Abstract
By homology with the unique plastid terminal oxidase (PTOX) found in plants, two genes encoding oxidases have been found in the Chlamydomonas genome, PTOX1 and PTOX2. Here we report the identification of a knockout mutant of PTOX2. Its molecular and functional characterization demonstrates that it encodes the oxidase most predominantly involved in chlororespiration in this algal species. In this mutant, the plastoquinone pool is constitutively reduced under dark-aerobic conditions, resulting in the mobile light-harvesting complexes being mainly, but reversibly, associated with photosystem I. Accordingly, the ptox2 mutant shows lower fitness than wild type when grown under phototrophic conditions. Single and double mutants devoid of the cytochrome b6f complex and PTOX2 were used to measure the oxidation rates of plastoquinols via PTOX1 and PTOX2. Those lacking both the cytochrome b6f complex and PTOX2 were more sensitive to light than the single mutants lacking either the cytochrome b6f complex or PTOX2, which discloses the role of PTOX2 under extreme conditions where the plastoquinone pool is overreduced. A model for chlororespiration is proposed to relate the electron flow rate through these alternative pathways and the redox state of plastoquinones in the dark. This model suggests that, in green algae and plants, the redox poise results from the balanced accumulation of PTOX and NADPH dehydrogenase.
Keywords: Chlamydomonas reinhardtii, state transitions, chloroplast, photosynthesis
Almost 40 y ago, Diner and Mauzerall (1) reported that oxygen had a positive feedback on photosynthetic oxygen evolution, and they attributed this to an oxygen-dependent oxidation of the intersystem electron carriers. Chlororespiration was subsequently described as a light-independent electron transport pathway in chloroplasts, involving plastoquinones as electron carriers in the oxygen-mediated oxidation of NADPH (2). Originally, chlororespiration was proposed to involve two membrane-bound proteins, a plastoquinol (PQH2) oxidase and an NAD(P)H dehy-drogenase (NDH). A dehydrogenase component has recently been identified and attributed to a type II NADPH dehydrogenase (NDA2) in Chlamydomonas (3). A gene encoding the plastid terminal oxidase (PTOX) was unexpectedly identified upon characterization of the IMMUTANS (im) mutant from Arabidopsis that showed a variegated and carotenoid-deficient phenotype (4, 5). The genetic and biochemical evidence for the existence of an oxidase in Chlamydomonas thylakoids was further substantiated using a specific antibody, raised against the mature sequence of the PTOX protein product from Arabidopsis, that recognized a 43-kDa protein present in thylakoid extracts (6).
Whereas studies of the PTOX-defective GHOST (gh) mutant from tomato, producing yellow fruit, pointed to the critical role of PTOX in carotenoid biosynthesis (7), two studies of marine phytoplankton with uneven photosystem II (PSII) to photosystem I (PSI) stoichiometries have recently highlighted the contribution of PTOX as an alternative electron sink (8, 9). As stressed by these authors, the importance of this mechanism may stem from the low iron environment of oligotrophic oceans compromising PSI and cytochrome b6f complex levels. It would contribute to photoprotection by maintaining an electron flow through PSII and PTOX, thereby building up a ΔpH and thus allowing for the production of ATP and the onset of nonphotochemical quenching (NPQ). This suggests that in stark contrast to most plants, marine species may have a PTOX activity that can compete with a compromised linear electron flow, albeit for the moment seen only in certain strains and under certain conditions. Still, a somewhat similar role for PTOX would be found in high mountain species, as part of a stress response, because their exposure to high light under chilling conditions results in an extremely high accumulation of PTOX (10, 11).
From the data presently available from genome sequencing projects, photosynthetic species have varying numbers of copies of the PTOX gene. Streptophytes appear to have only one PTOX, whereas prasinophytes, chlorophytes, diatoms, and red alga have two, sometimes significantly divergent, PTOX paralogues. Here we report the isolation and characterization of a knock-out mutant of PTOX2 in Chlamydomonas reinhardtii. We show that PTOX2 is the major oxidase involved in chlororespiration in Chlamydomonas, and we measure the in vivo oxidase activity of PTOX1 and PTOX2, taking advantage of the construction of a double mutant for PTOX2 and cytochrome b6f. Growth of mutants without PTOX2 and with or without the b6f complex under different light conditions allows us to discuss the functional significance of this thylakoid-bound oxidase.
Results
Isolation of a Chlamydomonas Mutant Devoid of Plastid Terminal Oxidase 2.
