Abstract
Recent work has identified a subset of cells resident in tumors that exhibit properties similar to those found in normal stem cells. Such cells are highly tumorigenic and may be involved in resistance to treatment. However, the genes that regulate the tumor initiating cell (TIC) state are unknown. Here, we show that overexpression of either of the nucleolar GTP-binding proteins nucleostemin (NS) or GNL3L drives the fraction of genetically defined tumor cells that exhibit markers and tumorigenic properties of TICs. Specifically, cells that constitutively express elevated levels of NS or GNL3L exhibit increased TWIST expression, phosphorylation of STAT3, expression of genes that induce pluripotent stem cells, and enhanced radioresistance; in addition, they form tumors even when small numbers of cells are implanted and exhibit an increased propensity to metastasize. GNL3L/NS forms a complex with the telomerase catalytic subunit [human telomerase reverse transcriptase (hTERT)] and the SWItch-Sucrose NonFermentable (SWI-SNF) complex protein brahma-related gene 1 (BRG1), and the expression of each of these components is necessary to facilitate the cancer stem cell state. Together, these observations define a complex composed of TERT, BRG1, and NS/GNL3L that maintains the function of TICs.
Both embryonic and organ-specific stem cells are characterized by the ability to self-renew and to differentiate into specialized cell types. Recent work has identified a subset of cells in tumors that exhibit properties similar to those found in normal stem cells (1–3). Such tumor initiating cells (TICs) or cancer stem cells are characterized by the capacity for unlimited self-renewal and the ability to differentiate into multiple cell types. Moreover, such cells are highly tumorigenic (4), show resistance to chemotherapy and/or radiotherapy (5–7), and may contribute to metastasis (8–11).
TICs are defined operationally. Specifically, such cells exhibit the ability to form tumors when placed in limiting numbers in animal hosts (4, 12). Although some cancer stem cells express particular cell surface receptors, the expression of such markers is not exclusive to TICs and no universal cancer stem cell markers have been identified. Indeed, recent evidence suggests that not all tumors may harbor such TICs (12, 13). Given the potential role for such cells in tumor initiation, progression, and response to treatment, defining the molecular alterations that program cancer stem cells is essential not only to identify such cells but to understand their contribution to malignant transformation.
The nucleolar GTP-binding protein nucleostemin (NS) and its closely related family member GNL3L are expressed at high levels in ES cells (14, 15), and NS has been proposed as a marker for TICs in highly aggressive brain tumors (16). NS has been reported to regulate cell proliferation through a direct interaction with p53 (14). Specifically, recent studies have shown that expression of NS and/or GNL3L delays the onset of cellular senescence by negatively regulating telomeric repeat-binding factor 1 (TRF1) stability (17) and that depletion of NS causes G1 arrest in a p53-dependent manner (18). However, other studies suggest that NS may also contribute to stem cell function independent of p53, because blastocysts derived from NS null mice failed to enter S phase even in the absence of p53 (19).
We hypothesized that NS may contribute directly to formation of cancer stem cells. Here, we show that the expression of NS/GNL3L increases the fraction of tumorigenic human cells that exhibit TIC properties.
Results
NS and GNL3L Regulate TIC Behavior.
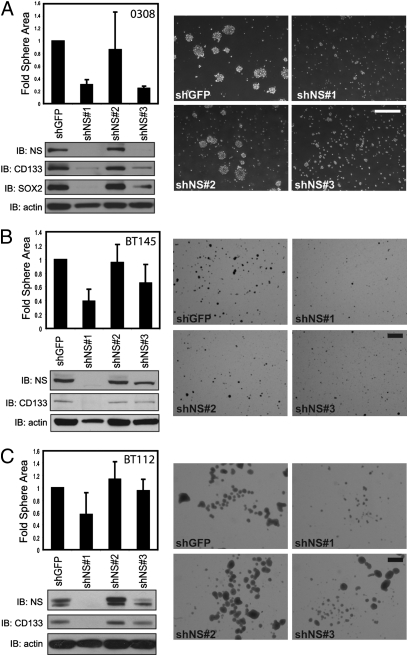
To examine whether endogenous NS was essential for the behavior of established TICs, we evaluated the consequences of suppressing NS in well-characterized TIC lines derived from glioblastomas (GBMs; 0308, BT145, and BT112) (20). To determine whether suppression of NS in these GBM TICs affected clonogenic neurosphere formation, a phenotype tightly correlated with tumorigenicity (21), cells were infected with either a control (short hairpin GFP) or three distinct NS-specific shRNAs [short hairpin NS (shNS) 1–3; Fig. 1]. We performed single-cell neurosphere formation assays and found that cells expressing the control shRNA formed neurospheres comparable to unmanipulated GBM TICs. In contrast, 0308 cells expressing NS-specific shRNAs displayed altered sphere-forming activity that corresponded to the degree of NS suppression. Specifically, cells expressing NS-specific shRNAs that produced the highest degree of NS suppression (shNS1 and shNS3) exhibited a significant decrease in average sphere size (P < 0.02; Fig. 1A, Upper Left). These cells also displayed decreased levels of neural stem cell markers CD133 and SOX2 (Fig. 1A, Lower Left). To eliminate the possibility that the observed effects were attributable to off-target effects of RNAi, we created a shRNA specific for the NS 3′UTR (shNS4) and showed that expression of this shRNA induced similar effects on the proliferation of MCF7 cells as the other NS-specific shRNAs and could be rescued by the expression of ectopic NS (Fig. S1 A–C). Moreover, suppression of NS in two other TIC lines, BT145 and BT112 (Fig. 1 B and C), also resulted in a decrease in sphere size and expression of the neural stem cell marker CD133 proportional to the degree of NS suppression. Together, these observations indicate that suppression of NS in 0308, BT145, and BT112 cells decreases the ability of these cells to form clonogenic neurospheres, and likely their tumor initiating capacity.
