonuclease may be important for telomere localization (66).
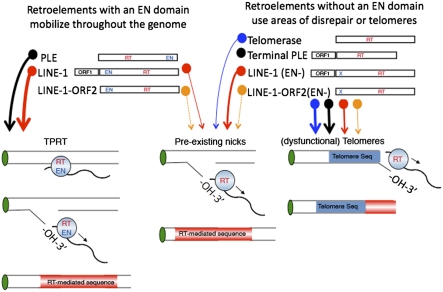
More recently, terminal Penelope-like element (PLE) retrotransposons have been found at chromosomal ends in several organisms representing four kingdoms of life (67) (Fig. 5). Like telomerase, terminal PLEs lack a canonical EN domain and have sequences within the 3′ end of their RNA that are complementary to telomere sequences of their respective organisms. However, terminal PLEs encode two ORFs, and it has been suggested that ORF1p may be important for directing them to telomere ends (68).
Fig. 5.
Model of EN-independent retrotransposition at telomere ends: The model builds on that put forth by Curcio and Belfort (68). Retrotransposons with intact EN domains, such as LINE-1 and PLEs, can retrotranspose to internal genomic locations (conventional retrotransposition). L1s can also integrate at preexisting genomic lesions using an exposed 3′-OH for reverse transcription (unconventional retrotransposition). These retrotransposition events lack structural hallmarks, such as target-site duplications. Only retrotransposons without a canonical EN domain (L1 EN mutants and terminal PLEs) are restricted to retrotransposing to preexisting free 3′-OH groups and/or telomeric ends. Thick arrows denote a major retrotransposition pathway. Thin arrows represent a less common retrotransposition pathway. Dashed arrows represent unconfirmed retrotransposition events.
We determined that ORF1p is not strictly required for ENi L1 retrotransposition at dysfunctional telomeres in V3 CHO cells. Although the ENi retrotransposition efficiency of constructs that either lack ORF1 or contain debilitating missense mutations in ORF1 is reduced compared with EN mutant constructs that contain a functional ORF1p, the relative proportion of ENi integration events at dysfunctional telomeres occurs at similar efficiencies (e.g., compare Fig. 1C with Fig. 4). These data suggest that activities contained within ORF2p are sufficient to direct ENi L1 retrotransposition to dysfunctional telomeres, although the presence of ORF1p clearly enhances the efficiency of ENi retrotransposition. It will be interesting to determine whether ORF1p from terminal PLEs functions in a similar manner to L1 ORF1p in this regard.
We also identified a nuclease activity in crude L1 RNPs that can process oligonucleotide LEAP adapters before their use as substrates for L1 reverse transcription. Whether this nuclease is encoded by L1 or is associated with a cellular protein contained within L1 RNPs requires additional study. However, previous studies suggest that a nuclease activity associated with the L1 RT domain may be important for TPRT (60). Similarly, although telomerase lacks an apparent EN domain, it is associated with a nuclease activity that can process telomeric adapter sequences before their use as substrates for telomere addition (69–71). It is tempting to speculate that the occult L1 nuclease activity may process dysfunctional telomeres in vivo before they can serve as substrates for ENi retrotransposition. Indeed, such an activity could explain why we observe perfect L1 poly (A)/telomere junctions associated with ENi retrotransposition events in V3 CHO cells.
There are striking similarities between the ENi L1 retrotransposition pathway and the action of telomerase (Fig. 5). L1 ORF2p and telomerase are both RTs that are prebound to a specific RNA, require a free 3′-OH for reverse transcription, are associated with a noncanonical nuclease activity, and can prime reverse transcription from telomeric ends (72–75). Interestingly, telomerase can also apparently reverse transcribe telomere RNA to internal chromosomal sites (75), suggesting that telomerase can “retrotranspose” its associated RNA to new chromosomal locations. Telomerase also may bind other mRNAs (76).
