Abstract
Stress pathways monitor intracellular systems and deploy a range of regulatory mechanisms in response to stress. One of the best-characterized pathways, the unfolded protein response (UPR), is responsible for maintaining endoplasmic reticulum (ER) homeostasis. The highly conserved Ire1 branch regulates hundreds of gene targets by activating a UPR-specific transcription factor. To understand how the UPR manages ER stress, a unique genetic approach was applied to reveal how the system corrects disequilibria. The data show that the UPR can address a wide range of dysfunctions that are otherwise lethal if not for its intervention. Transcriptional profiling of stress-alleviated cells shows that the program can be modulated, not just in signal amplitude, but also through differential target gene expression depending on the stress. The breadth of the functions mitigated by the UPR further supports its role as a major mechanism maintaining systems robustness.
Keywords: chaperones, signal transduction, protein folding, protein degradation, glycosylation
Robustness of biological systems is characterized by the reproducibility of biological processes, despite variability in genetic composition or external environment. This quality lies in cells having molecular circuits that produce precise and reliable outputs in the face of internal or external perturbations. Many examples of robust systems are known but the exact molecular mechanisms for ensuring robustness are still not well understood (1). In some cases where overlapping pathways exist, redundancy might confer robustness, whereas in other cases, a form of system control may be used in which positive/negative feedback allows the input signal to be modulated according to the output signal (2).
Stress pathways respond to systemic perturbations by regulating a wide range of functions. In this way, they are specialized mechanisms designed to monitor and maintain intracellular homeostasis. The unfolded protein response (UPR) is one of the best-studied stress pathways with the Ire1 branch being the most highly conserved among eukaryotes (3). It can be triggered by the abnormal accumulation of unfolded proteins in the endoplasmic reticulum (ER) caused by genetic or environmental changes. In budding yeast, the pathway initiates with Ire1p, an ER membrane protein that acts as the sole stress sensor and signal transducer (4, 5). Upon stress, activated Ire1p splices the pre-mRNA of HAC1 to initiate synthesis of Hac1p, the UPR-specific transcription factor (6). Hac1p then translocates into the nucleus to up-regulate the expression of UPR target genes (7).
An early indicator of the UPR's importance in cellular homeostasis came from transcriptional profiling experiments that identified ∼381 UPR target genes in budding yeast (8). Not only was the expression of expected ER chaperones elevated, but also the expression of genes involved in diverse functions including protein trafficking and quality control, metabolism, and cell wall biosynthesis. Strikingly, a recent study systematically analyzing 4,500 yeast deletion mutants revealed ∼10% displayed significant UPR up-regulation (9). Taken together, these studies show the remarkable breadth of functions both regulated and monitored by the UPR. Although the UPR term originated from studies using potent chemical inducers to disrupt protein folding, it is now known that various stresses caused by disease, infection, metabolic imbalance, genetic mutation, and even normal development can physiologically activate the pathway (10). It is therefore not surprising that UPR deficiencies can have severe consequences for health. Although many physiological inputs are now known, the key question of how the UPR output alleviates ER stress remains unclear. The lack of clarity is due in part to pleiotropic effects of most inducers along with the complexity of the UPR program.
In principle, the problem can be made tractable by exploiting a class of yeast mutants that physiologically activate the UPR as a requirement for viability. This characteristic reflects the direct link between genetically defined stress and the responding UPR (11). The advantage over other methods is each mutant specifies a form of stress that is also a measurable biochemical dysfunction. Unfortunately, the intrinsic synthetic lethality with the regulatory circuit makes analyses in the absence of the UPR, although experimentally critical, difficult with existing methodologies. To overcome this obstacle, we developed a unique genetic class, termed conditional synthetic lethality, which allows analysis in the absence of the UPR by temperature shift. Using this approach, we demonstrate that the UPR acts as a broad-spectrum compensatory mechanism, a quality that makes it particularly well suited in its role to maintain intracellular homeostasis. Interestingly, transcriptional profiling of stress-adapted cells reveals customized regulation of UPR target genes contingent on the form of stress. These studies reveal the remarkable breadth of the UPR in alleviating stress and surprising complexity in the regulation of its targets.