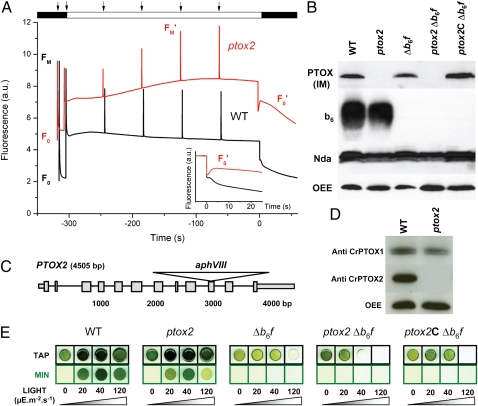
Mutants were generated by transformation with an aphVIII selectable marker cassette, with aphVIII under the constitutive control of the HSP70A-RBCS2 tandem promoter and with the RBCS2 gene transcriptional terminator. Initial selection of transformed clones was by their resistance to paromomycin. Over 12,000 resistant Chlamydomonas clones were screened for unusual chlorophyll fluorescence phenotypes by an in vivo fluorescence imaging setup (12) able to differentiate large numbers of clones from their chlorophyll fluorescence kinetics. From over 80 mutants showing a fluorescence response differing from the wild type (WT), we selected one that showed multiple traits expected for a highly reduced plastoquinone (PQ) pool in the dark. Fig. 1A compares the fluorescence kinetics of this mutant (red), hereafter called ptox2 (see below), with the wild-type strain (black). The fluorescence pattern in the mutant showed four major differences compared with the wild type: (i) After normalization to the maximum fluorescence yield (FM) probed with a saturating pulse (5,000 μE⋅m−2⋅s−1), the fluorescence yield of the dark-adapted state (F0) was higher in the mutant than in the wild type; (ii) at the onset of illumination (150 μE⋅m−2⋅s−1), the photosystem II photochemical rate φPSII (13) was much lower in the mutant than in the WT, yet it increased in the mutant throughout a 5-min period of illumination (Table S1); (iii) maximum fluorescence yield throughout illumination (FM′) rose steadily with time in the mutant, whereas FM′ decreased in the WT; and (iv) both strains showed a fast (t < 1 s) transient decrease of the fluorescence yield in the dark after the illumination period (F0′; Fig. 1A, Inset), probably due to the equilibration of the PQ pool with oxidized plastocyanin (PC) and cyt f, but the mutant showed a pronounced increase of F0′ (t < 5 s; Fig. 1A, Inset). This transient increase in F0′ in the mutant is cancelled in far-red light (Fig. S1), supporting its attribution to the reduction of the PQ pool in the dark by NADPH dehydrogenase (3).
Fig. 1.
Screening of insertion mutants and characterization of ptox2. (A) Fluorescence kinetics of ptox2 versus WT. The fluorescence rise observed after the period of illumination is indicative of a dark reduction of the PQ pool more pronounced in ptox2 than in the WT (Inset). (B) Western blot showing WT, ptox2, Δb6f, ptox2 Δb6f, and ptox2C Δb6f (the complemented double mutant) total cell proteins, reacted with antibodies: Arabidopsis (IM) anti-PTOX, cytochrome b6, and NDA2, and oxygen evolving complex (OEE) from PSII was used as a loading control (for other controls of PSII content in ptox2, see Fig. S2). (C) Scheme of aphVIII insertion in PTOX2. (D) Western blot with thylakoid extracts from WT and ptox2 reacted against CrPTOX1 and CrPTOX2 purified antibodies. (E) Double mutants are more light-sensitive than single mutants.
All of these fluorescence characteristics can be accounted for by the PQ pool being reduced in the dark. A high F0 and a low φPSII at the beginning of the illumination period show that, when dark-adapted, only a fraction of PSII is active as an efficient excitation trap. The smaller fraction of active PSII in the mutant reflects the presence of closed centers (in the QA− state), rather than a decrease in PSII accumulation (Fig. S2), and points to the PQ pool being significantly reduced in the dark-adapted state. This conclusion is further supported by the less pronounced sigmoidal shape of the fluorescence rise in the presence of 3(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) in the mutant (Fig. S3A and references therein). It reflects a lower connectivity between PSII centers that remain active at the onset of illumination, indicative of a smaller number of open centers. Finally, the increase in FM′ throughout the period of illumination (also observed in the presence of DCMU in Fig. S3B) reflects a transition from state 2 to state 1: A highly reduced PQ pool in darkness, which promotes state 2 (larger PSI antenna size), is progressively reoxidized by the turnover of PSI, accounting for the progressive increase in φPSII in the light and a rise in FM′ corresponding to the transition to state 1 (Fig. 1A); 77 K fluorescence emission spectra (Fig. S4) further support that contrary to the WT, ptox2 is in state 2 in the dark.
Due to this fluorescence phenotype, we hypothesized that chlororespiration was affected in the mutant. Thus, we analyzed the protein content of the thylakoid membrane for the presence of PTOX and NADPH dehydrogenase (NDA2) by immunoblotting. The Arabidopsis anti-PTOX (IM) antibody recognized a specific band in the WT that was absent from the mutant strain (Fig. 1B). This showed that the mutant lacked the PTOX protein. In contrast, the accumulation of NDA2 (3) and of its substrate, NADPH (Fig. S5), was similar in the two strains, suggesting that the net reduction of the PQ pool in the mutant is due solely to the absence of PTOX rather than to an enhanced NDA2 activity.
The mutant was backcrossed to the wild-type line and 72 progeny were tested for segregation between their chlorophyll fluorescence phenotype and antibiotic resistance. All individuals resistant to the antibiotic also showed the fluorescence phenotype described above, indicating that insertion of the aphVIII cassette was linked to the phenotype and that there was a unique insertion. Using PCR primers designed to the gene annotated in Joint Genome Institute Chlamydomonas Genome Version 4.0 (http://genome.jgi-psf.org/chlre4) as PTOX2, we walked along the gene until we were unable to amplify a region in the vicinity of the 10th exon of the PTOX2 gene. Further analysis by Southern hybridization using restriction enzymes that cut the PTOX2 gDNA or the PTOX2 gDNA and the aphVIII cassette (Fig. S6) confirmed the scheme presented for the insertion site in Fig. 1C, and the mutant was named ptox2.