ENi retrotransposition is most easily observed in p53-deficient NHEJ-deficient CHO cells. Clearly, ENi retrotransposition is not a preferred L1 retrotransposition pathway. Instead, we likely have serendipitously identified an experimental model whereby we can efficiently detect this alternative mode of L1 retrotransposition. We propose that ENi retrotransposition may reflect an ancestral pathway of retrotransposition associated with non-LTR retrotransposons before the acquisition of a canonical EN domain and may be considered as a type of RNA-mediated DNA repair (Fig. 5) (55, 67, 68, 77). It will be interesting to determine whether host cellular proteins interact with the ENi L1 retrotransposition machinery to target it to areas of disrepair and/or dysfunctional telomeres. Certainly, the emerging body of data emphasizes the importance of RNA in DNA maintenance and the evolving roles of reverse transcriptases.
Materials and Methods
Cell Lines.
HeLa cells were maintained in high-glucose DMEM (Invitrogen) with 10% FBS, 100 U/mL penicillin-streptomycin, and 0.29 mg/mL l-glutamine. AA8 and V3 CHO cell lines were maintained in low-glucose DMEM (Invitrogen) with 10% FBS, 100 U/mL penicillin-streptomycin, 0.29 mg/mL l-glutamine, and 0.1 mM nonessential amino acids. Cell lines were maintained at 37 °C with 7% CO2.
Plasmid Constructs.
The recombinant plasmids used here contain L1.3 (accession no. L19088) DNA cloned into pCEP4 (Invitrogen). L1 plasmids contain either the neomycin phosphotransferase indicator cassette (mneoI) or the blasticidin deaminase indicator cassette (mblastI).
The plasmid pJM102/L1.3 was previously described (55).
The plasmid pJJ101/L1.3 is similar to pJM101/L1.3 (78) but contains the mblastI cassette in the 3′ UTR. pJJ105/L1.3 is identical to pJJ101/L1.3 but contains a missense mutation (D702A) in the ORF2p RT domain. The pJJN14A/L1.3, pJJD145A/L1.3, pJJD205A/L1.3, and pJJH230A/L1.3 plasmids are identical to pJJ101/L1.3 but contain an N14A, D145A, D205A, and H230A mutation, respectively, in the ORF2p EN domain. The above mutations also were made in pAJ101/L1.3, which lacks ORF1 and contains an mneoI cassette.
The plasmid pJBM101/L1.3 is similar to pJJ101/L1.3 except it contains a T7 epitope tag on ORF1p. pJBM105/L1.3 is identical to pJBM101/L1.3 but contains two missense mutations R261A/R262A in ORF1p. pJBM205/L1.3 is identical to pJBM105/L1.3 but contains a missense mutation (H230A) in the ORF2p EN domain.
Cultured Cell Retrotransposition Assay.
The cultured cell retrotransposition assay was previously described (59) and is detailed in SI Materials and Methods.
Telomere-L1 Southern Blot Analysis.
CHO cells were transfected for the cultured cell retrotransposition assay as previously described (55), and the telomere-L1 Southern blot analysis is detailed in SI Materials and Methods.
LEAP Assay.
The LEAP assay was previously described (52) and is detailed in SI Materials and Methods. Sequences of the adapters are noted in Figs. 2 and 3 and are shown in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the J.V.M. Laboratory for critical reading of the manuscript and discussions during the course of this study, and the University of Michigan DNA Sequencing Core for sequencing services. H.C.K. was supported by the American Cancer Society Grant PF-07-059-01-GMC. J.V.M. is supported by National Institutes of Health Grant GM060518 and is an investigator in the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: J.V.M. is an inventor on a patent, “Compositions and methods of use of human retrotransposons,” application no. 60/006,831, issued November 2000. He receives no money from the patent, and the patent does not influence the results/interpretations in this paper. J.V.M. discloses this voluntarily; it should not represent a conflict of interest.
This article is a PNAS Direct Submission.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Telomerase and Retrotransposons: Reverse Transcriptases that Shaped Genomes,” held September 29–30, 2010, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at www.nasonline.org/telomerase_and_retrotransposons.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100275108/-/DCSupplemental.