Results
Mutant genes displaying synthetic lethality to UPR regulatory genes define functions monitored by the pathway (11). More importantly, because pathway activation reverses otherwise lethal dysfunctions, they encompass the minimum functional repertoire governed by the UPR. Using the classical approach, linkage analysis indicated that the mutant class is larger than is practical to identify all genes (11). Recently, a high-throughput yeast synthetic lethality screen called synthetic genetic arrays (SGA) was carried out that queried 1,712 mutants against deletion mutants of most nonessential genes (12). From this dataset, >100 genes were found, displaying synthetic lethality against the UPR regulatory genes IRE1 and HAC1 (Table S1). The results reveal an unexpected range in number and functional diversity. To analyze how the UPR might compensate for cellular defects, we focused on three nonredundant genes involved in different aspects of ER function: LHS1, ALG5, and SCJ1. Lhs1p is a member of the Hsp70 family and is involved in the translocation of presecretory proteins into the ER (13). ALG5 encodes UDP-glucose:dolichyl-phosphate glucosyltransferase, which catalyses the transfer of the glucose moiety from the donor UDP-glucose to dolichyl-phosphate, forming the glucose donor for the synthesis of core oligosaccharide Glc3Man9GlcNAc2 used in N-linked glycosylation of proteins (14). SCJ1, on the other hand, encodes for an ER-localized Hsp40/DnaJ protein that has been implicated in protein folding and ER-associated degradation (ERAD) (15, 16). These genes are believed to play important roles in the biosynthesis of secretory proteins but strikingly, their genomic deletions cause only mild phenotypes (Figs. 1 and 2B).
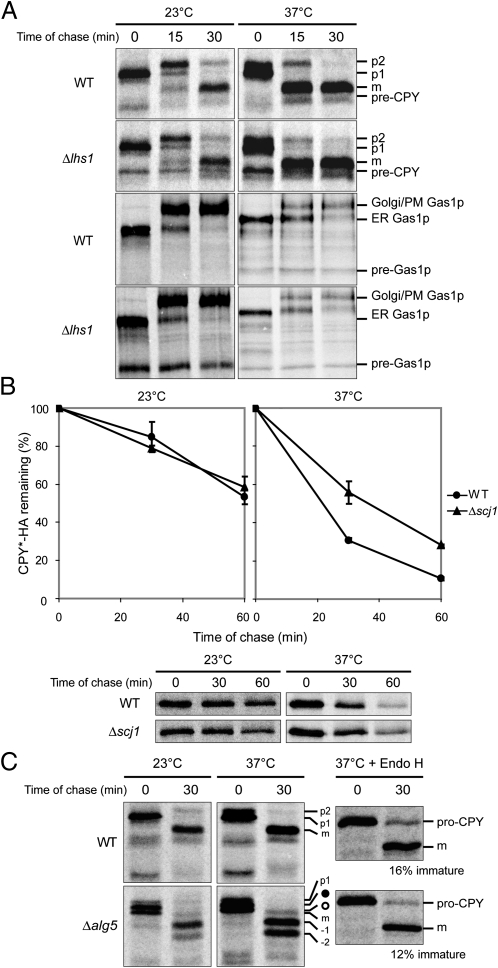
Fig. 1.
Δlhs1, Δscj1, and Δalg5 single deletion mutants exhibit mild phenotypes. (A) CPY and Gas1p biosynthesis was examined in wild-type (WT) and Δlhs1 strains by pulse-chase analysis. Positions of the nontranslocated, ER, Golgi, and mature forms of CPY are indicated by pre-CPY, p1, p2, and m, respectively. (B) The degradation of ERAD substrate, CPY*-HA, was monitored in WT and Δscj1 cells by a pulse-chase experiment. The graph shown is the mean ± SD of three independent experiments. (C) CPY biogenesis was examined in WT and Δalg5 strains as described in A. The underglycosylated ER/Golgi forms are indicated with ● and ○, and the underglycosylated mature CPY species are labeled −1 and −2. Treatment with Endoglycosidase H (Endo H) was performed after immunoprecipitation for the relevant samples. (Right) Immature CPY was expressed as a percentage of total CPY and is indicated below each section.
