Abstract
Long interspersed element-1 (L1) retrotransposons compose ∼20% of the mammalian genome, and ongoing L1 retrotransposition events can impact genetic diversity by various mechanisms. Previous studies have demonstrated that endogenous L1 retrotransposition can occur in the germ line and during early embryonic development. In addition, recent data indicate that engineered human L1s can undergo somatic retrotransposition in human neural progenitor cells and that an increase in human-specific L1 DNA content can be detected in the brains of normal controls, as well as in Rett syndrome patients. Here, we demonstrate an increase in the retrotransposition efficiency of engineered human L1s in cells that lack or contain severely reduced levels of ataxia telangiectasia mutated, a serine/threonine kinase involved in DNA damage signaling and neurodegenerative disease. We demonstrate that the increase in L1 retrotransposition in ataxia telangiectasia mutated-deficient cells most likely occurs by conventional target-site primed reverse transcription and generate either longer, or perhaps more, L1 retrotransposition events per cell. Finally, we provide evidence suggesting an increase in human-specific L1 DNA copy number in postmortem brain tissue derived from ataxia telangiectasia patients compared with healthy controls. Together, these data suggest that cellular proteins involved in the DNA damage response may modulate L1 retrotransposition.
Long interspersed element-1 (L1) retrotransposons are the only autonomously active retrotransposons in the human genome, and they mobilize (i.e., retrotranspose) by a “copy-and-paste” mechanism termed target-site primed reverse transcription (TPRT) (1, 2). Although the vast majority of human L1 sequences are retrotransposition defective, ∼80–100 full-length retrotransposition-competent L1s (RC-L1s) persist in the genome (3, 4). RC-L1s are 6 kb in length and contain two ORFs that encode proteins required for their mobility (5). ORF1 encodes a protein (ORF1p) with RNA binding and nucleic acid chaperone activity (6, 7), whereas ORF2 encodes a protein (ORF2p) with endonuclease (8) and reverse transcriptase (9) activities. L1 retrotransposition occasionally can lead to disease and can impact human genome structural variation by various mechanisms (1, 10, 11). Heritable L1 insertions must occur in the germ line or during early embryonic development (11). However, engineered human L1s can undergo somatic retrotransposition in the mammalian nervous system, and previous studies have demonstrated an increase in the DNA copy number of human-specific L1s in the brains of normal individuals compared with other somatic tissues (12, 13).
Host DNA repair processes may also impact L1 retrotransposition. For example, DNA repair pathways may either inhibit L1 retrotransposition or lead to L1 5′ truncation of de novo insertions (14, 15). Studies of cultured cells and comparative genomics analyses have further demonstrated that L1 retrotransposition events are associated with various genomic structural DNA rearrangements, which include intrachromosomal deletions, intrachromosomal duplication/inversions, and perhaps interchromosomal translocations (11, 16–20). Finally, mutations in genes required for the nonhomologous end-joining (NHEJ) pathway of DNA repair allow for an alternate, endonuclease-independent pathway of L1 retrotransposition (ENi) in select p53-deficient Chinese hamster ovary (CHO) cell lines (14, 21). ENi retrotransposition may occur at areas of DNA disrepair or at dysfunctional telomeres, and the resultant retrotransposition events generally lack canonical L1 structural hallmarks (14, 21).
The ataxia telangiectasia mutated (ATM) gene encodes a 350-kDa serine/threonine kinase that is a sensor of cellular DNA damage. ATM is activated by double-strand DNA breaks and subsequently phosphorylates downstream substrates, such as CHK2, p53, BRCA1, and the MRN complex (MRE11, Rad50, and NBS1), leading to the activation of a DNA damage checkpoint and cell cycle arrest (22). The damaged DNA subsequently is repaired, or the cell may undergo p53-mediated apoptosis (22). In humans, autosomal recessive mutations that inactivate ATM result in ataxia telangiectasia (AT), and AT patients exhibit progressive neurodegeneration and eventual death in the second or third decade of life (22). Gene knockout studies indicate that ATM-deficient mice are viable (23) and model human AT, exhibiting evidence of neurodegeneration, T-cell deficits, growth retardation, infertility, and sensitivity to gamma radiation (23).
Here, we examined L1 retrotransposition in the setting of ATM deficiency. Using a cell culture-based retrotransposition assay (5, 24) and a transgenic mouse model, we consistently observed a modest two- to fourfold increase in L1 retrotransposition in ATM-deficient cells compared with ATM-proficient controls. This increase in L1 retrotransposition likely occurs by conventional TPRT and may result in longer, or perhaps more, L1 retrotransposition events in cells deficient for ATM. We also present evidence for an increase in L1 DNA content in postmortem hippocampal brain tissues derived from human AT patients when compared to healthy age-matched controls. These findings strongly suggest that the ATM-signaling pathway plays a role in regulating L1 retrotransposition.
