Abstract
Variation in food quality and abundance requires animals to decide whether to stay on a poor food patch or leave in search of better food. An important question in behavioral ecology asks when is it optimal for an animal to leave a food patch it is depleting. Although optimal foraging is central to evolutionary success, the neural and molecular mechanisms underlying it are poorly understood. Here we investigate the neuronal basis for adaptive food-leaving behavior in response to resource depletion in Caenorhabditis elegans, and identify several of the signaling pathways involved. The ASE neurons, previously implicated in salt chemoattraction, promote food-leaving behavior via a cGMP pathway as food becomes limited. High ambient O2 promotes food-leaving via the O2-sensing neurons AQR, PQR, and URX. Ectopic activation of these neurons using channelrhodopsin is sufficient to induce high food-leaving behavior. In contrast, the neuropeptide receptor NPR-1, which regulates social behavior on food, acts in the ASE neurons, the nociceptive ASH neurons, and in the RMG interneuron to repress food-leaving. Finally, we show that neuroendocrine signaling by TGF-β/DAF-7 and neuronal insulin signaling are necessary for adaptive food-leaving behavior. We suggest that animals integrate information about their nutritional state with ambient oxygen and gustatory stimuli to formulate optimal foraging strategies.
Keywords: food-leaving decision, Marginal Value Theorem, sensory neurons
For most animals food is distributed unpredictably in patches of heterogeneous quality and quantity. As a food patch is depleted an animal must decide when it is advantageous to seek better feeding opportunities elsewhere. Understanding this decision is an important aim in behavioral ecology. Charnov's Marginal Value Theorem (MVT) suggests an animal's chosen strategy will reflect the energetic rewards gained upon encountering a better quality patch elsewhere, weighed against the energy expended (in locomotion and cessation of feeding) in traveling between patches (1). The MVT yields the “giving up time,” the point in time when an animal should leave. Although optimal foraging behavior has been studied in many organisms, the neural basis for these decisions is not understood in any animal.
Caenorhabditis elegans feeds on microorganisms that grow on rotting material (2) and is adapted to exploit transient resources rapidly. Its simple nervous system and powerful genetics provide opportunities to define mechanisms underlying food patch-leaving choice at both molecular and circuitry levels. Several aspects of C. elegans foraging have been studied previously. Shtonda and Avery showed that C. elegans exercises food choice, selecting high quality food and leaving “harder-to-eat” bacteria (3). Wild strains of C. elegans can sense where food is thickest and aggregate there (“social” behavior) (4). These behaviors are attenuated in the N2 laboratory reference strain because of a gain-of-function mutation in the neuropeptide receptor npr-1 gene that arose during laboratory domestication (5, 6). The gene npr-1 also affects dispersal behavior in the presence of abundant food. Social worms disperse between food patches more readily than solitary worms (7). Recently, a polymorphism in tyra-3, encoding a G protein-coupled receptor for tyramine and octopamine, was shown to modify exploratory leaving behavior (8). Interestingly, wild isolates of C. elegans increase their dispersal as food concentration decreases (9), suggesting that these animals continuously assess the value of the patch. The value an animal places on a given patch is determined not only by its intrinsic nutritional value, but also by the costs involved in acquiring the food. Such costs may include exposure to environmental hazards and predation. How sensory inputs associated with these aversive cues modify food-leaving behavior is not understood mechanistically in any organism. Although our understanding of C. elegans ecology is rudimentary (10), recent work suggests that two environmental cues that modify foraging are ambient levels of O2 and CO2 (11–14). C. elegans avoids ambient O2 concentrations close to 21%, a response that may ensure it avoids the surface with its associated hazards. Avoidance of high ambient O2 is promoted by a cGMP signaling pathway in the AQR, PQR, and URX O2-sensing neurons (11, 12, 15, 16). C. elegans also escapes environments with elevated CO2 (13, 14). Several neurons have been implicated in this behavior, including the ciliated head neurons BAG, AFD, and ASE (14, 17, 18). CO2 responses in these three neurons also involve cGMP signaling.
Here we establish a paradigm to study C. elegans food-leaving behavior specifically in response to resource depletion, in line with the MVT. Using targeted transgenic rescue of well-characterized mutants, we identify signaling pathways and sensory neurons regulating this behavior.
Results
Food-Leaving Is an Adaptive Exploratory Behavior Induced as Food Becomes Limiting.
To investigate behavioral responses to resource depletion in C. elegans, we developed an assay to study food-leaving behavior (Methods). We used a dense bacterial lawn that, over 20 h, was gradually depleted by feeding animals. We filmed young adults to follow their behavior as food diminished (Fig. 1). Initially animals accumulated at the border, where food was thickest, and reversed to remain on food if the head, the tip of which is rich in chemoreceptors and mechanoreceptors, emerged from the food lawn. Over time, wild-type animals exhibited higher food-leaving as the food became scarcer (Fig. 1 A and B). This pattern of food-leaving is predicted by the adaptive benefits of leaving a diminishing food resource in search of a better quality patch.
Fig. 1.
Adaptive food-leaving in wild-type animals. (A) Food-leaving probability increases over time as animals deplete their food source. Food-leaving probability is calculated as the number of worms leaving per minute divided by the total number of worms on the food at start of that minute, averaged over 15 min (3). Error bars show SD. (B) Single frames from 1-, 6-, and 12-h movies and schematic drawings. The arrows point at animals outside of food or at the outer edge of the food border. (C) Speed on food and at the border, and the proportion of animals encountering the border that leave food increased over time (D). Error bars denote SEM (C and D). ***P < 0.005, **P < 0.01, *P < 0.05. n = 6 or more per strain.