We raised antibodies against peptides specific to either PTOX2 or PTOX1. The translated proteins of the gene models after removal of the putative chloroplast targeting sequence gave an expected molecular mass of 44 kDa for PTOX2 and 49 kDa for PTOX1. Fig. 1D shows that the CrPTOX2 antibody recognized a protein of expected mass in wild-type thylakoids that is absent in the ptox2 mutant, whereas the CrPTOX1 antisera recognized a protein of equal abundance in both WT and ptox2 at the expected mass. Interestingly, the PTOX1 antibody reveals a double band when the proteins are separated on a urea/polyacrylamide gel system, which may be due to some presently unknown posttranslational modification of the PTOX1 protein.
In our attempt to complement the ptox2 mutation as proof of phenotype, we looked for a situation where restoration of plastoquinol oxidation by PTOX could be used in a positive selection screen. On solid media, we found no difference in growth rates between WT and ptox2 under mixotrophic conditions (Fig. 1E); neither did we find a pigmentation phenotype as observed for plants (Table S2). Light improved the growth of WT as well as ptox2 in acetate but not on minimal media (Fig. 1E).
We reasoned that if ptox2 mutants were subjected to an additional block in reoxidation of the PQ pool, they should be particularly light-sensitive even at low light intensity. Double mutants lacking both PTOX2 and the cytochrome b6f complex, denoted ptox2 Δb6f, were thus constructed. Progeny were screened by fluorescence video imaging (for Δb6f and ptox2 Δb6f kinetics, see below) and verified to be lacking both proteins by immunoblotting (Fig. 1B). Expectedly, the double mutant ptox2 Δb6f proved more photosensitive than the single mutants Δb6f and ptox2. As shown in Fig. 1E, whereas the Δb6f mutant still grows at 40 μE⋅m−2⋅s−1, the double mutant ptox2 Δb6f does not survive.
We used the light sensitivity of Δb6f ptox2 as a convenient means to complement the ptox2 mutation. We transformed the double-mutant cells with PTOX2 cDNA and placed the transformants in the light. We selected those clones that had the ability to grow under these illumination conditions (see growth test in Fig. 1E) and then screened for a restoration of Δb6f fluorescence kinetics (see below for typical fluorescence profiles). Complemented mutants, still resistant to paromomycin, denoted ptox2C Δb6f, showed a restored accumulation of the PTOX2 protein (Fig. 1B). These complementation experiments confirmed that the mutation we identified in the PTOX2 gene was responsible for the overreduction of the PQ pool in the dark, as in the light for the double mutant.
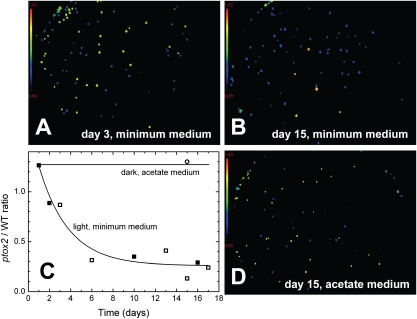
To test whether the inactivation of PTOX2 impacts fitness, we assessed the capacity for survival of the ptox2 strain in competition with the WT at medium light intensity (40 μE⋅m−2⋅s−1). The two strains were mixed in equal quantities (Fig. 2A) and grown under phototrophic conditions. WT cells rapidly became the predominant population (Fig. 2 B and C, where the ratio in cell count ptox2:WT is plotted against time), whereas under heterotrophic conditions in the dark the two populations remained evenly distributed (Fig. 2D). This demonstrates that, in Chlamydomonas, PTOX2 activity provides a fitness advantage in the light.
Fig. 2.
Loss in fitness in the light for ptox2 cells compared with WT cells. (A and B) ptox2 (yellow) and WT (blue) clones analyzed by video imaging after growth in liquid medium. (C) Ratio of cell count ptox2:WT plotted against time; open and closed symbols are the results of two independent experiments. D and circles in C are a control experiment under heterotrophic growth conditions in the dark.
Kinetics of Plastoquinone Pool Oxidation in the Dark.