Results
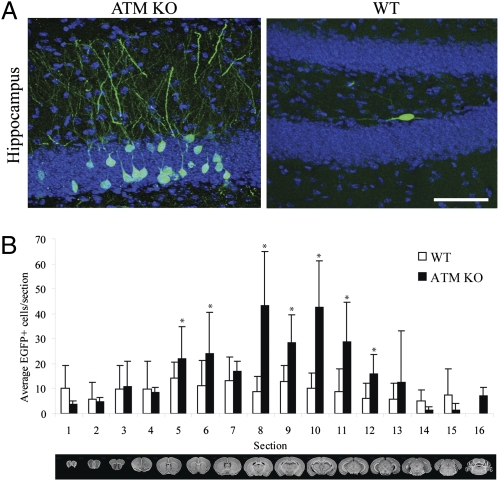
We previously demonstrated that an engineered human RC-L1 (L1RP) (25) containing a retrotransposition indicator cassette could retrotranspose in the brain of transgenic mice (13, 25). L1RP–enhanced green fluorescent protein (EGFP) transgenic mice harbor a retrotransposition-competent human L1 driven by its native promoter (13, 24). The 3′ UTR of the L1 also contains a retrotransposition indicator cassette (mEGFPI) that can be activated only upon L1 retrotransposition (13, 24). To analyze the role of ATM in L1 retrotransposition in vivo, we generated ATM knockout (KO) mice containing the L1RP–EGFP transgene (Fig. S1A) (13, 23, 26). We then used genotyping to screen for mice that contained the L1RP–EGFP transgene but lacked detectable de novo germ-line retrotransposition events (Fig. S1A). Comparisons of the brain samples derived from 3-mo-old wild-type (WT) and ATM KO mice revealed a statistically significant increase in the number of EGFP-positive cells in ATM KO mice compared with WT animals (Fig. 1 A and B). The most marked increase was observed in the hippocampus (Fig. 1A), and EGFP-positive cells were not apparent in other somatic tissues (Fig. S1B). Interestingly, EGFP-positive cells were not seen in the testes of ATM-deficient animals; however, as described previously, rare EGFP-positive cells were noted in WT testes (Fig. S1C) (13, 27). These data suggest an increased rate of L1 retrotransposition in ATM KO animals. The L1 insertions may occur during embryonic development or during adult hippocampal neurogenesis.
Fig. 1.
ATM modulates neuronal L1 retrotransposition in vivo. (A) L1–EGFP retrotransposition occurs in the brains of wild type (WT) and ATM KO animals. (Left) Images of maximal numbers of EGFP-positive cells (indicative of L1 retrotransposition) in ATM KO mice brain. (Right) An image of EGFP-positive cells in an ATM-proficient control mouse brain; the most marked increase was found in the hippocampus. (Scale bar, 50 μm.) (B) Quantification of brain sections in an ATM KO transgenic mouse background revealed more EGFP-positive cells compared with a WT control (six age-matched animals per group). Error bars indicate SEM. *P < 0.05.
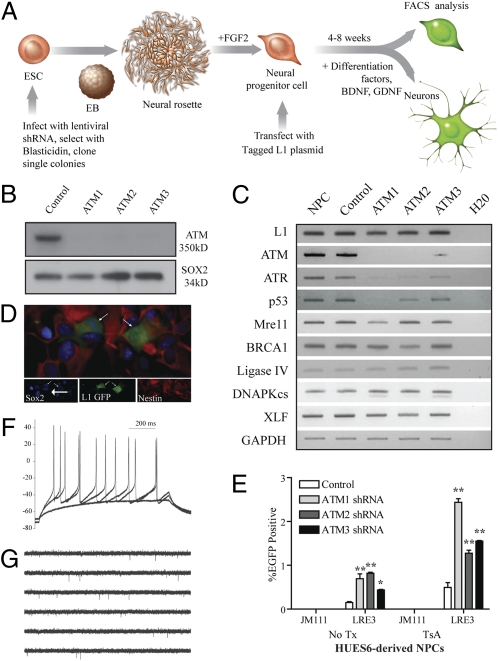
We next examined L1 retrotransposition in ATM-deficient human neural progenitor cells (NPCs). Briefly, we transduced human embryonic stem cells (HUES6-hESCs) with three different lentiviral vectors harboring small hairpin RNAs (shRNAs) designed to knock down ATM expression (Fig. 2A). Each shRNA suppressed ATM expression, whereas a scrambled control shRNA did not (Fig. 2B and Fig. S2 C and D). The derived hESC lines were karyotypically stable (Fig. S3 A–D) and exhibited marked reductions in both ATM expression (Fig. 2C) and protein levels (Fig. 2B). Notably, RT-PCR experiments revealed minor alterations in the expression of p53 and ataxia telangiectasia and Rad3-related protein (ATR) in ATM-deficient cells (Fig. 2C).
Fig. 2.
ATM modulates L1 retrotransposition in HUES6-derived NPCs. (A) Diagram was adapted from ref. 12. HUES6-hESCs were infected with shRNA-expressing lentiviruses and selected with blasticidin. HESC colonies with a strong knockdown of ATM were differentiated to embryoid bodies, then neural rosettes, and were next manually dissected. The resulting NPCs were propagated with fibroblast growth factor 2 (FGF2). NPCs were transfected with an engineered human L1 and were assayed for retrotransposition and/or their ability to differentiate. (B) Western blot analysis indicates robust depletion of ATM expression with ATM 1-3 shRNAs lentiviruses in HUES6-derived NPCs. SOX2 is a loading control. (C) RT-PCR of ATM-deficient HUES6-derived NPCs for genes involved in DNA damage signaling, exhibiting an effect of ATM knockdown on ATR and, to a lesser extent, on p53. (D) ATM-deficient NPCs accommodate L1 retrotransposition; cells exhibiting EGFP expression continued to express neural progenitor markers SOX2 and nestin (arrows indicate colabeled cells). (E) ATM-deficient HUES6-derived NPCs exhibit increased L1 retrotransposition when transfected with an engineered human L1. The addition of trichostatin A (TsA) revealed epigenetic silencing of some engineered L1 insertions. Error bars indicate SEM. *P < 0.05 and **P < 0.01 are a result of a one-way ANOVA with Bonferroni correction. (F and G) LRE3–EGFP-positive ATM-deficient HUES6-derived neurons exhibit suprathreshold responses to somatic current injections from a current clamped potential of −70 mV (F) and can exhibit spontaneous action potentials (G).