We next sought to deconstruct food-leaving into more elemental behavioral components. Using videotracking software we quantified how the speed of animals, their reversal frequency all over the food patch and specifically at the border, their rate of arrival at the food border, the time they spend at the border, and the proportion of animals that leave food per encounter with the border, changed as they depleted food. Animals increased their speed from 25 to 35 μm/s as food decreased (Fig. 1C). This increase in speed did not increase the rate at which animals encountered the food border (Fig. 1D); however, the proportion of animals that left food per border encounter progressively increased from 0.1 to 0.5 over time (Fig. 1D) as reversals at the border decreased (Fig. S1A).
These time-course data focused our subsequent studies on the behavior of animals that have been foraging on our assay plates for 6 h, because this represented an intermediate frequency of food-leaving that allowed us to measure both enhancement and suppression of food-leaving behavior.
Gustatory Neuron ASE, the O2-Sensing Neurons AQR, PQR, and URX, and the CO2-Sensing Neuron BAG Promote Adaptive Food-Leaving.
Evaluation of a food source is likely to reflect integration of two kinds of sensory cues: external cues, associated with the food itself and its environmental context, and internal cues, such as neuroendocrine signals that communicate the animal's nutritional status. To initiate our circuitry dissection we sought first to disrupt sensory input from external cues.
Chemosensory transduction in C. elegans is generally mediated either by transient receptor potential V-like ion channels encoded by osm-9 and its associated subunits encoded by the ocr genes (19, 20), or by cGMP-gated channels that incorporate subunits encoded by the tax-4 and tax-2 genes (21, 22). We asked whether mutations in these genes altered food-leaving behavior.
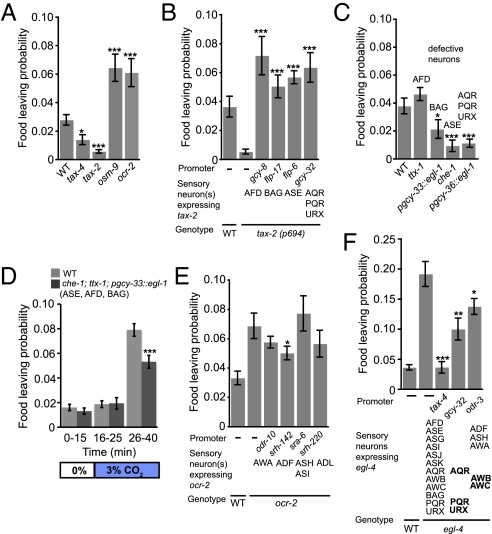
Loss-of-function mutations in either tax-4 or tax-2 reduced food-leaving (Fig. 2A). The tax-2 mutants moved at speeds similar to wild-type and encountered the border at a similar rate in our assay (Fig. S1B); however, the proportion of animals at the border that left the food per edge encounter was reduced compared with wild-type (Fig. S1C). In contrast, mutations in osm-9 or ocr-2 increased food-leaving (Fig. 2A). In osm-9 mutants the proportion of animals leaving food per edge encounter was not altered compared with wild-type controls, but animals showed elevated speed and increased border arrival rate, accounting for their higher food-leaving rate (Fig. S1C).
Fig. 2.
Adaptive food-leaving is promoted by ASE, BAG, and the body cavity neurons AQR, PQR, and URX and suppressed by ADF neurons. (A) The tax-4 and tax-2 mutants show lower food-leaving than wild-type animals, whereas osm-9 and ocr-2 mutants show higher food-leaving. (B) Increased food-leaving of tax-2(p694) animals can be rescued by expressing tax-2 cDNA in AFD (pgcy-8), BAG (pflp-17), ASE (an ASE-specific pflp-6 fragment), and AQR, PQR, and URX (pgcy-32). (C) The ttx-1 mutants behave similarly to wild-type animals with respect to food-leaving. In contrast, animals in which BAG neurons are ablated using a pgcy-33::egl-1 transgene, as well as che-1 mutants defective for ASE function, and animals genetically ablated for AQR, PQR, and URX neurons using egl-1 expression from the gcy-36 promoter stayed strongly on food. (D) After 15 min of airflow at 21% O2, wild-type animals and che-1;ttx-1 pgcy-33::egl-1 animals (which are defective in the CO2 sensing neurons ASE, AFD, and BAG) were subjected to a shift to 3% CO2 (21% O2). This shift elicited a substantial increase in food-leaving after about 10 min that was partially suppressed in che-1;ttx-1 pgcy-33::egl-1 animals. (E) High food-leaving in ocr-2 mutants can be partially rescued by expressing ocr-2 cDNA in ADF neurons. (F) The egl-4 loss-of-function mutants show higher food-leaving than wild-type. Wild-type levels of food-leaving can be restored by expressing egl-4 from the tax-4 promoter. Expressing egl-4 from the gcy-32 promoter, which drives expression specifically in AQR, PQR, and URX neurons, or the odr-3 promoter, which drives expression in AWA, AWB, AWC, ASH, and ADF neurons, results in partial rescue. Error bars denote SEM. ***P < 0.005, **P < 0.01, *P < 0.05. n = 6 or more per genotype.