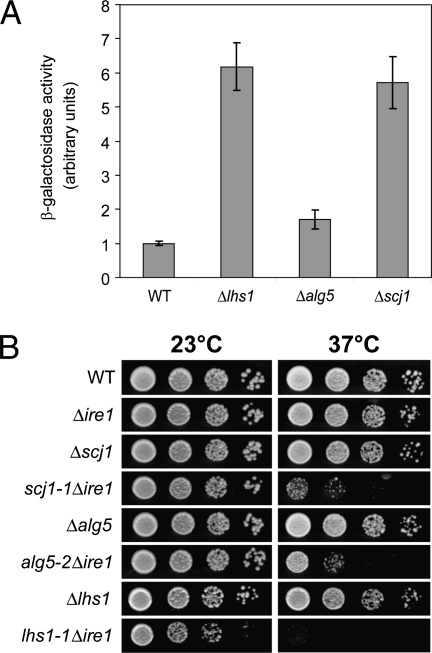
Fig. 2.
The UPR is required for viability of LHS1-, SCJ1-, and ALG5-deficient cells. (A) UPR induction was measured for the indicated cells using a β-galactosidase reporter assay. Data shown are the mean ± SD of three independent experiments. (B) The strains were grown at 23 °C and serial dilutions of the culture were spotted onto plates. These plates were incubated at the indicated temperature until the appearance of colonies.
When examined by pulse-chase analysis, the Δlhs1 single deletion strain exhibits only slight defects in the translocation of posttranslational translocation substrates, carboxypeptidase Y (CPY), and Gas1p at both 23 °C and 37 °C (Fig. 1A). Likewise, Δscj1 cells are proficient in the degradation of a well-characterized ERAD substrate, CPY* (16). In Δscj1 cells, CPY* degrades as rapidly as wild type at 23 °C and only slightly slower at 37 °C (Fig. 1B). In the case of ALG5, the absence of the gene produces core glycan donors lacking glucose residues, which are transferred to protein substrates at reduced efficiency, forming underglycosylated proteins (14). This was easily observed for CPY biogenesis in Δalg5 cells with the initial appearance of underglycosylated pro-CPY (p1) (Fig. 1C, ● and ○), which matured into underglycosylated CPY with most containing only two or three of the normal four glycans (Fig. 1C, “−2” and “−1”). Even with this defect, CPY still trafficked to the vacuole as efficiently as in wild-type cells after in vitro glycan cleavage to differentiate CPY precursors from the vacuolar processed mature form (Fig. 1C). Thus, the impact of eliminating LHS1, SCJ1, or ALG5 seems to be minimal even though they can play crucial roles in the biogenesis of some secretory proteins.
Typically, UPR synthetic lethal mutants constitutively activate the UPR in response to ER stress (11). Accordingly, LHS1, SCJ1, or ALG5 mutants display constitutively activated UPRs as measured by the UPRE-LacZ reporter assay (Fig. 2A) (6). Together, these data suggest that the UPR activation might actively compensate for the loss of these functions and mask severe deficiencies that are otherwise lethal. To date, there is no direct evidence that UPR activation can widely compensate for biochemical dysfunctions. To determine whether the UPR performs this function, we designed a strategy to examine the effects of genetically defined ER stress with the UPR muted. Here, unique alleles of LHS1, SCJ1, and ALG5 were isolated that are temperature-sensitive (ts) lethal only in cells lacking a functional UPR (Δire1) (Fig. S1 and Fig. 2B).