NPC lines expressing the shRNA vectors were transfected with an LRE3–EGFP construct, and L1 retrotransposition was monitored by flow cytometry (EGFP-positive cells) 14 d post transfection (Fig. 2A; Methods). An L1 construct containing a pair of missense mutations in ORF1p (JM111/L1RP) served as a negative control (5, 24). We reliably detected an approximately twofold increase in L1 retrotransposition in ATM-deficient NPCs compared with controls (Fig. 2E; n = 3). Notably, the number of EGFP-positive cells was more pronounced upon the addition of the histone deacetylase inhibitor trichostatin A (500 nM) (Fig. 2E), suggesting that some engineered L1 retrotransposition events in NPCs are subject to epigenetic silencing (12, 13, 28).
Experiments were next conducted to examine the integrity of ATM-deficient NPCs. EGFP-positive, ATM-deficient NPCs expressed the neural progenitor markers Nestin and Sox2 (Fig. 2D) and could be differentiated into both neuronal (Map2a+b, βIII tubulin, and synapsin) and glial (glial fibrillary acidic protein-positive) cell fates (Fig. S4 A–E). Characterization of differentiated EGFP-positive, ATM-deficient neurons revealed that some expressed subtype-specific neuronal markers such as the dopaminergic marker tyrosine hydroxylase (Fig. S5A), the cholinergic marker choline acetyltransferase (Fig. S5A), and the inhibitory neuronal marker GABA (γ-aminobutyric acid) (Fig. S4D). Finally, whole-cell perforated patch-clamp recording demonstrated that certain EGFP-positive, ATM-deficient neurons exhibited both responses to somatic current injection and spontaneous action potentials that were similar to ATM-proficient HUES6-derived neurons containing the control, scrambled shRNA (n = 5) (Fig. 2 F and G and Fig. S6 A–E). Thus, WT and ATM-deficient NPCs accommodate engineered human L1 retrotransposition and can be differentiated into functional, mature neurons.
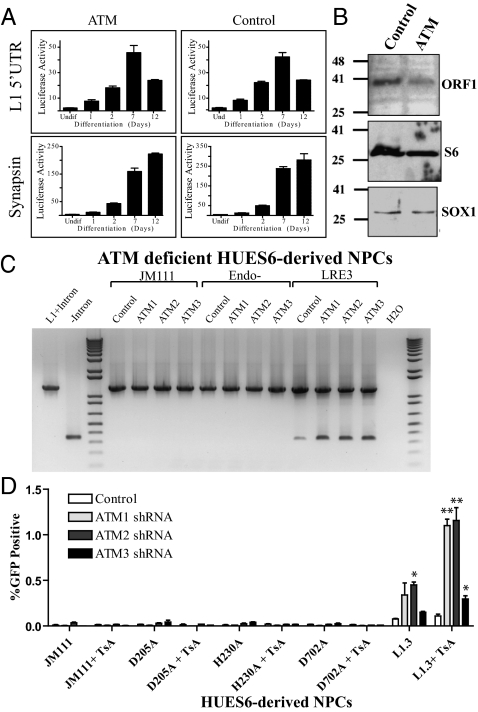
We next investigated why L1 retrotransposition is increased in ATM-deficient NPCs. A recent study reported that loss of MeCP2 leads to an increase in both L1 promoter activity and engineered L1 retrotransposition (27). Thus, we assessed L1 promoter strength, as well as L1 ribonucleoprotein particle (RNP) levels, in ATM-proficient and ATM-deficient hESC-derived NPCs. We did not observe significant differences in L1 promoter activity upon the differentiation of ATM-proficient or ATM-deficient NPCs (Fig. 3A); similarly, the synapsin promoter showed that the differentiation potential and activity of a cellular promoter were similar in the two cell types (Fig. 3A). Moreover, we did not observe marked differences in the level of endogenous ORF1p present in RNPs isolated from ATM-proficient or ATM-deficient hESC-derived NPCs or hESCs (Fig. 3B and Fig. S7D). L1 expression has been reported to induce cellular toxicity (29). Thus, ATM-deficient cells may have an increased tolerance for L1-induced toxicity, allowing for increased survival of cells containing L1 events. Finally, we observed similar cell cycle profiles and cell division rates, as well as similar 8-d growth and survival rates between ATM-proficient and ATM-deficient hESCs and NPCs (Fig. S2 A, B, and E).
Fig. 3.