Both tax-4 and tax-2 are coexpressed in about 12 sensory neurons (21, 22). The tax-2 allele we used, tax-2(p694), is a promoter deletion that disrupts the function of only a subset of these sensory neurons, namely AQR, PQR, URX, ASE, AFD, and BAG (17, 21, 23). This finding suggested that one or more of these neurons promoted food-leaving behavior. To identify these neurons, we sought to rescue the food-leaving defect in tax-2(p694) mutants by expressing tax-2 cDNA using cell-specific promoters. Expressing tax-2 in AFD alone, BAG alone, ASE alone, or in AQR, PQR, and URX restored food-leaving to tax-2 mutants (Fig. 2B and Fig. S1E). These data suggested that each of these neurons can promote food-leaving behavior.
To extend our studies, we examined the consequences of functionally ablating each neuron identified above (Fig. 2C). We genetically disrupted AFD neurons using a mutation in ttx-1; ttx-1 encodes a homeodomain transcription factor of the otd/otx subclass that is specifically required to specify AFD neurons (24). The ttx-1 mutants exhibited wild-type food-leaving behavior. Animals in which the BAG neurons were ablated by cell-specific expression of the egl-1 cell-death gene exhibited significantly less food-leaving than wild-type animals. Strikingly, animals defective in che-1, a zinc-finger transcription factor required to specify the ASE neurons (25), or lacking AQR, PQR, and URX because of specific expression of egl-1 in these O2-sensing neurons, resulted in animals that stayed strongly on food (Fig. 2C). Thus, our cell-disruption experiments, like our tax-2 rescue data, suggest that BAG, ASE, and one or more of the AQR, PQR, and URX neurons inhibit food-leaving. Interestingly, whereas the rescue experiments suggested that any one of the BAG, ASE, or AQR, PQR, or URX neurons was sufficient to restore food-leaving behavior to tax-2(p694) mutants, our cell-ablation/mis-specification experiments suggested that each of these neurons or neuron groups was necessary for wild-type food-leaving rates. One explanation for this difference was that the tax-2(p694) mutation disrupted a neural input that suppresses food-leaving probability. The likeliest candidate for this role was AFD. Consistent with this finding, disrupting the function of AFD using a ttx-1 mutation suppressed the food-leaving phenotypes associated with loss of BAG or ASE (Fig. S2 A and B). These data suggest that AFD can act antagonistically to BAG and ASE to inhibit food-leaving behavior. Although these results contrast with our earlier tax-2 rescue data implicating AFD in promoting food-leaving, recent work has shown that the AFD neurons can elicit opposite behavioral outcomes, depending on the level of activation (26).
Our data on the role of ASE, BAG, and AFD in food-leaving were highly reminiscent of a recent study implicating these neurons in CO2 avoidance (13). All three neurons are CO2 sensors. The neurons are activated by elevated CO2 via a cGMP signaling pathway and promote avoidance of CO2, although under some circumstances AFD can also inhibit avoidance of high CO2. These parallels prompted us to examine if high ambient CO2 levels altered food-leaving probability. Switching CO2 concentration from 0 to 3% induced a high rate of food-leaving (Fig. 2D and Fig. S2C). This increased food-leaving in response to CO2 was reduced in ASE-, AFD-, and BAG-defective animals. Thus, one way that BAG, ASE, and AFD neurons may regulate food-leaving is by responding to elevated CO2 levels associated with microbial food or worm respiration.
We next investigated sensory neurons that prevent animals from leaving a food patch prematurely. The osm-9 and ocr-2 mutants left food more than wild-type. Both ocr-2 and osm-9 are coexpressed in six pairs of sensory neurons: ADF, ADL, ASH, AWA, PHA, and PHB (19, 20). Expressing ocr-2 cDNA from neuron-specific promoters in four of these neurons individually implicated ADF neurons in food-leaving behavior (Fig. 2E), although selective expression of ocr-2 in ADF rescued the ocr-2 phenotype only partially. These results suggest that ocr-2 functions in ADF neurons to inhibit food-leaving, but that other ocr-2–expressing neurons are also required for the wild-type food-leaving pattern.
The cGMP-dependent protein kinase, encoded in C. elegans by the egl-4 gene, is a conserved modulator of food-related behaviors (27). The egl-4(lf) mutants spend more time roaming on food than wild-type animals (28). EGL-4 is also required for odor adaptation in the olfactory neuron AWC (29). We found that egl-4(lf) mutants exhibited increased food-leaving (Fig. 2F). This phenotype was associated with elevated locomotory activity on food, higher encounter frequency with the bacterial lawn border, reduced border reversal rate, and increased probability of food-leaving per border encounter (Fig. S2 D–H). Expressing egl-4 cDNA under the control of the tax-4 promoter rescued all these phenotypes (Fig. S2 D–H). Expressing egl-4 in only AQR, PQR, and URX partially rescued these phenotypes. Expressing egl-4 under the control of the odr-3 promoter, which drives expression in AWA, AWB, AWC, ASH, and ADF also partially rescued these phenotypes. In summary, our results suggest that cGMP signaling in the ASE, AQR, PQR, URX, and BAG neurons mediated by a cyclic nucleotide channel containing the TAX-2 subunit promotes food-leaving, and that ocr-2 and egl-4 prevent premature food-leaving in ADF and in tax-4–expressing neurons (in particular AQR, PQR, and URX), respectively.