References
- 1.Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 4.Scott AF, et al. Origin of the human L1 elements: Proposed progenitor genes deduced from a consensus DNA sequence. Genomics. 1987;1:113–125. doi: 10.1016/0888-7543(87)90003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 6.Holmes SE, Singer MF, Swergold GD. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J Biol Chem. 1992;267:19765–19768. [PubMed] [Google Scholar]
- 7.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 8.Hohjoh H, Singer MF. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Januszyk K, et al. Identification and solution structure of a highly conserved C-terminal domain within ORF1p required for retrotransposition of long interspersed nuclear element-1. J Biol Chem. 2007;282:24893–24904. doi: 10.1074/jbc.M702023200. [DOI] [PubMed] [Google Scholar]
- 10.Khazina E, Weichenrieder O. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc Natl Acad Sci USA. 2009;106:731–736. doi: 10.1073/pnas.0809964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolosha VO, Martin SL. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc Natl Acad Sci USA. 1997;94:10155–10160. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolosha VO, Martin SL. High-affinity, non-sequence-specific RNA binding by the open reading frame 1 (ORF1) protein from long interspersed nuclear element 1 (LINE-1) J Biol Chem. 2003;278:8112–8117. doi: 10.1074/jbc.M210487200. [DOI] [PubMed] [Google Scholar]
- 13.Martin SL. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol. 2010;7:706–711. doi: 10.4161/rna.7.6.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin SL, et al. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J Mol Biol. 2005;348:549–561. doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Doucet AJ, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ergün S, et al. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J Biol Chem. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 18.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 19.Martín F, Marañón C, Olivares M, Alonso C, López MC. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: Homology of the first ORF with the ape family of DNA repair enzymes. J Mol Biol. 1995;247:49–59. doi: 10.1006/jmbi.1994.0121. [DOI] [PubMed] [Google Scholar]
- 20.Hattori M, Kuhara S, Takenaka O, Sakaki Y. L1 family of repetitive DNA sequences in primates may be derived from a sequence encoding a reverse transcriptase-related protein. Nature. 1986;321:625–628. doi: 10.1038/321625a0. [DOI] [PubMed] [Google Scholar]
- 21.Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 22.Kazazian HH, Jr, et al. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 23.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 24.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 25.Hancks DC, Kazazian HH., Jr SVA retrotransposons: Evolution and genetic instability. Semin Cancer Biol. 2010;20:234–245. doi: 10.1016/j.semcancer.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buzdin A, et al. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of l1. Genomics. 2002;80:402–406. doi: 10.1006/geno.2002.6843. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 2007;17:602–611. doi: 10.1101/gr.5870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber MJ. Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genet. 2006;2:e205. doi: 10.1371/journal.pgen.0020205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 31.Tchénio T, Segal-Bendirdjian E, Heidmann T. Generation of processed pseudogenes in murine cells. EMBO J. 1993;12:1487–1497. doi: 10.1002/j.1460-2075.1993.tb05792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei W, et al. Human L1 retrotransposition: Cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck CR, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hormozdiari F, et al. Alu repeat discovery and characterization within human genomes. Genome Res. 2011;21:840–849. doi: 10.1101/gr.115956.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang CR, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iskow RC, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills RE, et al. 1000 Genomes Project Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witherspoon DJ, et al. Mobile element scanning (ME-Scan) by targeted high-throughput sequencing. BMC Genomics. 2010;11:410. doi: 10.1186/1471-2164-11-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 42.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 43.Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004;32:3846–3855. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum Mol Genet. 2005;14:3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 46.Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol. 1991;11:4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 2006;20:210–224. doi: 10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McMillan JP, Singer MF. Translation of the human LINE-1 element, L1Hs. Proc Natl Acad Sci USA. 1993;90:11533–11537. doi: 10.1073/pnas.90.24.11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubo S, et al. L1 retrotransposition in nondividing and primary human somatic cells. Proc Natl Acad Sci USA. 2006;103:8036–8041. doi: 10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cost GJ, Boeke JD. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- 51.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 53.Christensen SM, Eickbush TH. R2 target-primed reverse transcription: Ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Mol Cell Biol. 2005;25:6617–6628. doi: 10.1128/MCB.25.15.6617-6628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu T, Miller CM, Ridder GM, Aardema MJ. Characterization of p53 in Chinese hamster cell lines CHO-K1, CHO-WBL, and CHL: Implications for genotoxicity testing. Mutat Res. 1999;426:51–62. doi: 10.1016/s0027-5107(99)00077-9. [DOI] [PubMed] [Google Scholar]
- 55.Morrish TA, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 56.Morrish TA, et al. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 57.Weichenrieder O, Repanas K, Perrakis A. Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon. Structure. 2004;12:975–986. doi: 10.1016/j.str.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 58.Repanas K, et al. Determinants for DNA target structure selectivity of the human LINE-1 retrotransposon endonuclease. Nucleic Acids Res. 2007;35:4914–4926. doi: 10.1093/nar/gkm516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei W, Morrish TA, Alisch RS, Moran JV. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal Biochem. 2000;284:435–438. doi: 10.1006/abio.2000.4675. [DOI] [PubMed] [Google Scholar]
- 60.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abad JP, et al. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol Biol Evol. 2004;21:1620–1624. doi: 10.1093/molbev/msh180. [DOI] [PubMed] [Google Scholar]
- 62.Biessmann H, et al. HeT-A, a transposable element specifically involved in “healing” broken chromosome ends in Drosophila melanogaster. Mol Cell Biol. 1992;12:3910–3918. doi: 10.1128/mcb.12.9.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- 64.Pardue ML, DeBaryshe PG. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu Rev Genet. 2003;37:485–511. doi: 10.1146/annurev.genet.38.072902.093115. [DOI] [PubMed] [Google Scholar]
- 65.Sheen FM, Levis RW. Transposition of the LINE-like retrotransposon TART to Drosophila chromosome termini. Proc Natl Acad Sci USA. 1994;91:12510–12514. doi: 10.1073/pnas.91.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gladyshev EA, Arkhipova IR. A subtelomeric non-LTR retrotransposon Hebe in the bdelloid rotifer Adineta vaga is subject to inactivation by deletions but not 5′ truncations. Mob DNA. 2010;1:12. doi: 10.1186/1759-8753-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gladyshev EA, Arkhipova IR. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci USA. 2007;104:9352–9357. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Curcio MJ, Belfort M. The beginning of the end: Links between ancient retroelements and modern telomerases. Proc Natl Acad Sci USA. 2007;104:9107–9108. doi: 10.1073/pnas.0703224104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huard S, Autexier C. Human telomerase catalyzes nucleolytic primer cleavage. Nucleic Acids Res. 2004;32:2171–2180. doi: 10.1093/nar/gkh546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oulton R, Harrington L. A human telomerase-associated nuclease. Mol Biol Cell. 2004;15:3244–3256. doi: 10.1091/mbc.E04-03-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niu H, Xia J, Lue NF. Characterization of the interaction between the nuclease and reverse transcriptase activity of the yeast telomerase complex. Mol Cell Biol. 2000;20:6806–6815. doi: 10.1128/mcb.20.18.6806-6815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feng J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 73.Lingner J, et al. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 74.Shippen-Lentz D, Blackburn EH. Functional evidence for an RNA template in telomerase. Science. 1990;247:546–552. doi: 10.1126/science.1689074. [DOI] [PubMed] [Google Scholar]
- 75.Wells RA, Germino GG, Krishna S, Buckle VJ, Reeders ST. Telomere-related sequences at interstitial sites in the human genome. Genomics. 1990;8:699–704. doi: 10.1016/0888-7543(90)90257-u. [DOI] [PubMed] [Google Scholar]
- 76.Maida Y, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Voliva CF, Martin SL, Hutchison, CA, 3rd, Edgell MH. Dispersal process associated with the L1 family of interspersed repetitive DNA sequences. J Mol Biol. 1984;178:795–813. doi: 10.1016/0022-2836(84)90312-7. [DOI] [PubMed] [Google Scholar]
- 78.Sassaman DM, et al. Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.