Examination of L1 retrotransposition in ATM-deficient HUES6-derived NPCs. (A) The L1 5′ UTR is rapidly induced upon HUES6-derived NPC differentiation in both ATM-deficient and ATM-proficient shRNA-infected cells, as is synapsin, a marker of differentiation. The x axis indicates days postdifferentiation; the y axis indicates luciferase fold activity. (B) RNP particles were isolated from control, and ATM-deficient NPCs and were subjected to Western blotting using anti-ORF1p and antiribosomal S6 antibodies. SOX1 is a whole-cell lysate control. (C) PCR of genomic DNA confirmed L1 retrotransposition. The 1,243-bp product corresponds to the EGFP retrotransposition indicator cassette (lane 1, L1 + intron); the 342-bp product is diagnostic for intron loss (lane 2, control EGFP no intron). (D) L1 retrotransposition (EGFP-positive cells) is detected in HUES6-derived NPCs transfected with a WT L1 (L1.3–EGFP), but not in cells transfected with mutant L1 constructs [L1.3/JM111–EGFP (ORF1 mutant), L1.3/D205A–EGFP, or L1.3/H230A–EGFP (endonuclease mutants) or L1.3/D702A–EGFP (reverse transcriptase mutant)]. Notably, L1.3–EGFP retrotransposition is modestly increased in ATM-deficient cells compared with controls. Also, the addition of the histone deacetylase inhibitor trichostatin A (TsA) enhanced the ability to detect retrotransposition. Error bars indicate SEM. *P < 0.05 and **P < 0.01 are a result of a one-way ANOVA with Bonferroni correction.
Previous studies revealed an alternative pathway of L1 retrotransposition (ENi), which occurs in select CHO cell lines that were defective NHEJ (14, 21). Therefore, we next analyzed whether ENi was responsible for the increase in L1 retrotransposition observed in ATM-deficient cells. As above, we observed an increase in the L1.3–EGFP retrotransposition efficiency in ATM-deficient NPCs compared with ATM-proficient controls (Fig. 3D). By comparison, we did not observe retrotransposition from constructs containing missense mutations in the ORF1p RNA-binding domain (RR261-262AA; JM111/L1RP) (5, 24), the ORF2p RT domain (L1.3/D702A) (30), or the ORF2p endonuclease domain (L1.3/D205A and L1.3/H230A) (5, 8, 14) (Fig. 3D and Fig. S7A). Similar results were obtained from experiments conducted with ATM-deficient and ATM-proficient human fetal brain stem cells (hCNS-SCns) (31) and with HUES6 hESCs (32) (Fig. S7 B and C). Finally, PCR amplification experiments conducted on genomic DNA derived from ATM-deficient, HUES6-derived NPCs revealed splicing of the intron from the mEGFPI reporter cassette in cells transfected with a WT L1 (LRE3–EGFP) but not from cells transfected with ORF1p or endonuclease-deficient mutant L1 constructs (Fig. 3C). Thus, ENi retrotransposition does not appear to be responsible for the increase in engineered L1 retrotransposition observed in ATM-deficient cells.
Using inverse PCR, we characterized engineered L1 retrotransposition events from either ATM-proficient (3 events) or ATM-deficient (13 events) NPCs (12–14, 28) (Table S1). Comparisons of the integration sequences revealed that each retrotransposition event occurred in an actual or inferred L1 endonuclease consensus cleavage site (5′-TTTT/A and derivatives) (14). Three L1 insertions were fully characterized and the remaining 13 insertions were characterized only at their 3′ ends, each ending in a poly(A) tail that ranged in size from ∼22 to > ∼130 bp. Thus, the majority of L1 retrotransposition events in ATM-deficient cells likely occur by conventional TPRT.
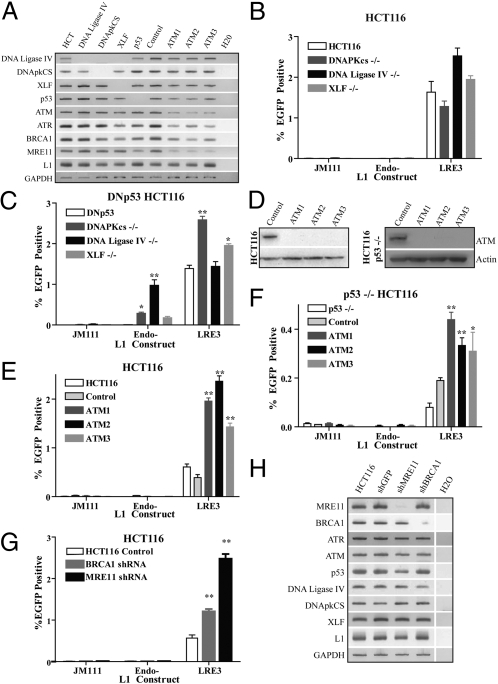
Given the discrepancy between our findings and those of a previous study (33), we sought to independently analyze the role of ATM in L1 mobilization in a nonembryonic/neuronal cell type. We also investigated possible interactions between NHEJ and the ATM DNA repair pathways. We obtained a parental human HCT116 colorectal cancer cell line (34) as well as isogenic mutant HCT116 derivatives that lacked the p53 gene or lacked genes important for NHEJ (DNA-PKcs, XRCC4-like factor (XLF), or the DNA Ligase IV genes, respectively) (Fig. 4A). Notably, each of the NHEJ-deficient cell lines assayed supported LRE3–EGFP retrotransposition but did not accommodate ENi retrotransposition (Endo-, LRE3–H230A–EGFP) (Fig. 4B).