Insulin and TGF-β, Mediators of Nutritional State and Environmental Conditions, Respectively, Modulate Food-Leaving.
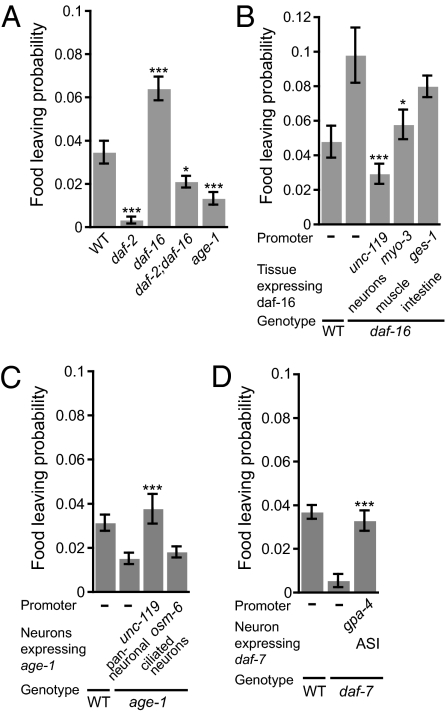
We hypothesized that the food-leaving decision would be regulated by postingestive inputs (e.g., nutritional state or feeding history). Insulin signaling has previously been linked to perception of feeding state and food-related behavioral plasticity (30–33). This finding prompted us to analyze adaptive food-leaving in mutants defective in the insulin pathway. Mutants in the daf-2 insulin receptor (34) stayed on food more than wild-type (Fig. 3A). Mutants in the PI3K age-1, one of the known downstream targets of insulin signaling, behaved similarly (35), suggesting AGE-1 mediated the affect of the DAF-2 receptor, although we cannot exclude that AGE-1 is activated by other insulin independent pathways. In contrast, mutants in daf-16, encoding the Forkhead box O transcription factor inhibited by insulin-like signaling (36), exhibited higher food-leaving behavior (see Fig. S3 for behavioral parameters of daf-2 and daf-16 mutants). The daf-2;daf-16 double-mutants had a phenotype closer to wild-type. The incomplete epistasis of daf-16 over daf-2 suggests that daf-2 promotes food-leaving by mechanisms that are only partly dependent on daf-16. To determine the site of action of DAF-16, we restored its expression in different tissues to daf-16 mutant animals. Neuronal expression of daf-16, but not intestinal expression, conferred wild-type food-leaving to daf-16 mutants (Fig. 3B). There was also partial rescue by expression of daf-16 in body muscle in the daf-16 mutants. Similarly, neuronal expression of an age-1 transgene in age-1 mutants restored wild-type food-leaving behavior (Fig. 3C). In contrast, expressing age-1 in all ciliated neurons using the osm-6 promoter (37), using a transgene previously shown to rescue other osm-6 phenotypes, did not rescue the food-leaving defect. These data support a neuronal role for insulin signaling in food-leaving behavior, possibly in interneurons, although our negative results with the posm-6::age-1 transgene does not allow us to rigorously exclude a role in sensory neurons.
Fig. 3.
Neuronal insulin and TGF-β signaling from the ASI neurons promote food-leaving. (A) The daf-2 and age-1 mutants exhibit reduced food-leaving compared with wild-type animals. The daf-16 mutants exhibit higher food-leaving. daf-2 suppresses the high food-leaving phenotype of daf-16 mutants. (B) Expression of daf-16 in all neurons from the unc-119 promoter restores wild-type food-leaving. Expression of daf-16 in muscle from the myo-3 promoter shows partial rescue. Intestinal expression of daf-16 from the ges-1 promoter does not rescue the food-leaving phenotype of daf-16 mutants. (C) age-1 expression from the unc-119 promoter, but not the osm-6 promoter, restores wild-type food-leaving to age-1 mutants. (D) The daf-7 mutants exhibit reduced food-leaving. Wild-type food-leaving can be restored by expressing daf-7 in the ASI neurons, using the gpa-4 promoter. Error bars denote SEM. ***P < 0.005, *P < 0.05. n = 6 or more per strain.
Signaling via the TGF-β–like ligand DAF-7 transduces external conditions. Food shortage, high population density, and dauer pheromone inhibit daf-7 expression and promote dauer formation (38, 39). As adults, daf-7 mutants accumulate more fat and have a reduced pharyngeal pumping rate (40). Adaptive food-leaving was strongly reduced in daf-7 mutants (Fig. 3D); daf-7 is expressed in one pair of head sensory neurons, ASI (38, 39). Reintroducing daf-7 expression in ASI in daf-7 mutants completely restored wild-type food-leaving. Thus, TGF-β signaling from the ASI neurons promotes food-leaving behavior.
Neuropeptide Signaling via NPR-1 in ASH and RMG Control Food-Leaving Behavior.