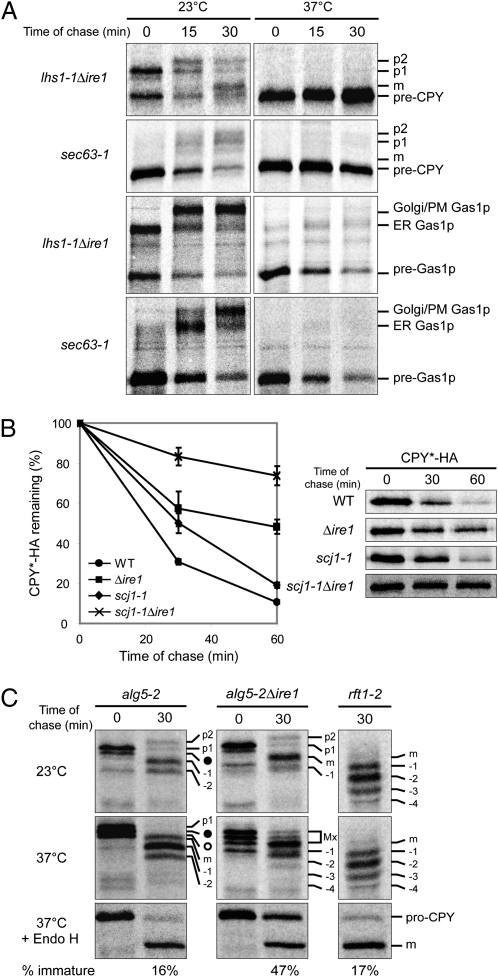
First, we examined the loss of LHS1 function in the absence and presence of UPR activation. Pulse-chase analysis was performed on lhs1-1Δire1 and lhs1-1 cells at permissive and restrictive temperatures. Translocation of the posttranslational substrates CPY and Gas1p displayed minor or no defects at 23 °C in both strains as expected (Fig. 3A and Fig. S2A). However, at 37 °C, the difference was dramatic depending on the status of the UPR. CPY and Gas1p translocation was nearly halted in lhs1-1Δire1 cells whereas only a slight delay was observed in lhs1-1 cells (Fig. 3A and Fig. S2A, “37 °C”). Interestingly, the cotranslational substrate DPAP B was processed proficiently in both strains at 37 °C, suggesting that Lhs1p functions primarily in a posttranslational mode of translocation (Fig. S3B).
Fig. 3.
Maintaining the UPR resting state reveals severe defects in LHS1-, SCJ1-, and ALG5-deficient strains. (A) CPY and Gas1p biogenesis was analyzed in lhs1-1Δire1 and sec63-1 strains at 23 °C and 37 °C as described in Fig. 1A. (B) Pulse-chase analysis was performed at 37 °C to examine the degradation of CPY*-HA in WT, Δire1, scj1-1, and scj1-1Δire1 cells. The graph represents the mean ± SD of three independent experiments. (C) The biosynthesis of CPY was monitored in alg5-2Δire1 as described in Fig. 1C. rft1-2 cells were included to indicate positions of underglycosylated mCPY, which are denoted as “−1”, “−2”, “−3”, and “−4”, representing triply-, doubly-, singly-, and nonglycosylated species, respectively (11). “Mx” denotes the portion of the gel composed of p1, p2, and mCPY forms that are not easily differentiated. The other labels are described as in Fig. 1C. The immature form of CPY after Endo H digestion was expressed as a percentage of the total and is indicated.
Similarly, the absence of UPR induction in SCJ1-deficient cells resulted in nearly complete impairment of ERAD, as demonstrated by CPY* stabilization (Fig. 3B). The partial stabilization of CPY* in Δire1 cells could not be avoided as applied stress, in the form of misfolded protein expression, is necessary to analyze ERAD. Nonetheless, the severity of the scj1-1Δire1 phenotype compared with scj1-1 indicates that UPR activation efficiently alleviates the ERAD defect in SCJ1-deficient cells. Notably, the biogenesis of three different substrates, CPY, DPAP B, and Gas1p, was unaffected in the scj1-1Δire1 cells (Fig. S4). Despite its identity as an ER DnaJ homolog, these data suggest that its function may be restricted to ERAD.