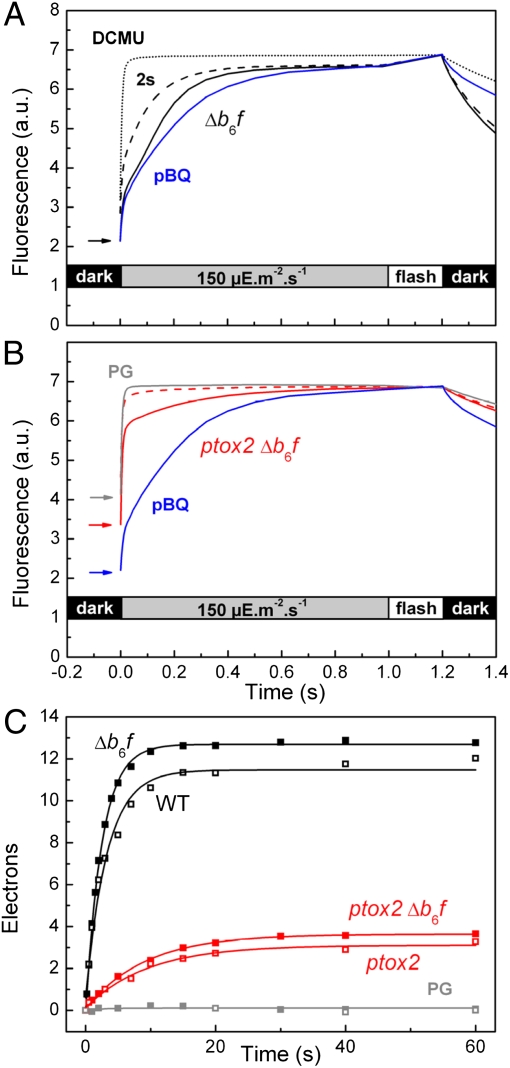
We then assessed quantitatively the redox state of the PQ pool in the dark by comparing the time course of fluorescence changes in ptox2 Δb6f and Δb6f strains. In cells devoid of the cytochrome b6f complex, the light-induced PSII turnover progressively reduces the PQ pool, and this can be followed as a fluorescence rise (Fig. 3A, black solid line). In addition, the area over the fluorescence curve is proportional to the number of PSII electron acceptors (Fig. S7 and references therein). The comparison of Fig. 3A (black solid line) and Fig. 3B (red solid line) shows that the area over the curve is much smaller in ptox2 Δb6f than in the Δb6f mutant, thus showing that the PQ pool is more reduced in the ptox2 Δb6f mutant than it is in the single Δb6f mutant. Under fully oxidized conditions [50 μM p-benzoquinone (pBQ); see blue lines in Fig. 3 A and B], the total pool sizes were similar in both strains, ruling out a perturbation in quinone to PSII stoichiometry associated with the ptox2 mutation.
Fig. 3.
In vivo fluorescence monitoring of PTOX2 and PTOX1 activity in Chlamydomonas cells. (A) Fluorescence rises obtained with the Δb6f strain during the first illumination (solid line), a second illumination 2 s later (dashed line), and treated with 10 μM DCMU (dotted line) or 50 μM p-benzoquinone (blue line). (B) Same as A but for the double mutant ptox2 Δb6f. Addition of 100 μM propylgallate (gray line) gives a fluorescence rise similar to that observed in DCMU in A. (C) Kinetics of reoxidation of the PQ pool after a preillumination (solid symbols are for cells devoid of cytochrome b6f; open symbols are for cells submitted to a hyperosmotic stress); see text for details. Solid lines are the result of a fit with a first-order equation, yielding fluxes of 5 e−⋅s−1⋅PSII−1 for the Δb6f mutant and 0.4 e−⋅s−1⋅PSII−1 for the ptox2 Δb6f double mutant. In the presence of PG, the PQ pool stays reduced.
Successive fluorescence measurements in cells devoid of cytochrome b6f give access to the kinetics of PQH2 oxidation by PTOX. The first illumination fully reduces the PQ pool, then a second illumination, for example 2 s later (see dashed lines in Fig. 3 A and B), probes the number of PQs that have been regenerated through PTOX activity during this 2-s dark period. Dashed lines in Fig. 3 A and B show that the area over the fluorescence rise is much smaller for ptox2 Δb6f than for Δb6f. By varying the delay time between two successive illuminations, one can then obtain the detailed kinetics of PQH2 reoxidation through PTOX (Fig. 3C). This rate can be expressed in terms of electrons transferred per s and per PSII after normalization to the area obtained in the presence of DCMU (one electron borne by QA). We estimate this reoxidation rate at 5 e−⋅s−1 per PSII in the b6f mutant, in good correlation with previous reports (14), and at 0.4 e−⋅s−1 per PSII in the ptox2 Δb6f mutant (Fig. 3C, closed symbols). We performed similar experiments with the WT and ptox2 single mutant, placed under hyperosmotic stress in 1M sucrose, which prevents PQ pool oxidation by PSI (15), and obtained similar values for PTOX activities (Fig. 3C, open symbols). These functional measurements are in agreement with the similar accumulation of the PTOX2 protein in WT and b6f mutant (Fig. 1B) and of the PTOX1 protein in WT and ptox2 mutant (Fig. 1D). Indeed, we tentatively attribute such a residual oxidase activity in ptox2 Δb6f to PTOX1 because propylgallate (PG), a quinone analog acting as an inhibitor of PTOX (16), results in a full reduction of the PQ pool detectable as a fluorescence rise similar to that obtained in DCMU (see gray line in Fig. 3B). Interestingly, a 2-min incubation with 100 μM PG was sufficient to obtain the full reduction of the PQ pool in the ptox2 Δb6f mutant (this work), whereas a 10-min incubation with 1 mM PG had no effect on the redox state of the PQ pool in Δb6f (14). This suggests that PG has a much higher affinity for the residual oxidase, PTOX1, than for PTOX2.
We then assessed the turnover rate of PTOX2 in the light. In the double mutant ptox2 Δb6f, the steady-state fluorescence FS obtained after 1 s of medium light (150 μE⋅m−2⋅s−1) is equal to FM probed after a 250-ms saturating pulse (5,000 μE⋅m−2⋅s−1) (Fig. 3B), whereas FS is lower than FM in the single Δb6f mutant (Fig. 3A). As previously hypothesized (12), this shows that under moderate light, PTOX2 can partially relieve the reducing pressure exerted by PSII on the PQ pool. Based on the (FM − FS)/FM value (13) of 4.4% and the light intensity of 140 electrons transferred per PSII per s (determined from the fluorescence rise time in the presence of DCMU), we estimate the flow sustained by PTOX2 at about 6 e−⋅s−1, in good agreement with the above estimate of 5 e−⋅s−1.