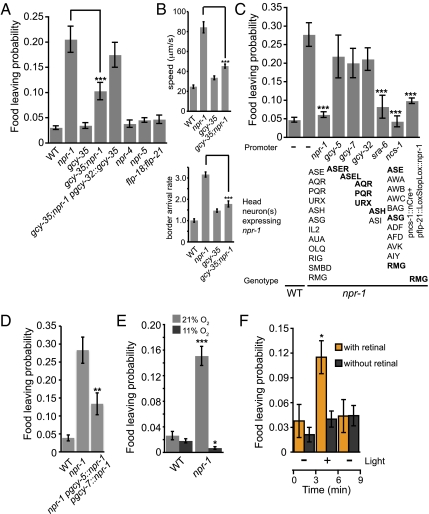
The neuropeptide receptor NPR-1 regulates food-related behaviors, such as aggregation (social feeding) (4), and has been implicated in C. elegans dispersal (7). The npr-1 mutants had much higher food-leaving than wild-type (Fig. 4A). This phenotype was associated with much higher speed and border-arrival rates (Fig. 4B; see Fig. S4 for more behavioral parameters). The food-leaving defect in npr-1 mutants, as well as the speed and border-arrival rate phenotypes, were suppressed by a mutation in gcy-35, which encodes an atypical soluble guanylate cyclase that mediates O2 sensing in the body cavity neurons AQR, PQR, and URX (11, 41). Restoring gcy-35 expression in the body cavity neurons reverted food-leaving behavior of gcy-35;npr-1 to npr-1 behavior, suggesting that the high food-leaving in npr-1 mutants requires gcy-35 activity in AQR, PQR, and URX (Fig. 4A).
Fig. 4.
The neuropeptide receptor NPR-1 functions in ASE, ASH, and RMG neurons to inhibit food-leaving and high ambient O2 or ectopic activation of AQR, PQR, and URX induces high food-leaving. (A) The npr-1 mutants show higher food-leaving than wild-type animals. gcy-35 partially suppresses npr-1 food-leaving. Expression of gcy-35 from the gcy-32 promoter in gcy-35; npr-1 mutants shows that gcy-35 acts in AQR, PQR, and URX to suppress the high food-leaving of npr-1 mutants. Neither a flp-18; flp-21 double-mutant nor mutants in their additional receptors npr-4 and npr-5 (50) showed higher food-leaving than wild-type. (B) The npr-1 mutants show increased speed and border-arrival rate, which is suppressed in gcy-35;npr-1 mutants. (C) Wild-type food-leaving can be restored in npr-1 mutants by expressing npr-1 specifically in ASH (and ASI) from the sra-6 promoter, and in RMG using Cre-mediated recombination (51). Full rescue is also achieved from the ncs-1 promoter, driving expression in RMG and 10 other neurons. (D) Expression of npr-1 in both ASEL and ASER partially rescue food-leaving in npr-1 mutants. (E and F) Food-leaving is context-dependent. (E) Wild-type and npr-1 mutants were recorded at 21% oxygen and then switched to 11% oxygen in a Perspex chamber. Food-leaving of npr-1 mutants was reduced at low oxygen tension, when the AQR, PQR, and URX neurons are less active (15, 16). (F) Ectopic stimulation of AQR, PQR, and URX neurons in npr-1 lite-1 mutants by channelrhodopsin activation is sufficient to induce high food-leaving in animals kept at 11% oxygen. Animals were filmed 3 min before and after light activation. The orange bars denote the addition of the necessary cofactor, retinal. Error bars denote SEM. ***P < 0.005, **P < 0.01, *P < 0.05. n = 6 or more.
FMRF-amide (Phe-Met-Arg-Phe-NH2)-related peptides encoded by the flp-18 and flp-21 genes have been implicated as NPR-1 ligands (42, 43). However, a double-mutant flp-18; flp-21 did not show higher food-leaving than wild-type, suggesting that neuropeptides other than FLP-18 and FLP-21 regulate food-leaving mediated by NPR-1.
To identify the cells where npr-1 activity is required for adaptive food-leaving, we expressed npr-1 using neuron-specific promoters in npr-1 mutants. The ncs-1 promoter drives expression in a subset of npr-1–expressing neurons: ASE, ASG, PHA, PHB, and RMG (44). Expression of npr-1 from this promoter restored wild-type food-leaving behavior in npr-1 mutants (Fig. 4C), as did expression of npr-1 in the inter/motor neuron RMG and in the polymodal nociceptive neuron ASH. Expression of npr-1 in either ASEL or ASER, from the gcy-7 and gcy-5 promoters, respectively, did not rescue npr-1 mutants, but expression in both ASEL and ASER did (Fig. 4D). Expression of npr-1 in the body cavity neurons did not significantly rescue the adaptive food-leaving behavior. This result implicates the ASE, ASH, and RMG neurons in the regulation of adaptive food-leaving behavior. We conclude that npr-1 prevents food-leaving in ASE, ASH, and RMG neurons and that in the absence of npr-1, high food-leaving partly reflects the activity of the gcy-35 O2 sensor in the AQR, PQR, and URX neurons.
Activation of the Body Cavity Neurons Is Sufficient to Induce Food-Leaving.
Ca2+ imaging studies have shown that AQR, PQR, and URX are activated by high ambient O2 (15, 16). To investigate the role of the O2-sensing body cavity neurons AQR, PQR, and URX in adaptive food-leaving, we first examined whether increased food-leaving by npr-1 mutants was dependent on ambient O2 levels. We found that npr-1 mutants left food less when ambient O2 was at 11%, consistent with AQR, PQR, and URX being less active at low O2 (Fig. 4E). This result, together with our observation that mutations in the gcy-35 O2 sensor suppressed the food-leaving phenotype of npr-1 animals, suggests that activation of the O2 signaling pathway in AQR, PQR, and URX when animals are in 21% ambient O2 promotes food-leaving.