In ALG5-deficient cells, the absence of UPR induction revealed increased underglycosylation of the p1 form, indicating that glycosylation is less efficient in the absence of UPR induction (Fig. 3C, compare 0-min lanes). The dearth of corresponding mature forms after a 30-min chase suggested CPY maturation is defective in this strain (Fig. 3C). This effect can be quantified after deglycosylation with Endo H, showing that 47% of CPY fails to reach the vacuole in alg5-2Δire1 cells compared with 16% in alg5-2 and 12% in Δalg5 (Figs. 1C and 3C and Fig. S2B). The increased immature fraction persisted even after a long chase, indicating that retention is a terminal event (Fig. S5B). The block is not caused by a general trafficking defect because the transport of Wsc1p, a COPII cargo protein not subject to ER quality control, is unaffected under the same conditions (Fig. S5C) (17). Instead, it could be due to a folding defect because endogenous CPY must fold for transport. Consistent with this notion, an assay based on chemical modification of unpaired cysteine residues shows that alg5-2Δire1 cells impair formation of native CPY disulfide bonds, a process dependent on correct protein folding (Fig. S5D) (17). Although UPR activation serves to improve protein glycosylation in ALG5-deficient cells, another important function may be to improve the folding of underglycosylated proteins.
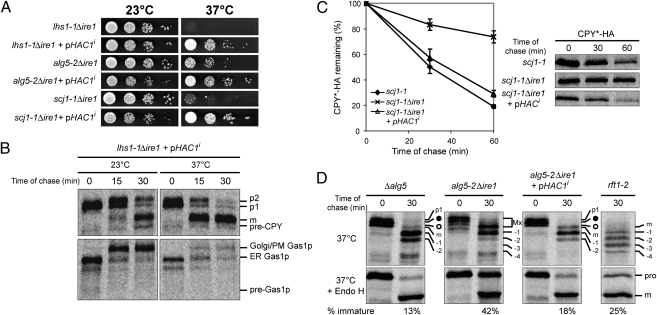
To confirm that UPR activation is responsible for alleviating these cellular defects, we introduced an active form of the downstream effector, Hac1ip into the temperature-sensitive strains (4). As shown in Fig. 4A, Hac1i suppressed the temperature-sensitive phenotype and alleviated the genetic defect of each strain (Fig. 4 B–D).
Fig. 4.
The constitutive UPR activator HAC1i alleviates defects in the ts strains. (A) Cells with or without HAC1i-bearing plasmid were grown at 23 °C and serial dilutions of the culture were spotted onto duplicate selective SC plates. These plates were incubated at the indicated temperature until the appearance of colonies. (B) The synthesis of CPY and Gas1p was examined in lhs1-1Δire1 containing a HAC1i-bearing plasmid as described in Fig. 1A. (C) The degradation of CPY*-HA was analyzed in scj1-1, scj1-1Δire1, and scj1-1Δire1 with a HAC1i-containing plasmid at 37 °C. The graph was obtained from the mean ± SD of three independent experiments. (D) CPY biogenesis was monitored in Δalg5, alg5-2Δire1, alg5-2Δire1, and rft1-2 carrying HAC1i-bearing plasmid at 37 °C and labeled as described in Figs. 1C and 3C. The percentage of immature CPY compared to the total is shown below each section.