Discussion
Depending on the photosynthetic species, plastid terminal oxidases have been ascribed two major functions: in carotenoid biosynthesis and in photoprotection, acting as a safety valve when the plastoquinone pool is overreduced (17). The former role was substantiated by studies of the carotenoid-deficient mutants IMMUTANS in Arabidopsis and GHOST in tomato, which have provided a model to understand the role of plastoquinone as a substrate in chloroplast and chromoplast development (18). The major role attributed to PTOX in Arabidopsis is as a central regulator of excitation pressure during chloroplast biogenesis (19). Developing plastids have a threshold of excitation pressure that in im plants results in variegation. White sections are due to plastids bearing overreduced thylakoids, whereas green sections have plastids with underthreshold excitation pressure, resulting in normal plastid development. The physiology of unicellular algae is widely different because no cell differentiation develops during growth that occurs through continuous rounds of mitotic division. Here we demonstrate that Chlamydomonas PTOX2 provides a major contribution to chlororespiration that is reflected in the lower fitness of the ptox2 mutant when mixed with the WT during phototrophic growth (Fig. 2). Based on the critical contribution of state transitions to the flexibility of photosynthetic electron flow upon metabolic changes (20), we consider that this lower fitness likely stems from the restricted capability of the ptox2 mutant to undergo state transitions (20) due to the overreduction of its plastoquinone pool.
Indeed, we found that PTOX2 is the major oxidase able to pull out electrons from the plastoquinone pool. Based on the kinetics of pool oxidation in Δb6f and ptox2 Δb6f, we assessed the maximum flow that can be sustained by PTOX2 (5 e−⋅s−1) and a residual oxidase (0.4 e−⋅s−1). Importantly, this latter activity proved to be highly sensitive to propylgallate, an inhibitor specific to alternative oxidases, making PTOX1, the second plastid terminal oxidase encoded by green alga genomes, a very likely candidate.
Redox Model.
The determination of the electron flows through these two oxidases, together with the estimation of the redox state of the PQ pool in the dark, allows the assessment of the redox equilibration model proposed earlier (1, 2, 21) and represented in Fig. 4. In the dark, the plastoquinone pool reaches a steady state, at which its oxidation flow rate Fox is identical to its reduction flow rate Fred. These relative flows are dependent upon the redox state of the pool:
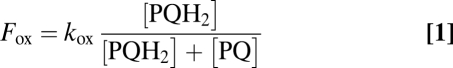 |
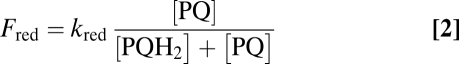 |
Fig. 4.
Model for chlororespiration in the dark involving the NADPH dehydrogenase (NDA2) and PTOX1 and PTOX2 (A) or only PTOX1 (B). Electron transfer rates kred (22) and kox (this work) allow us to calculate the oxidation state of the PQ pool from Eq. 3, here shown in %. Arrows’ thickness is proportional to the electron flow F through the enzymes, expressed per photosystem unit, and calculated from Eqs. 1 and 2.
where kox and kred are the rate constants of pool oxidation and reduction, respectively. Reciprocally, the redox state of the pool can be directly derived from kinetics parameters as follows:
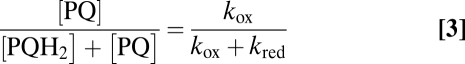 |
As stressed in ref. 21, the rate constants kox and kred are equal to the oxidizing and reducing flow rates Fox and Fred when the pool is fully reduced [PQH2/(PQ + PQH2) = 1] or oxidized [PQ/(PQ + PQH2) = 1], respectively. The rate constant kred depends upon starch breakdown and glycolysis and has been estimated to be about 2 s−1 (22) (Fig. S5). The rate constant kox is found here to be about 5 s−1 with both PTOXs and 0.4 s−1 with PTOX1 only (this work). Thus, the PQ pool is expected to be 72% oxidized in the presence of PTOX2 and 17% oxidized in its absence. These estimations nicely match the direct estimation of the pool sizes of PSII electron acceptors from the areas over the fluorescence rise: A total of 17.7 is estimated from the pBQ-treated sample (Fig. 3 A and B), 12.6 for the Δb6f strain, and 3.6 for the ptox2 Δb6f mutant (see amplitudes in Fig. 3C). After the discount of one electron borne by QA, we obtained a redox state of the PQ pool of 69% and 16% oxidation, respectively. This satisfying agreement with the above estimates of 72% and 17% shows that the redox state of the quinone pool is indeed determined by the simple combination of the electron inflow and outflow predicted by the chlororespiration model.
PTOX2 and Carotenoid Biosynthesis.
We found no evidence for an alteration in carotenoid biosynthesis in the ptox2 mutant (Table S2). The lack of a carotenoid phenotype suggests that the remaining PTOX1 activity is sufficient to regenerate the oxidized PQ required for carotenoid biosynthesis. In the Chlorophyceae Haematococcus pluvialis, PTOX1 rather than PTOX2 has been associated with production of the carotenoid astaxanthin (23). Furthermore, in our hands, the varying sensitivities of the two oxidases to propylgallate point to likely structural differences. Still, the molecular mechanism of the respective enzymes appears to be the same, that is, oxidizing PQH2, the only difference being the flows they can sustain.