Fig. 4.
The pursuit of endonuclease-independent L1 retrotransposition in HCT116-derived cell lines. (A) RT-PCR analysis confirmed the knockout of NHEJ genes in each of the isogenic HCT116 cell lines. ATM shRNA-infected cells showed a decrease in ATM expression compared with controls. (B) Isogenic HCT116 lines defective for NHEJ, but harboring a WT p53, exhibited robust LRE3–EGFP retrotransposition; however, they did not accommodate the retrotransposition of mutant L1 constructs [ORF1 mutant (JM111–LRE3) and endonuclease mutant (LRE3–H230A–EGFP)]. (C) LRE3–EGFP retrotransposition in HCT116-derived cell lines expressing a dominant-negative p53 (DNp53). Control DNp53 transfection shows robust L1 retrotransposition similar to isogenic p53−/− lines (F), but no ENi retrotransposition was observed (white bar). However, the combined loss of NHEJ (DNA-PKcs, DNA Ligase IV, XLF) and p53 function led to detectable ENi retrotransposition. (D) Western blot analyses in HCT116 (Left) and HCT116 p53−/− (Right) cells infected with control and ATM shRNA lentiviruses. (E) HCT116 cells infected with ATM shRNAs exhibit increased LRE3–EGFP retrotransposition compared with cells infected with control shRNA. (F) In the p53−/− background, knockdown of ATM does not result in ENi retrotransposition activity. However, a modest increase in LRE3–EGFP retrotransposition activity is observed in the ATM shRNA p53−/− cell lines. (G) HCT116 cells were infected with shRNAs against BRCA1 and MRE11, which led to slight increases in engineered L1 retrotransposition. Error bars indicate SEM. *P < 0.05 and **P < 0.01 are a result of a one-way ANOVA with Bonferroni correction. (H) RT-PCR analysis of HCT116 cells transfected with shRNA vectors targeting MRE11 and BRCA1. The expression of other NHEJ genes was not severely affected.
Because human HCT116 cells possess WT p53 and CHO cells lack p53 expression (35), we investigated whether the loss of both NHEJ and p53 signaling is required for efficient ENi L1 retrotransposition in HCT116 cells. To accomplish this goal, we used stable transfection to introduce a dominant-negative p53 construct (DNp53) into the NHEJ-deficient HCT116 lines (36). Remarkably, the resultant cell lines accommodated readily detectable levels of ENi retrotransposition when transfected with an endonuclease-deficient L1 construct (LRE3–H230A–EGFP) (Fig. 4C). Thus, consistent with previous reports, efficient ENi retrotransposition in mammalian cells seems to require mutations that inactivate both NHEJ and p53 functions (14, 21). Notably, in contrast to a previous report (37), we observed efficient L1 retrotransposition in both p53-proficient and p53-deficient HCT116 cells (Fig. 4 B and C).
We next used lentiviral shRNAs to knock down ATM expression in both parental and isogenic p53-deficient HCT116 cells (Fig. 4D). As above, ATM-deficient HCT116 cells transfected with LRE3–EGFP exhibited an approximately twofold increase in L1 retrotransposition (Fig. 4E). However, we did not observe ENi retrotransposition in ATM/p53-deficient cells (Fig. 4F). Finally, we demonstrated that shRNAs directed against the ATM-signaling partners MRE11 and BRCA1 led to slight increases in LRE3–EGFP retrotransposition (Fig. 4 G and H). These data confirm that ATM deficiency leads to a modest increase in engineered L1 retrotransposition.
We next sought to explain why L1 retrotransposition is enhanced in ATM-deficient cells. Notably, most genomic L1 insertions are 5′ truncated (1, 38). Because the L1 ORF2p RT has been shown to be highly processive in vitro (39), we hypothesized that cellular DNA repair and damage sensing proteins may impact L1 5′ truncation (15, 17). If so, the loss of ATM may lead to longer, or perhaps more, L1 insertions in ATM-deficient cells.
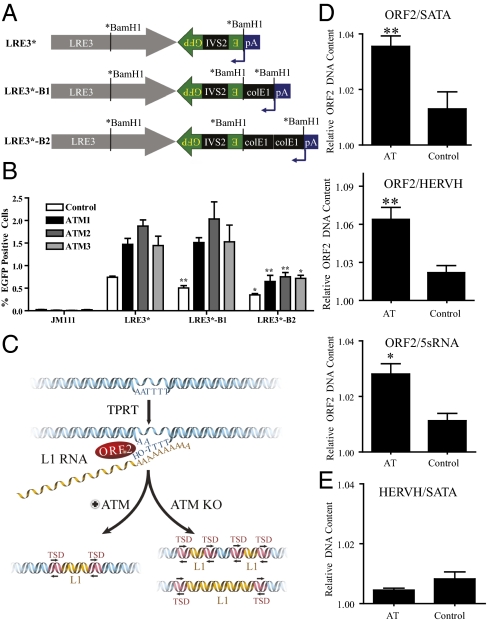
As an initial test of the above hypothesis, we generated constructs that contained either one or two 500-bp spacer regions (ColE1) between the LRE3 polyadenylation site and the mEGFPI retrotransposition indicator cassette (Fig. 5A). A retrotransposition of 1.6 kb (no spacer; LRE3*–EGFP), 2.1 kb (one spacer; LRE3*–B1–EGFP), or 2.6 kb (two spacers; LRE3*–B2-EGFP) would be required to activate EGFP expression (24). Notably, the ColE1 spacer sequence does not prohibit engineered L1 retrotransposition but, when located 3′ to the mneoI indicator, reduces the detection of L1 retrotransposition (16). The apparent decrease in L1 retrotransposition likely reflects the increased length of retrotransposed products that are needed to allow the expression of the retrotransposed mneoI indicator cassette.