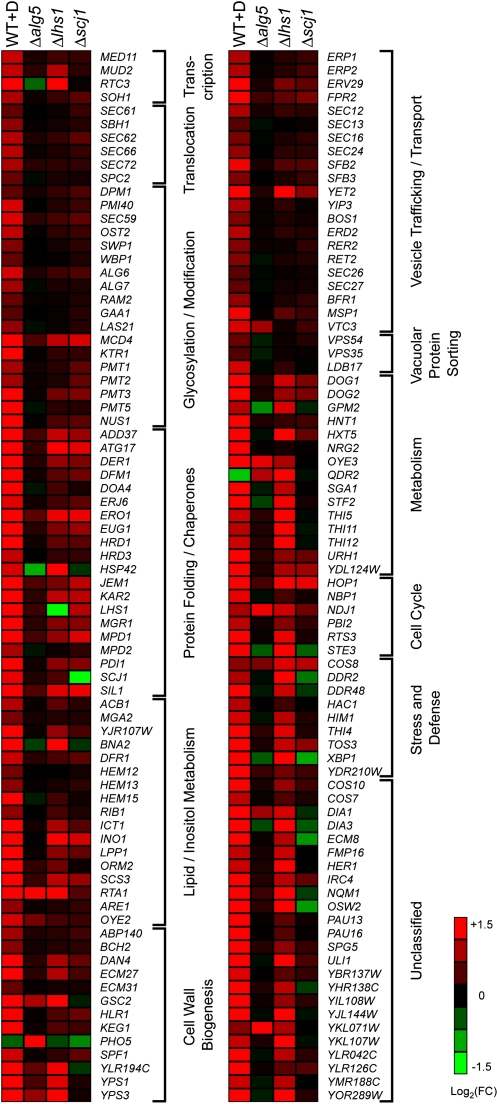
Taken together, these data show that UPR activation effectively compensates for diverse biochemical dysfunctions to aid survival. We next examined how the UPR program is deployed against these different forms of ER stress. For this, DNA microarray analysis was performed using wild type, Δlhs1, Δscj1, and Δalg5 strains. These strains were chosen because they are well adapted to the loss of these functions through UPR activation (Figs. 1 and 2). Consistent with results of the UPRE-LacZ assay, the activation level of UPR target genes in Δalg5 cells is low. Interestingly, the only genes showing consistent up-regulation, albeit modest, are those involved in protein folding (Fig. 5). This finding supports the observed enhancement of ER protein maturation in ALG5-deficient cells. Up-regulation of glycosylation genes was not observed, suggesting that glycosylation enhancement is through a different mechanism. Because N-glycosylation sites must be unstructured for modification, increased chaperone concentrations could explain the enhancement because of their role in preventing inappropriate structures in nascent polypeptides (18). For Δlhs1 and Δscj1 strains the pattern was particularly intriguing. Although both strains display strongly activated UPRs, activation of individual UPR targets is dramatically different. In Δscj1 cells, genes involved in protein folding and quality control are most consistently up-regulated (Fig. 5). Given that most genes annotated for “protein folding” are also involved in ERAD (19), the Δscj1 transcriptional pattern displays a high degree of functional specificity. Δlhs1 cells display the greatest range of up-regulated UPR target genes, but still fewer than cells treated with the chemical inducer DTT. Surprisingly, target genes encoding components of the translocation pore complex are not up-regulated, suggesting that it does not become limiting when Lhs1p is eliminated. Instead, ER chaperones are strongly up-regulated, consistent with a posttranslational translocation defect when they are limiting. Indeed, it was reported that overexpression of the ER chaperone Sil1p can partially suppress the synthetic lethality of a Δlhs1Δire1 double mutant (20). Why genes involved in cell wall biogenesis and metabolism are also broadly up-regulated remains unclear. Perhaps they reflect sensitivity to compromised ER protein translocation, a critical prerequisite for nearly all proteins of the endomembrane system. The analysis of three distinct forms of ER stress reveals that the UPR program is not one-dimensional and can be remodeled differentially according to the needs of the cell.
Fig. 5.
Up-regulated genes by the UPR in Δalg5, Δlhs1, and Δscj1 differ from genes by chemically induced UPR. Shown is a heat map of DNA microarray data for three biological replicates of WT+D (WT cells preincubated 1 h with 2 mM DTT before RNA extraction), Δalg5, Δlhs1, and Δscj1 cells. (Right) Inset assigns heat map colors to the gene expression fold change (FC) compared with WT cells on a log2 scale. The genes shown were selected from previously reported UPR-regulated genes (8).