A role for PTOX1 in regenerating oxidized PQ for phytoene desaturation is further supported by examining the flow sustained by the enzyme. In ptox2, one should not underestimate the flow through PTOX1 (0.4 e− per PSII) based on the simple finding that its rate is 12-fold smaller than that of PTOX2 (5 e− per PSII). Indeed, as shown by Eq. 1, one has to take into account the 3-fold more reduced state of the plastoquinone pool in the absence of PTOX2. Without PTOX2, the chlororespiration flow is F = 2 × 17% = 0.34 e−⋅s−1 per PSII whereas in its presence it is F = 2 × 72% = 1.44 e−⋅s−1 per PSII, that is, 4-fold larger (Fig. 4). This is due to the 3-fold stimulation of the flow through PTOX1 in ptox2 mutants and shows that PTOX1 is competent for the regeneration of oxidized PQ.
Broader Perspective on Chlororespiration.
Our chlororespiration model links the redox state of the PQ pool to the inflow (NDA2) and outflow (PTOX) of electrons. We have shown in Chlamydomonas that the rate constants of reduction and oxidation are of similar magnitude, poising the PQ pool in a moderately oxidized state. Interestingly, this redox poise of the chloroplast in the dark appears to be a conserved trait in photosynthetic species. This may stem from the consequences of large changes in the redox state of the pool on the supramolecular organization of the thylakoid membranes demonstrated here by our comparative study of state transitions in the WT and ptox2 (SI Text). Then what does our model imply for other organisms? In plants, where the PQ pool is almost fully oxidized in the dark, the only in vivo quantitative estimation of the flow through PTOX pointed to a slow dark oxidation (t1/2 = 20 s) of PSII electron acceptors (12 electron equivalents), yielding 0.4 e−⋅s−1 per PSII (24). This is slower than in Chlamydomonas, where a flow of this magnitude is only observed in the ptox2 mutant. In plants, such a slow oxidation is sufficient to keep the pool oxidized, so that it must be paralleled by an even slower reduction rate. Simply overexpressing PTOX does not provide the expected benefits in Arabidopsis (25) or tobacco (26), whereas the leaf variegation phenotype of im is alleviated at low light intensities by inactivation of NDH and PGR5 (27). Alpine species not only accumulate higher levels of PTOX but also detoxification enzymes (10, 11) and NDH (28). This suggests a concerted adjustment of the electron inflow and outflow. In the framework of the present redox model for chlororespiration, these different observations show that just as photosynthetic organisms prevent the overreduction of the PQ pool in the light, they also avoid its complete oxidation in the dark, as though PQH2 would be a substrate or a signal for other physiological processes (for a review on plastid retrograde signaling, see ref. 29), just like PQ is required for carotenoid biosynthesis (30).
Materials and Methods
Cell Culture.
C. reinhardtii wild-type, mutant, and complemented strains were cultivated in media containing Tris⋅acetate⋅phosphate (TAP) or lacking acetate (minimum). The wild-type strain Jex4 mt−, possessing a cell wall, was selected for its ability to produce numerous clones when transformed and its competence for crossing. Jex4 was used as recipient strain for nuclear transformation experiments, generating the ptox2 mutant strain. Crosses were performed with WT T222 mt+ strain or a strain devoid of cytochrome b6f complex, ΔpetB.
Transformation.
Generation of mutant library.
The aphVIII cassette, used to transform the cells, was obtained by digestion of the pBC1 plasmid derived from vector pSI103 reported in ref. 31 with SacI and KpnI, and was gel-purified (Qiagen). Wild-type cells from 2 × 106 cells⋅mL−1 cultures were transformed by electroporation (32) using 100 ng of the aphVIII cassette. Selection was performed on solid TAP medium + 10 μg.⋅mL−1 paromomycin under illumination of 1 μE⋅m−2⋅s−1 at 25 °C.
For complementation.
The linear sequence of PTOX2 was generated by digestion of cDNA AV395349 (Kazusa Institute) by KpnI. Double mutant ptox2 Δb6f cells were transformed by electroporation (32) in the presence of 2 μg of PTOX2 sequence and 40 μg of carrier DNA. Selection was performed under 80 μE⋅m−2⋅s−1 at 25 °C. Negative controls were used to validate that complementation was not the result of a reversion event.
Protein Analysis.
Protein isolation was performed on cell cultures and grown on TAP at low light. Whole-cell extracts denatured in SDS/carbonate buffer were loaded onto a 12% SDS/PAGE system. Thylakoids were isolated on sucrose gradients as in ref. 33. Immunodetection was carried out using an enhanced chemiluminescence (ECL plus) method (Amersham International) according to the protocol supplied by the manufacturer. PTOX2 at the top of Fig. 1B was detected by using a 1:1,000 polyclonal rabbit antibody against A. thaliana PTOX (IM) (Uniplastomic). NDA2 was detected by a 1:15,000 diluted polyclonal rabbit antibody (3). Cytochrome b polyclonal antibody was used at a 1:20,000 dilution. Loading was controlled using the OEE2 polyclonal rabbit antibody diluted 10,000-fold. In Fig. 1D, PTOX1 was detected using antisera diluted 5,000-fold raised against the PTOX1 peptide WDREHAREQSGRGVET, and PTOX2 was detected used at a 1:500 dilution of antibody raised and purified against PTOX2 peptide CADYGFRSGSGRLYQE.