Fig. 1.
NS is required for TIC function. (A, Upper Left) Suppression of NS in 0308 cells results in decreased neurosphere size. The 0308 cells expressing control or NS-specific shRNAs were grown under neurosphere promoting conditions. Triplicate images of each cell population for three independent experiments were analyzed for average neurosphere size per area (μm2). Error bars indicate SD for three independent experiments. (Lower Left) Control (short hairpin GFP) and NS-specific shRNAs (shNS1–3) were introduced into 0308 cells, and endogenous NS suppression was measured by immunoblotting. IB, immunoblot; shGFP, short hairpin GFP. The expression of CD133 and SOX2 was also assessed; β-actin is shown as a loading control. (Right) Representative images of GBM-derived 0308 TICs containing a control or NS-specific shRNAs. (Scale bar = 250 μm.) As described in A performed on BT145 cells (B) or BT112 cells (C). (Scale bar = 1 mm in representative images.)
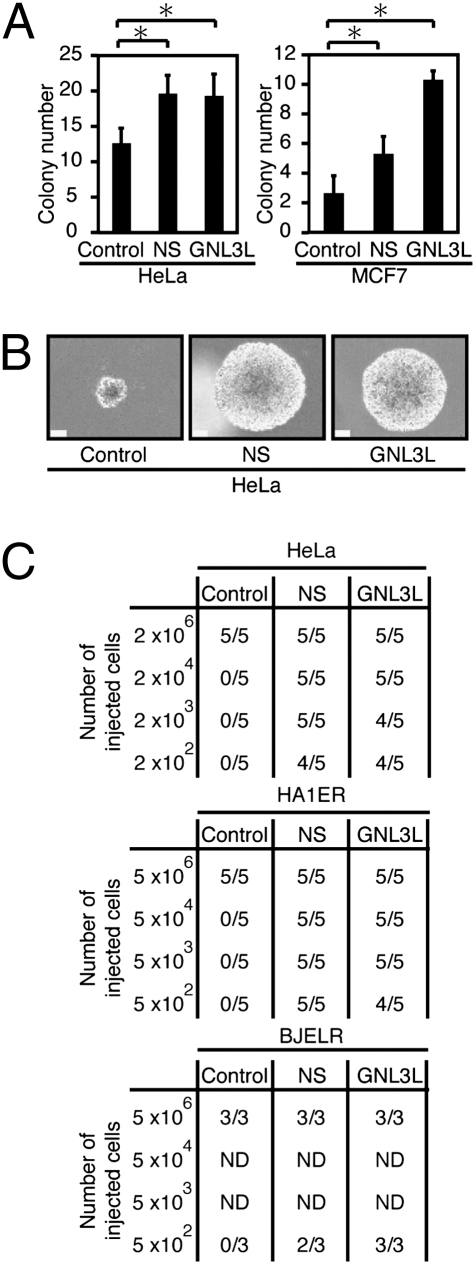
Although tumorigenic, human cancer cell lines derived from tumors or engineered by the expression of specific genetic elements (22) contain only a small fraction of cells that exhibit TIC behavior (4, 10, 23). To determine the effects of NS or GNL3L on the tumorigenicity of either engineered tumorigenic fibroblast [BJELR (full name: BJEHEcR-LT-Ras-ST)] and kidney epithelial (HA1ER) cells expressing the SV40 large and small T antigens, hTERT, and oncogenic H-RAS (22) or two established human cancer lines (HeLa or MCF7), we expressed NS or GNL3L at levels found in GBM-derived neurospheres and analyzed several phenotypes associated with TICs. We found that expression of either NS or GNL3L conferred the ability to form statistically larger and more numerous anchorage-independent colonies (Fig. 2 A and B) and induced enhanced tumorigenicity (Fig. 2C). Specifically, we observed tumor formation after the s.c. inoculation of a small number of cells overexpressing NS or GNL3L (≤500 cells), whereas cells expressing a control vector failed to form tumors. TICs have also been reported to exhibit relative resistance to ionizing radiation (5). When we irradiated BJ-hTERT, MCF7, or HeLa cells expressing a control vector, NS, or GNL3L, we found that cells expressing NS or GNL3L were more resistant to γ-irradiation (Fig. S2). These observations suggest that expression of NS or GNL3L alters the fraction of TICs present in both established and engineered cancer cell lines.