To begin analyzing how UPR outputs alleviate stress, we constructed overexpression vectors containing ADD37, COS8, DER1, EUG1, FPR2, JEM1, KAR2, and MPD1 genes, which encompass the major UPR targets activated in scj1 mutant cells (Fig. 5 and Table S2). When transformed into scj1-1Δire1 cells, only JEM1 or KAR2 overexpression partially suppressed the ts phenotype (Fig. S6A). This result was intriguing because Kar2p is the ER Hsp70 homolog and Jem1p is an ER DnaJ class protein whose function may overlap with Scj1p. Each of these proteins has been implicated in ERAD (16). Although KAR2-mediated suppression was stronger, only elevated JEM1 reduced the UPR response in Δscj1 cells. However, neither one rescued the ERAD defect in scj1-1Δire1 cells (Fig. S6C). Taken together, these data support the physiological relevance of UPR output data and that full compensation requires the activation of multiple UPR targets.
Discussion
By muting the UPR, severe functional defects were revealed for LHS1-, SCJ1-, and ALG5-deficient cells. Because the UPR is quiescent in unstressed cells, the severity of the phenotypes reflects the importance of these genes under normal conditions (6). Through this approach, we provide direct evidence that the UPR can alleviate stress by reversing severe dysfunctions as diverse as protein translocation, glycosylation, and ERAD. For LHS1 and SCJ1 deficiencies, UPR activation compensates for their primary functions. Both being ER chaperones, up-regulation of multiple chaperones with functional overlaps seem to be sufficient to compensate for their loss (Fig. 5). Cells lacking ALG5, on the other hand, remain completely deficient in oligosaccharide glucosylation (14). The UPR compensates for its indirect defects in protein glycosylation and in the folding of underglycosylated proteins.
These studies provide unique insight in how the UPR program is deployed to maintain homeostasis. Instead of blanket up-regulation of its nearly 400 target genes, the network displays unexpected plasticity according to the specific needs of the cell. This additional level of regulation cannot be explained by the current Ire1p-Hac1p paradigm and suggests unique signaling mechanisms emanating from the ER to modulate individual or subsets of UPR target genes. However, there is evidence that the UPR Ire1p-Hac1p signaling mechanism acts more like a hair trigger for rapid activation (21). This action would be important under conditions of rapid, acute stress (e.g., exposure to chemical perturbants), which is generally the manner under which the UPR is studied. The Δlhs1, Δscj1, and Δalg5 null mutants, on the other hand, experience chronic forms of stress. Because these strains were grown for many generations, they represent cells thriving at a new homeostatic equilibrium of their internal systems. It may be this mode that allows the clearest view of target gene modulation.
The spectrum of biological processes the UPR can compensate is wide, indicated by the range of mutants displaying synthetic lethality with UPR regulatory genes. Indeed, the UPR is so broadly effective that scores of “nonessential” genes would be entirely essential if not for the compensatory effect of UPR activation. The effect might help explain the paradox of why most yeast genes are nonessential (∼80%), even as many are known to perform important functions. Because of scant redundancy of the yeast haploid genome, there are likely numerous strategies of functional compensation at play that can cover lost or impaired genes.
The ability of the UPR to buffer flaws in diverse processes might contribute to the evolution of cells. By supporting survival of negative mutations potentially beneficial for adaptation to new conditions, the fitness of a population could be enhanced by increased genetic diversity. In this way, the UPR can play the role of “capacitor of phenotypic variation,” a concept first put forth by Lindquist and coworkers for the molecular chaperone Hsp90 (22). It was observed that Drosophila lines could bear morphological mutations whose phenotypes are masked through the activity of Hsp90, thus providing a biochemical mechanism that expands genetic diversity.
A conceptual advance from these studies could have practical implications for experimental genetics. The phenotype, or lack thereof, of any UPR synthetic lethal mutant is the sum of the genetic loss-of-function and the activation of various UPR target genes. For example, weak functional defects in Δlhs1 and Δscj1 strains do not reflect the contributions of corresponding genes under normal conditions, when the UPR is quiescent. Instead, their importance was revealed only through the use of conditional synthetic lethal mutants that muted the UPR to better replicate the normal regulatory environment (Fig. 3A). By extension, mild phenotypes of mutants in other systems could be explained by masking through their own compensatory pathways. Thus, disabling the effects of these pathways could improve functional studies when coupled to genetic analysis.