Fig. 5.
An assay to detect changes in L1 insertion sizes in ATM-deficient, HUES6-derived NPCs and multiplex qPCR analysis of L1 DNA copy number. (A) A derivative of LRE3–EGFP was created where the BamH1 site in ORF2 is silently mutated (*BamH1). A second BamH1 site immediately upstream of the polyadenylation sequence (pA) was used to insert one or two copies of a ColE1 spacer. (B) Transfection of the LRE3–EGFP constructs with zero, one, or two ColE1 spacers resulted in decreasing L1–EGFP expression. There was a statistically significant decrease in EGFP expression between constructs with zero and one copy of ColE1 in control shRNA-infected cells; however, there was no change in the amount of EGFP expression in ATM-deficient cells regardless of the ATM shRNA used. Using a one-way t test, we compared the decreases between LRE3 and LRE3*–B1 or between LRE3*–B1 and LRE3-B2 for each sample (n = 6). (C) A schematic model for the role of ATM in L1 retrotransposition. L1 retrotransposition through TPRT involves first-strand nicking by the ORF2p endonuclease and priming by the exposed 3′ hydroxyl group for reverse transcription and likely is followed by second-strand cleavage and second-strand L1 cDNA synthesis. In our model, ATM is involved in recognizing the DNA breakage intermediate created during L1 integration and is implicated in its resolution and repair. Loss of ATM leads to either more or longer insertions as a result. (D) Results from the multiplex qPCR analysis of L1 sequences from the hippocampus. The ratio of ORF2 to internal control represents the amount of L1 ORF2 DNA sequence in each sample relative to the amount of another multicopy DNA control, such as 5S RNA sequence, HERVH, and α-satellite. Under these conditions, the copy number of L1 ORF2 sequences was higher in the AT hippocampal neurons (NeuN-positive) compared with those in control hippocampal neurons. (E) Multiplexing of control HERVH primers with satellite α (SATA) primers indicated no significant change in copy number; P ≤ 0.15. Error bars in all panels indicate SEM. *P < 0.05 and **P < 0.005 were obtained using a one-way t test.
We first tested the spacer constructs for retrotransposition in both HCT116 and HeLa cells. The addition of one spacer reduced retrotransposition by ∼3-fold in HCT116 cells and by ∼2.5-fold in HeLa cells compared with the parental construct (Fig. S8 A and B). Similarly, the addition of two spacers resulted in an ∼5-fold and an ∼3.5-fold reduction of L1 retrotransposition in HCT116 cells and HeLa cells, respectively (Fig. S8 A and B) (16). The presence of two spacers resulted in an incremental decrease in EGFP-positive cells compared with one spacer; however, this decrease was not statistically significant in a direct comparison between one and two spacers (Fig. S8 A and B). Control PCR reactions with primers flanking the inserted cassette (Fig. S8E, primers cF, cR) confirmed the increased insertion size of the vector and retrotransposed products containing the additional spacers (Fig. S8C). Finally, PCR reactions with primers flanking the intron in the EGFP indicator cassette (Fig. S8E, primers dF, dR) confirmed the presence of the retrotransposed products in each sample (Fig. S8D).
Given the above results, we hypothesized that the spacer constructs could be used as a proxy to determine the overall length of L1 retrotransposition events in ATM-proficient and ATM-deficient NPCs. As expected, the addition of one or two spacers resulted in decreased L1 retrotransposition (∼1.5- and ∼2- to 3-fold, respectively) in ATM-proficient NPCs (Fig. 5B, white bars). In contrast, we did not observe a significant decrease in the retrotransposition efficiency in ATM-deficient NPCs transfected with the construct that contained one spacer (1.6% LRE3* vs. 1.7% LRE3*–B1 EGFP-positive cells) (Fig. 5B). Notably, we observed a similar trend in data using the construct with two spacers (LRE3*–B2–EGFP), although both control and ATM-deficient NPCs also showed statistically significant drops in EGFP expression (Fig. 5B). These data are consistent with the hypothesis that the resultant L1 retrotransposition insertions are longer in ATM-deficient NPCs compared with ATM-proficient controls. Notably, a similar finding indicated that deficiencies in NHEJ resulted in longer insertions when assaying a zebrafish LINE element in chicken DT40 cells (15). Thus, we speculate that ATM may recognize intermediates that are generated during the process of L1 integration as DNA damage and thereby reduce the length of the resultant retrotransposition events (Fig. 5C).