Gene Mapping.
Total DNA from ptox2 strain cells was isolated using phenol:chloroform. One hundred nanograms of isolated DNA was used to amplify PTOX2 gene fragments by PCR (Qiagen). Various primers were designed from the PTOX2 coding sequence to amplify overlapping regions of the gene.
Fluorescence and Absorption Spectroscopy.
A new instrument for chlorophyll fluorescence imaging (12) was used for the screening of mutant algal colonies plated directly onto Petri dishes. Fluorescence measurements were further monitored with a JTS-10 spectrophotometer (BioLogic). For spectroscopic experiments, cells were resuspended in 20 mM Hepes (pH 7.2) plus Ficoll 20% to avoid sedimentation. Nonactinic short light pulses (<100 μs) were used to probe the fluorescence yield. Light excitation of 150 μE⋅m−2⋅s−1 was provided by green light-emitting diodes. FM was detected after a 250-ms saturating light pulse of 5,000 μE⋅m−2⋅s−1.
Measurement of PTOX Activity in Vivo.
There is a linear relationship between PSII photochemical rate, monitored by oxygen evolution, and the variable part of fluorescence. The area over the fluorescence rise curve, which is a product of photochemical rate and time, represents the pool size of PSII electron acceptors. In the presence of DCMU, this area corresponds to one electron (transferred to QA). In mutants devoid of the cytochrome b6f complex, plastocyanine, or PSI, this area reflects the number of oxidized quinones before illumination (2). For a graphical illustration, see Fig. S7 and references therein. When one varies the time interval between two successive illuminations, one can probe the kinetics of plastoquinol oxidation by PTOX (14).
A full oxidation of the PQ pool in the dark was achieved as follows: The cells were treated with 50 μM p-benzoquinone 30 min before centrifugation, washed in fresh medium, and pelleted again for resuspension in buffer for the measurements. Commercially available p-benzoquinone was recrystallized through sublimation before dilution in fresh 50 mM ethanol stock solutions.
Supplementary Material
Acknowledgments
We are grateful to Pierre Cardol (Liège University) for sharing NDA2 antisera and for HPLC analysis of pigment extracts. We thank Niaz Ahmad and Peter J. Nixon (Imperial College, London) for helping screen PTOX1 antisera. Sophie Landier is acknowledged for the large media preparation necessitated by the generation and screening of the mutant collection. We are also indebted to Pierre Roussel and Rémi Dubois (Ecole Supérieure de Physique et de Chimie Industrielles, Paris) for image-processing algorithms used in the mutant screen. Agence Nationale pour la Recherche ANR-08-BIOE-002 ALGOMICS, European Commission EUFP7 SUNBIOPATH-GA-245070, and the Fondation Pierre-Gilles de Gennes pour la Recherche supported this work. L.H.-V. is supported by a fellowship from the Région Ile de France.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110518109/-/DCSupplemental.
References
- 1.Diner B, Mauzerall D. Feedback controlling oxygen production in a cross-reaction between two photosystems in photosynthesis. Biochim Biophys Acta. 1973;305:329–352. doi: 10.1016/0005-2728(73)90180-1. [DOI] [PubMed] [Google Scholar]
- 2.Bennoun P. Evidence for a respiratory chain in the chloroplast. Proc Natl Acad Sci USA. 1982;79:4352–4356. doi: 10.1073/pnas.79.14.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jans F, et al. A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc Natl Acad Sci USA. 2008;105:20546–20551. doi: 10.1073/pnas.0806896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carol P, et al. Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell. 1999;11(1):57–68. doi: 10.1105/tpc.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu D, Wright DA, Wetzel C, Voytas DF, Rodermel S. The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell. 1999;11(1):43–55. doi: 10.1105/tpc.11.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cournac L, et al. Electron flow between photosystem II and oxygen in chloroplasts of photosystem I-deficient algae is mediated by a quinol oxidase involved in chlororespiration. J Biol Chem. 2000;275:17256–17262. doi: 10.1074/jbc.M908732199. [DOI] [PubMed] [Google Scholar]
- 7.Giuliano G, Bartley GE, Scolnik PA. Regulation of carotenoid biosynthesis during tomato development. Plant Cell. 1993;5:379–387. doi: 10.1105/tpc.5.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey S, et al. Alternative photosynthetic electron flow to oxygen in marine Synechococcus. Biochim Biophys Acta. 2008;1777:269–276. doi: 10.1016/j.bbabio.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Cardol P, et al. An original adaptation of photosynthesis in the marine green alga Ostreococcus. Proc Natl Acad Sci USA. 2008;105:7881–7886. doi: 10.1073/pnas.0802762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Streb P, Aubert S, Gout E, Bligny R. Cold- and light-induced changes of metabolite and antioxidant levels in two high mountain plant species Soldanella alpina and Ranunculus glacialis and a lowland species Pisum sativum. Physiol Plant. 2003;118(1):96–104. doi: 10.1034/j.1399-3054.2003.00099.x. [DOI] [PubMed] [Google Scholar]