To directly test this hypothesis we expressed channelrhodopsin [ChR2] in the body cavity neurons of npr-1 mutants and light-activated these neurons while animals were at 11% oxygen. Light stimulation induced a dramatic increase in food-leaving (Fig. 4F). This finding is consistent with the tax-2 rescue experiments in AQR, PQR, and URX and suggests that activation of AQR, PQR, and URX is sufficient to induce high food-leaving behavior.
Discussion
A central question in behavioral ecology is what controls the timing of an animal's decision to leave a depleting resource patch. We find that C. elegans conforms to expectations, increasing its food-leaving probability as food becomes depleted, when a negative energy budget may increase the adaptive value of such risky behaviors (1, 45). This behavior, which we term “adaptive food-leaving,” has the hallmarks of an optimal patch-leaving strategy. We identify a small set of neurons and signaling molecules that control adaptive food-leaving as food becomes limited (see Fig. 5 for our model).
Fig. 5.
Model for regulation of adaptive food-leaving behavior. The BAG, ASE, and AQR, PQR, URX neurons promote food-leaving via TAX-2/TAX-4. Food-leaving is also promoted by the ASI neurons via DAF-7 and by neuronal insulin signaling. High CO2 and O2 promote food-leaving. Food-leaving is suppressed by the ADF neuron via OCR-2, the AFD neuron and by NPR-1 neuropeptide signaling in ASE, ASH, and RMG neurons.
The ASE gustatory neurons promote food-leaving. In well-fed animals the ASE neurons mediate chemoattraction to water-soluble cues. However, several studies have shown that food withdrawal can turn this attraction to salt into repulsion (33, 46–48). Our work suggests this gustatory plasticity occurs not only upon food withdrawal but also in feeding animals experiencing a gradual decrease in food patch quality. The work on gustatory plasticity has shown that the switch in preference involves signaling by the daf-2 insulin receptor in ASER (33). Our data also implicate daf-2 signaling in promoting adaptive food-leaving, although we have not identified an anatomical focus for this action. The BAG sensory neurons, which are exquisitely sensitive to rises in CO2 (17, 18), also promote food-leaving. BAG neurons, as well as the ASE neurons, are tonically activated by elevated CO2 (17). One possibility is that adaptive food-leaving in our assay partly reflects animals avoiding CO2 generated by bacteria and other nematodes. Recent work has also implicated BAG as a modulator of food-leaving behavior, but suggests it inhibits food-leaving (8). However, these results were obtained in a different genetic background (in the presence of an npr-1 allele derived from the Hawaiian wild strain) and under significantly less food-restricted conditions (8), so the data are not directly comparable.
The body cavity neurons AQR, PQR, and URX modulate adaptive food-leaving behavior, promoting food-leaving at high ambient O2. These neurons are activated by high ambient [O2] (15, 16) and promote reversals and turns when O2 levels rise, as well as high locomotory activity (increased roaming) if the npr-1 neuropeptide receptor is defective (12, 16). These neurons promote food-leaving in both npr-1(N2) and npr-1(null) mutant backgrounds. Moreover, we find that ectopic activation of AQR, PQR, and URX neurons using channelrhodopsin rapidly induces food-leaving. The effects of AQR, PQR, and URX on food-leaving depend on GCY-35, an atypical soluble guanylate cyclase that mediates O2 responses (11) and is also required for the increased roaming of npr-1 mutants on food (16). In npr-1 animals, adaptive food-leaving can be directly modulated by altering ambient O2: high O2 is associated with rapid food-leaving, whereas low O2 is associated with low food-leaving. Regulation of food-leaving by ambient O2 may allow animals to balance their need to acquire food with avoiding a hazardous environment at the surface and accumulating in buried but food-containing environments.
The role of AFD neurons in food-leaving is more complex. AFD has been studied extensively for its role in temperature sensing, but recent data indicate it is also a CO2 sensor that promotes CO2 avoidance in some contexts and inhibits CO2 avoidance in others (17). Our data suggest that similarly, AFD can both promote and suppress food-leaving depending on the context. More specifically, we find that AFD promotes food-leaving when ASE and BAG neurons are defective, but inhibits food-leaving if either ASE or BAG neurons are functional. A recent optogenetic study has shown that the level of activation of AFD determines whether it evokes attractive or repulsive behavior (26). Strong activation of AFD results in weaker activation of AIY, a major postsynaptic target of AFD, and avoidance behavior. In contrast, attenuating the activation of AFD using halorhodopsin leads to strong activation of AIY and attraction behavior. AIY interneurons receive synaptic input not only from AFD but also from the ASE and BAG neurons (49). We suggest that this common circuitry explains why the behavioral effects of AFD on food-leaving depend on the activity state of ASE and BAG. One caveat in our experiments is that we are using a mutation in ttx-1 to genetically disrupt AFD, and we cannot be sure that this is equivalent to ablating the neuron.
In summary, we address the neuronal and molecular basis for food-leaving, a clearly defined and theoretically predictable aspect of animal behavior. Optimal forging has been widely studied at the behavioral level and modeled extensively; C. elegans provides an opportunity to test predictions made by these studies by manipulating its nervous system.
Methods
Strains.
Nematodes were grown at 20 °C under standard conditions. All strains used can be found as SI Methods.
Behavioral Assays.