Finally, we analyzed the copy number of endogenous, human-specific L1s using a previously described quantitative (Taqman-based) PCR (qPCR) methodology (12, 27). Briefly, we designed primers that amplify ∼4,500 genomic L1s (12, 27). The primers primarily target L1PA1/L1Hs elements (∼1,800 genomic elements); however, because of sequence homology, they also can amplify L1s from slightly older subfamilies (e.g., L1PA2 and L1PA3). Because hippocampal sections from AT patients exhibited distorted ultrastructures, we used laser capture for neuronal nuclei (NeuN)-stained nuclei. We then used qPCR to compare changes in L1 copy numbers (12, 27) in postmortem hippocampal samples from AT and age- and sex-matched control patients (n = 7/group, 8–28 y of age). qPCR was used to compare the number of L1 ORF2 sequences normalized to nonmobile repetitive DNA sequences (SATA, HERVH, 5sRNA gene). In all comparisons, we observed a statistically significant increase in ORF2 copy number in AT neurons compared with controls (Fig. 5D). Notably, we did not observe a marked difference between two other repeat sequences, HERVH and SATA (12, 27), between AT patients and controls (Fig. 5E).
It is worth pointing out caveats regarding the above analysis. First, we are measuring changes only in L1 copy number, and the characterization of endogenous retrotransposition events is needed to unequivocally prove de novo, somatic L1 retrotransposition. Second, and unfortunately, we could not obtain other somatic tissues from AT patients, which limited our ability to estimate changes in L1 copy number. With these caveats clearly stated, our findings suggest that L1 retrotransposition might be exacerbated in AT patients. Clearly, recent advances in DNA sequencing technology should help definitively show if L1 retrotransposition is indeed elevated in certain somatic cells in the brain.
Discussion
Using various strategies, we have shown that ATM can impact L1 retrotransposition. First, we observed an increase in the retrotransposition efficiency of engineered human L1s, likely by conventional TPRT, in ATM-deficient hESCs, hESC-derived NPCs, hCNS-SCns, and HCT116 cells. Second, we observed an increase in the retrotransposition efficiency of an engineered human L1 in ATM knockout transgenic mice. Third, we observed an increase in L1 DNA copy number in brain samples from AT patients.
Our initial experiments suggested that engineered L1 retrotransposition events in ATM-deficient cells may be of a longer length compared with ATM-proficient controls. However, we cannot exclude that ATM deficiency allows cells to accommodate more L1 retrotransposition events per cell. Finally, although we did not observe major changes in the cell cycle, growth, proliferation, or survival rates between ATM-proficient and ATM-deficient cells, it is formally possible that ATM deficiency could render cells more tolerant to L1-induced toxicity (29). Clearly, further experiments are required to determine why ATM deficiency results in higher levels of retrotransposition.
During the course of our studies, we also analyzed L1 retrotransposition in NHEJ- and p53-defective human cell lines. Although previous studies suggested that p53 limits L1 retrotransposition by inducing apoptosis (37), we observed similar L1 retrotransposition efficiencies in p53-proficient and p53-deficient HCT116 cells; these data compel a re-examination of the importance of p53 in L1 retrotransposition.
Consistent with previous studies in chicken DT40 cells (15), we did not observe an increase in ENi retrotransposition in DNA-PKcs, XRCC4-like factor (XLF), or DNA Ligase IV-deficient HCT116 cell lines. However, each of these HCT116 cell lines accommodated readily detectable levels of ENi retrotransposition when p53 function was abrogated by the expression of a dominant-negative allele of p53. The latter findings are consistent with previous experiments conducted in XRCC4 and DNA-PKcs deficient CHO cells that lack p53 function (14, 21). Thus, it is likely that the combined loss of p53 and NHEJ is required to allow L1 to use endogenous DNA lesions as substrates for the alternative ENi retrotransposition pathway in mammalian cells.
Notably, Gasior and colleagues (33) have previously reported that ATM is required for L1 retrotransposition in immortalized ATM-deficient human fibroblasts. Those data are in stark contrast to what we observed in hESCs, NPCs, hCNS-SCns, HCT116 cells, and ATM-KO mice; these discrepancies warrant further investigation. It is worth highlighting differences between our studies. First, Gasior and colleagues used an L1 retrotransposition assay based on the generation of G418-resistant foci; they noted a decrease in G418-resistant foci formation with the control neoR vector in ATM-deficient cells compared with a control cell line (33). Thus, ATM-deficient fibroblasts either may be more sensitive to experimental manipulation, or may respond differently to L1 retrotransposition compared with other cell types. Previous reports have indicated that L1 retrotransposition occurs at very low levels in fibroblasts (40), and we did not observe L1 retrotransposition in skin in our transgenic mouse model. Finally, it is possible that our use of shRNAs may provoke cellular perturbations that result in compensatory mechanisms; however, the finding of increased L1 retrotransposition in ATM-KO mice leads us to conclude that ATM is not strictly required for L1 retrotransposition.
Our data provide evidence that ATM has a role in L1 retrotransposition in neuronal and nonneuronal cell types. It is tempting to speculate that L1 retrotransposition may be involved in diverse neurological disease processes. Whether increased L1 retrotransposition contributes to the neurodegenerative disease process of AT remains to be determined, and may merely be a consequence, rather than an actor, in the course of disease.
Methods
Detailed methods and data collection are provided in SI Methods.
Animals.
The L1RP–EGFP transgenic mouse (13) was crossed into an ATM-deficient background (23).
Constructs and PCR.