Methods
Plasmids, yeast strains, and primers used in this study are listed in Tables S2, S3, and S4, respectively. Detailed descriptions of the materials used and the experimental conditions, cell labeling and immunoprecipitation, and the DNA microarray are provided in SI Methods.
Genetic Screen for ts Alleles.
A genetic screen for ts alleles of the nonessential genes ALG5, LHS1, and SCJ1 was carried out on the basis of yeast colony color phenotype, taking advantage of the yeast strain, DNY419, with an ire1 null background that was generated for the per screen (11). The ts alleles of LHS1, SCJ1, and ALG5 that were obtained had multiple point mutations and were designated lhs1-1, scj1-1, and alg5-2, respectively. Further details are given in SI Methods and Fig. S1.
β-Galactosidase Reporter Assay.
The strains were transformed with pJC31, which contains the UPRE-CYC1-LacZ reporter previously described (4). The assay was carried as previously described. Experimental details are provided in SI Methods.
Alkylation Sensitivity Folding Assay.
The alkylation sensitivity folding assay was carried out as described previously (17). In this study, the strains were preincubated at 37 °C for 1 h before the addition of 110 μCi of [35S]methionine/cysteine. After a 5-min pulse, cold methionine/cysteine was added to a final concentration of 2 mM. Samples were taken at the 30-min time point and treated with 5 mM Methoxypolyethyleneglycol 5000 maleimide (mPEG) (Fluka) where indicated.
Supplementary Material
Acknowledgments
We thank Michael Costanzo, Brenda Andrews, and Charlie Boone (University of Toronto) for providing Δhac1 synthetic genetic array data prior to publication. This work was supported by funds from the Temasek Trust and by grants from the Singapore Millennium Foundation (postdoctoral fellowship to G.T. and N.I.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The DNA microarray data discussed in this publication has been deposited in NCBI's Gene Expression Onmibus (GEO) under series number GSE33844.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117184109/-/DCSupplemental.
References
- 1.Barkai N, Shilo BZ. Variability and robustness in biomolecular systems. Mol Cell. 2007;28:755–760. doi: 10.1016/j.molcel.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 2.Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- 3.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 4.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 5.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 6.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 7.Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signalling from endoplasmic reticulum to nucleus: Transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- 8.Travers KJ, et al. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 9.Jonikas MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol. 2010;189:783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng DT, Spear ED, Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craven RA, Egerton M, Stirling CJ. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 1996;15:2640–2650. [PMC free article] [PubMed] [Google Scholar]
- 14.Heesen S, Lehle L, Weissmann A, Aebi M. Isolation of the ALG5 locus encoding the UDP-glucose:dolichyl-phosphate glucosyltransferase from Saccharomyces cerevisiae. Eur J Biochem. 1994;224:71–79. doi: 10.1111/j.1432-1033.1994.tb19996.x. [DOI] [PubMed] [Google Scholar]
- 15.Silberstein S, Schlenstedt G, Silver PA, Gilmore R. A role for the DnaJ homologue Scj1p in protein folding in the yeast endoplasmic reticulum. J Cell Biol. 1998;143:921–933. doi: 10.1083/jcb.143.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Ng DT. Evasion of endoplasmic reticulum surveillance makes Wsc1p an obligate substrate of Golgi quality control. Mol Biol Cell. 2010;21:1153–1165. doi: 10.1091/mbc.E09-10-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lizak C, Gerber S, Numao S, Aebi M, Locher KP. X-ray structure of a bacterial oligosaccharyltransferase. Nature. 2011;474:350–355. doi: 10.1038/nature10151. [DOI] [PubMed] [Google Scholar]
- 19.Vembar SS, Brodsky JL. One step at a time: Endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyson JR, Stirling CJ. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 2000;19:6440–6452. doi: 10.1093/emboj/19.23.6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rüegsegger U, Leber JH, Walter P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 2001;107:103–114. doi: 10.1016/s0092-8674(01)00505-0. [DOI] [PubMed] [Google Scholar]
- 22.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.