Low peptone [5% of regular amount of bactopeptone (4)] nematode growth media plates were seeded 2 d before assay with 25 μL of OP-50 saturated overnight culture (from the Caenorhabditis Genetics Center) in 2XYT. Twenty young adults were transferred to the assay plates and allowed to feed for 6 h [except for the timescale (Fig. 1) and the channelrhodopsin experiment], after which they were recorded for 15 min or placed in a Perspex chamber for the O2 and CO2 experiments. The food-leaving probability was calculated as described by Shtonda and Avery (3). Briefly: the ratio between number of worms leaving per minute divided by the total number of worms on the food at start of that minute was calculated and averaged over 15 min. A minimum of six assays was done per genotype. All statistical analyses were conducted using Minitab15 software. To test the validity of pooling data from different assay days, a Kruskal–Wallis test was conducted. Because food-leaving probability for each strain did not differ significantly between assay days (P > 0.05), statistical analyses were performed on pooled data sets used the t test.
Details can be found in SI Methods.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center, funded by the National Institutes of Health, for strains; A. Bretscher, M. Ezcurra, R. Gatsi, I. Mori, and B. Schafer for strains and constructs; H. Baylis and Q.-L. Ch'ng for critically reading the manuscript; and J. Chen for help with analyses. This work was funded by the Medical Research Foundation (to B.O. and K.M.) and the Swiss National Science Foundation (K.E.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106134109/-/DCSupplemental.
References
- 1.Charnov EL. Optimal foraging, the marginal value theorem. Theor Popul Biol. 1976;9:129–136. doi: 10.1016/0040-5809(76)90040-x. [DOI] [PubMed] [Google Scholar]
- 2.Kiontke K, Sudhaus W. Ecology of Caenorhabditis species. WormBook. 2006 doi: 10.1895/wormbook.1.37.1. ed The C. elegans Research Community, pp 1–14, 10.1895/wormbook.1.37.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shtonda BB, Avery L. Dietary choice behavior in Caenorhabditis elegans. J Exp Biol. 2006;209:89–102. doi: 10.1242/jeb.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Bono M, Bargmann CI. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell. 1998;94:679–689. doi: 10.1016/s0092-8674(00)81609-8. [DOI] [PubMed] [Google Scholar]
- 5.Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 2009;5:e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber KP, et al. Whole genome sequencing highlights genetic changes associated with laboratory domestication of C. elegans. PLoS ONE. 2010;5:e13922. doi: 10.1371/journal.pone.0013922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gloria-Soria A, Azevedo RB. npr-1 Regulates foraging and dispersal strategies in Caenorhabditis elegans. Curr Biol. 2008;18:1694–1699. doi: 10.1016/j.cub.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 8.Bendesky A, Tsunozaki M, Rockman MV, Kruglyak L, Bargmann CI. Catecholamine receptor polymorphisms affect decision-making in C. elegans. Nature. 2011;472:313–318. doi: 10.1038/nature09821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harvey SC. Non-dauer larval dispersal in Caenorhabditis elegans. J Exp Zoolog B Mol Dev Evol. 2009;312B:224–230. [Google Scholar]
- 10.Félix MA, Braendle C. The natural history of Caenorhabditis elegans. Curr Biol. 2010;20:R965–R969. doi: 10.1016/j.cub.2010.09.050. [DOI] [PubMed] [Google Scholar]
- 11.Gray JM, et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 12.Cheung BH, Cohen M, Rogers C, Albayram O, de Bono M. Experience-dependent modulation of C. elegans behavior by ambient oxygen. Curr Biol. 2005;15:905–917. doi: 10.1016/j.cub.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 13.Bretscher AJ, Busch KE, de Bono M. A carbon dioxide avoidance behavior is integrated with responses to ambient oxygen and food in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2008;105:8044–8049. doi: 10.1073/pnas.0707607105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallem EA, Sternberg PW. Acute carbon dioxide avoidance in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2008;105:8038–8043. doi: 10.1073/pnas.0707469105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmer M, et al. Neurons detect increases and decreases in oxygen levels using distinct guanylate cyclases. Neuron. 2009;61:865–879. doi: 10.1016/j.neuron.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Persson A, et al. Natural variation in a neural globin tunes oxygen sensing in wild Caenorhabditis elegans. Nature. 2009;458:1030–1033. doi: 10.1038/nature07820. [DOI] [PubMed] [Google Scholar]
- 17.Bretscher AJ, et al. Temperature, oxygen, and salt-sensing neurons in C. elegans are carbon dioxide sensors that control avoidance behavior. Neuron. 2011;69:1099–1113. doi: 10.1016/j.neuron.2011.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallem EA, et al. Receptor-type guanylate cyclase is required for carbon dioxide sensation by Caenorhabditis elegans. Proc Natl Acad Sci USA. 2011;108:254–259. doi: 10.1073/pnas.1017354108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colbert HA, Smith TL, Bargmann CI. OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J Neurosci. 1997;17:8259–8269. doi: 10.1523/JNEUROSCI.17-21-08259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tobin D, et al. Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron. 2002;35:307–318. doi: 10.1016/s0896-6273(02)00757-2. [DOI] [PubMed] [Google Scholar]