- 11.Streb P, Aubert S, Gout E, Bligny R. Reversibility of cold- and light-stress tolerance and accompanying changes of metabolite and antioxidant levels in the two high mountain plant species Soldanella alpina and Ranunculus glacialis. J Exp Bot. 2003;54:405–418. doi: 10.1093/jxb/erg048. [DOI] [PubMed] [Google Scholar]
- 12.Johnson X, et al. A new setup for in vivo fluorescence imaging of photosynthetic activity. Photosynth Res. 2009;102(1):85–93. doi: 10.1007/s11120-009-9487-2. [DOI] [PubMed] [Google Scholar]
- 13.Genty B, Briantais J-M, Baker NR. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta. 1989;990(1):87–92. [Google Scholar]
- 14.Bennoun P. Chlororespiration and the process of carotenoid biosynthesis. Biochim Biophys Acta. 2001;1506(2):133–142. doi: 10.1016/s0005-2728(01)00190-6. [DOI] [PubMed] [Google Scholar]
- 15.Cruz JA, Salbilla BA, Kanazawa A, Kramer DM. Inhibition of plastocyanin to P(700)(+) electron transfer in Chlamydomonas reinhardtii by hyperosmotic stress. Plant Physiol. 2001;127:1167–1179. [PMC free article] [PubMed] [Google Scholar]
- 16.Josse EM, Alcaraz JP, Labouré AM, Kuntz M. In vitro characterization of a plastid terminal oxidase (PTOX) Eur J Biochem. 2003;270:3787–3794. doi: 10.1046/j.1432-1033.2003.03766.x. [DOI] [PubMed] [Google Scholar]
- 17.McDonald AE, et al. Flexibility in photosynthetic electron transport: The physiological role of plastoquinol terminal oxidase (PTOX) Biochim Biophys Acta. 2011;1807:954–967. doi: 10.1016/j.bbabio.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 18.Josse EM, et al. A plastid terminal oxidase associated with carotenoid desaturation during chromoplast differentiation. Plant Physiol. 2000;123:1427–1436. doi: 10.1104/pp.123.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosso D, et al. Photosynthetic redox imbalance governs leaf sectoring in the Arabidopsis thaliana variegation mutants immutans, spotty, var1, and var2. Plant Cell. 2009;21:3473–3492. doi: 10.1105/tpc.108.062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cardol P, et al. Impaired respiration discloses the physiological significance of state transitions in Chlamydomonas. Proc Natl Acad Sci USA. 2009;106:15979–15984. doi: 10.1073/pnas.0908111106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alric J. Cyclic electron flow around photosystem I in unicellular green algae. Photosynth Res. 2010;106(1-2):47–56. doi: 10.1007/s11120-010-9566-4. [DOI] [PubMed] [Google Scholar]
- 22.Alric J, Lavergne J, Rappaport F. Redox and ATP control of photosynthetic cyclic electron flow in Chlamydomonas reinhardtii (I) aerobic conditions. Biochim Biophys Acta. 2010;1797(1):44–51. doi: 10.1016/j.bbabio.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Sommerfeld M, Hu Q. Occurrence and environmental stress responses of two plastid terminal oxidases in Haematococcus pluvialis (Chlorophyceae) Planta. 2009;230(1):191–203. doi: 10.1007/s00425-009-0932-4. [DOI] [PubMed] [Google Scholar]
- 24.Joliot P, Joliot A. Cyclic electron transfer in plant leaf. Proc Natl Acad Sci USA. 2002;99:10209–10214. doi: 10.1073/pnas.102306999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosso D, et al. IMMUTANS does not act as a stress-induced safety valve in the protection of the photosynthetic apparatus of Arabidopsis during steady-state photosynthesis. Plant Physiol. 2006;142:574–585. doi: 10.1104/pp.106.085886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heyno E, et al. Plastid alternative oxidase (PTOX) promotes oxidative stress when overexpressed in tobacco. J Biol Chem. 2009;284:31174–31180. doi: 10.1074/jbc.M109.021667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okegawa Y, Kobayashi Y, Shikanai T. Physiological links among alternative electron transport pathways that reduce and oxidize plastoquinone in Arabidopsis. Plant J. 2010;63:458–468. doi: 10.1111/j.1365-313X.2010.04252.x. [DOI] [PubMed] [Google Scholar]
- 28.Streb P, et al. Evidence for alternative electron sinks to photosynthetic carbon assimilation in the high mountain plant species Ranunculus glacialis. Plant Cell Environ. 2005;28:1123–1135. [Google Scholar]
- 29.Pogson BJ, Woo NS, Förster B, Small ID. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008;13:602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Norris SR, Barrette TR, DellaPenna D. Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell. 1995;7:2139–2149. doi: 10.1105/tpc.7.12.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sizova I, Fuhrmann M, Hegemann P. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene. 2001;277:221–229. doi: 10.1016/s0378-1119(01)00616-3. [DOI] [PubMed] [Google Scholar]
- 32.Shimogawara K, Fujiwara S, Grossman A, Usuda H. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics. 1998;148:1821–1828. doi: 10.1093/genetics/148.4.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chua NH, Bennoun P. Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: Wild-type and mutant strains deficient in photosystem II reaction center. Proc Natl Acad Sci USA. 1975;72:2175–2179. doi: 10.1073/pnas.72.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.