ATM lentiviral shRNA constructs were commercially available. Cells were transfected with L1s containing an EGFP retrotransposition cassette in a modified version of pCEP4 (Invitrogen) (24). Adult human tissues from AT patients and control samples were obtained from the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders (University of Maryland). Inverse PCR and PCR cycling conditions were described previously (12, 27, 32).
Cell Culture and Analysis.
HUES6 hESCs and NPCs were derived as described (12). Lentivirus was produced as previously described (13).
Supplementary Material
Acknowledgments
We thank J. Simon for schematics, M. L. Gage and N. Leff for editorial comments, Prof. M. Weitzman for the kind gifts of cell lines, K. Stecker for laser capture, E. Mejia for tissue assistance, and L. Randolph-Moore for molecular advice. F.H.G.'s laboratory is supported by the Mather's Foundation, California Institute for Regenerative Medicine, and National Institutes of Health/National Institute of Neurological Disorders and Stroke (MH088485) J.L.G.-P's laboratory is supported by Instituto Salud Carlos III-Consejeria de Salud Junta de Andalucia- Fondo Europeo Desarrollo Regional (EMER07/056); by Marie Curie International Reintegration Grant Action FP7-PEOPLE-2007-4-3-IRG; by Consejeria de Innovacion Ciencia y Economia- Fondo Europeo Desarrollo Regional (P09-CTS-4980); by Proyectos en Salud-FEDER PI-002 from Junta de Andalucia (Spain); and by the Spanish Ministry of Health (FIS-FEDER PI08171). A.R.M. is supported by the National Institutes of Health through the National Institutes of Health Director's New Innovator Award Program (1-DP2-OD006495-01) and by the Emerald Foundation. J.V.M. is supported by National Institutes of Health Grants GM06518 and GM082970. J.V.M. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Telomerase and Retrotransposons: Reverse Transcriptases That Shaped Genomes,” held September 29–30, 2010, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at www.nasonline.org/telomerase_and_retrotransposons.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100273108/-/DCSupplemental.
References
- 1.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 2.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 3.Beck CR, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 6.Hohjoh H, Singer MF. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 9.Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 10.Kazazian HH., Jr Mobile elements and disease. Curr Opin Genet Dev. 1998;8:343–350. doi: 10.1016/s0959-437x(98)80092-0. [DOI] [PubMed] [Google Scholar]
- 11.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 14.Morrish TA, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31(2):159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki J, et al. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 2009;5:e1000461. doi: 10.1371/journal.pgen.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Symer DE, et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 19.Han K, et al. Genomic rearrangements by LINE-1 insertion-mediated deletion in the human and chimpanzee lineages. Nucleic Acids Res. 2005;33(13):4040–4052. doi: 10.1093/nar/gki718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrish TA, et al. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 22.Shiloh Y. ATM (ataxia telangiectasia mutated): Expanding roles in the DNA damage response and cellular homeostasis. Biochem Soc Trans. 2001;29:661–666. doi: 10.1042/0300-5127:0290661. [DOI] [PubMed] [Google Scholar]
- 23.Barlow C, et al. Atm-deficient mice: A paradigm of ataxia telangiectasia. Cell. 1996;86(1):159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 24.Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH., Jr Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostertag EM, et al. A mouse model of human L1 retrotransposition. Nat Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 26.Muotri AR, Zhao C, Marchetto MC, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. 2009;19:1002–1007. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muotri AR, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Perez JL, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466:769–773. doi: 10.1038/nature09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace NA, Belancio VP, Deininger PL. L1 mobile element expression causes multiple types of toxicity. Gene. 2008;419(1–2):75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei W, et al. Human L1 retrotransposition: Cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uchida N, et al. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci USA. 2000;97:14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Perez JL, et al. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 33.Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Topaloglu O, Hurley PJ, Yildirim O, Civin CI, Bunz F. Improved methods for the generation of human gene knockout and knockin cell lines. Nucleic Acids Res. 2005;33(18):e158. doi: 10.1093/nar/gni160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moro F, et al. p53 expression in normal versus transformed mammalian cells. Carcinogenesis. 1995;16:2435–2440. doi: 10.1093/carcin/16.10.2435. [DOI] [PubMed] [Google Scholar]
- 36.Irwin M, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 37.Haoudi A, Semmes OJ, Mason JM, Cannon RE. Retrotransposition-competent human LINE-1 induces apoptosis in cancer cells with intact p53. J Biomed Biotechnol. 2004;2004(4):185–194. doi: 10.1155/S1110724304403131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pavlícek A, Paces J, Zíka R, Hejnar J. Length distribution of long interspersed nucleotide elements (LINEs) and processed pseudogenes of human endogenous retroviruses: Implications for retrotransposition and pseudogene detection. Gene. 2002;300(1–2):189–194. doi: 10.1016/s0378-1119(02)01047-8. [DOI] [PubMed] [Google Scholar]
- 39.Piskareva O, Schmatchenko V. DNA polymerization by the reverse transcriptase of the human L1 retrotransposon on its own template in vitro. FEBS Lett. 2006;580:661–668. doi: 10.1016/j.febslet.2005.12.077. [DOI] [PubMed] [Google Scholar]
- 40.Kubo S, et al. L1 retrotransposition in nondividing and primary human somatic cells. Proc Natl Acad Sci USA. 2006;103:8036–8041. doi: 10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.