- 21.Coburn CM, Bargmann CI. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron. 1996;17:695–706. doi: 10.1016/s0896-6273(00)80201-9. [DOI] [PubMed] [Google Scholar]
- 22.Komatsu H, Mori I, Rhee JS, Akaike N, Ohshima Y. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron. 1996;17:707–718. doi: 10.1016/s0896-6273(00)80202-0. [DOI] [PubMed] [Google Scholar]
- 23.Coates JC, de Bono M. Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans. Nature. 2002;419:925–929. doi: 10.1038/nature01170. [DOI] [PubMed] [Google Scholar]
- 24.Satterlee JS, et al. Specification of thermosensory neuron fate in C. elegans requires ttx-1, a homolog of otd/Otx. Neuron. 2001;31:943–956. doi: 10.1016/s0896-6273(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 25.Uchida O, Nakano H, Koga M, Ohshima Y. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development. 2003;130:1215–1224. doi: 10.1242/dev.00341. [DOI] [PubMed] [Google Scholar]
- 26.Kuhara A, Ohnishi N, Shimowada T, Mori I. Neural coding in a single sensory neuron controlling opposite seeking behaviours in Caenorhabditis elegans. Nat Commun. 2011;2:355. doi: 10.1038/ncomms1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reaume CJ, Sokolowski MB. cGMP-dependent protein kinase as a modifier of behaviour. Handb Exp Pharmacol. 2009;191:423–443. doi: 10.1007/978-3-540-68964-5_18. [DOI] [PubMed] [Google Scholar]
- 28.Fujiwara M, Sengupta P, McIntire SL. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron. 2002;36:1091–1102. doi: 10.1016/s0896-6273(02)01093-0. [DOI] [PubMed] [Google Scholar]
- 29.L'Etoile ND, et al. The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron. 2002;36:1079–1089. doi: 10.1016/s0896-6273(02)01066-8. [DOI] [PubMed] [Google Scholar]
- 30.Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- 31.Hasshoff M, Böhnisch C, Tonn D, Hasert B, Schulenburg H. The role of Caenorhabditis elegans insulin-like signaling in the behavioral avoidance of pathogenic Bacillus thuringiensis. FASEB J. 2007;21:1801–1812. doi: 10.1096/fj.06-6551com. [DOI] [PubMed] [Google Scholar]
- 32.Kodama E, et al. Insulin-like signaling and the neural circuit for integrative behavior in C. elegans. Genes Dev. 2006;20:2955–2960. doi: 10.1101/gad.1479906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomioka M, et al. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron. 2006;51:613–625. doi: 10.1016/j.neuron.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 34.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 35.Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 36.Ogg S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 37.Collet J, Spike CA, Lundquist EA, Shaw JE, Herman RK. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics. 1998;148:187–200. doi: 10.1093/genetics/148.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schackwitz WS, Inoue T, Thomas JH. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron. 1996;17:719–728. doi: 10.1016/s0896-6273(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 39.Ren P, et al. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- 40.Greer ER, Pérez CL, Van Gilst MR, Lee BH, Ashrafi K. Neural and molecular dissection of a C. elegans sensory circuit that regulates fat and feeding. Cell Metab. 2008;8:118–131. doi: 10.1016/j.cmet.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheung BH, Arellano-Carbajal F, Rybicki I, de Bono M. Soluble guanylate cyclases act in neurons exposed to the body fluid to promote C. elegans aggregation behavior. Curr Biol. 2004;14:1105–1111. doi: 10.1016/j.cub.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 42.Rogers C, et al. Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR-1. Nat Neurosci. 2003;6:1178–1185. doi: 10.1038/nn1140. [DOI] [PubMed] [Google Scholar]
- 43.Pocock R, Hobert O. Hypoxia activates a latent circuit for processing gustatory information in C. elegans. Nat Neurosci. 2010;13:610–614. doi: 10.1038/nn.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomez M, et al. Ca2+ signaling via the neuronal calcium sensor-1 regulates associative learning and memory in C. elegans. Neuron. 2001;30:241–248. doi: 10.1016/s0896-6273(01)00276-8. [DOI] [PubMed] [Google Scholar]
- 45.Bateson M. Perspectives in the study of food intake. Proc Nutr Soc. 2002;61:509–516. doi: 10.1079/pns2002181. [DOI] [PubMed] [Google Scholar]
- 46.Saeki S, Yamamoto M, Iino Y. Plasticity of chemotaxis revealed by paired presentation of a chemoattractant and starvation in the nematode Caenorhabditis elegans. J Exp Biol. 2001;204:1757–1764. doi: 10.1242/jeb.204.10.1757. [DOI] [PubMed] [Google Scholar]
- 47.Hukema RK, Rademakers S, Dekkers MP, Burghoorn J, Jansen G. Antagonistic sensory cues generate gustatory plasticity in Caenorhabditis elegans. EMBO J. 2006;25:312–322. doi: 10.1038/sj.emboj.7600940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hukema RK, Rademakers S, Jansen G. Gustatory plasticity in C. elegans involves integration of negative cues and NaCl taste mediated by serotonin, dopamine, and glutamate. Learn Mem. 2008;15:829–836. doi: 10.1101/lm.994408. [DOI] [PubMed] [Google Scholar]
- 49.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 50.Cohen M, et al. Coordinated regulation of foraging and metabolism in C. elegans by RFamide neuropeptide signaling. Cell Metab. 2009;9:375–385. doi: 10.1016/j.cmet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 51.Macosko EZ, et al. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009;458:1171–1175. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.