Fig. 2.
Expression of NS or GNL3L induces TICs. (A) Anchorage-independent growth. For each cell line, 100 cells were plated and colony numbers were counted after 1 mo. The mean ± SD for three independent experiments is shown. *P < 0.05. (B) Representative micrographs demonstrate colony sizes (magnification: 100×) of HeLa cells expressing NS, GNL3L, or a control vector (control). (Scale bar = 100 μm.) (C) For tumorigenicity assays, the indicated numbers of cells were injected s.c. into immunodeficient mice and are reported as the number of tumors formed/number of injection sites.
NS or GNL3L Expression Induces Markers and Pathways Associated with TICs.
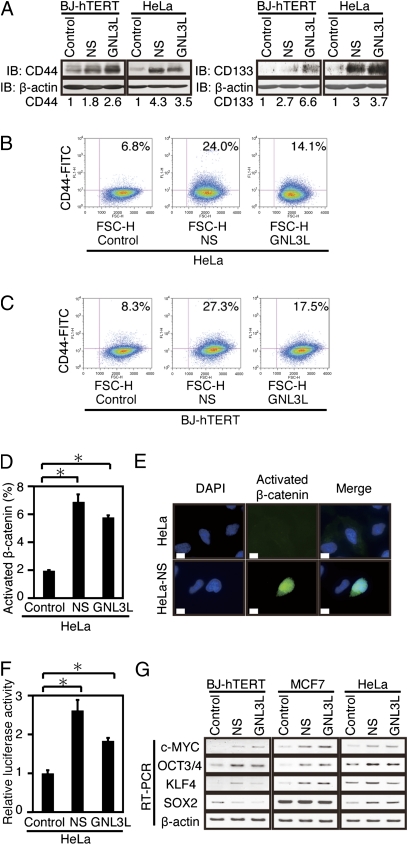
Several markers have been described to be expressed by TICs. When we analyzed cells overexpressing NS or GNL3L, we found increased expression of CD44 and CD133, markers associated with TICs (4, 24) (Fig. 3 A–C). Indeed, we found that the expression of NS or GNL3L in HeLa cells and BJ-hTERT cells induced a two- to fourfold increase in the percentage of cells expressing high levels of CD44 compared with cells expressing a control vector when we assessed either total CD44 levels (Fig. 3A) or CD44 expression in individual cells (Fig. 3 B and C). In addition, we confirmed that NS and GNL3L are significantly overexpressed in the subpopulation of several cancer cell lines previously shown (4, 23, 25) to be enriched in cells that express gene signatures related to TICs (CD44 high fraction of HeLa and CD44 high/CD24 low fraction of MDA-MB-157, MDA-MB-231, and MDA-MB-436 cells; Fig. S3 A and B).
Fig. 3.
Effects of NS or GNL3L on cancer stem cell markers. Effects of overexpressing NS or GNL3L on CD44 protein expression as assessed by immunoblotting (A) or flow cytometry (B and C). Cells were stained with FITC-conjugated anti-CD44 (Leu-44) antibody. IB, immunoblot. (B) Fractions of HeLa cells expressing high levels of CD44 were 6.8% (control vector), 24.0% (NS), and 14.1% (GNL3L). (C) Fractions of BJ-hTERT cells expressing high levels of CD44 were 8.3% (control vector), 27.3% (NS), and 17.5% (GNL3L). (D) Percentage of cells that harbor nuclear activated β-catenin in cells expressing a control vector, NS, or GNL3L is shown. *P < 0.05. (E) Effects of overexpressing NS or GNL3L on β-catenin function. Representative immunofluorescence images are shown. HeLa cells (Upper) and HeLa cells expressing NS (Lower) were stained with an antibody that recognizes unphosphorylated (active) β-catenin and visualized with Alexa Fluor 488 (Invitrogen)-conjugated anti-mouse IgG (green, magnification: 600×); DAPI (blue) indicates DNA. (Scale bar = 10 μm.) (F) TOPFLASH-Luc luciferase reporter activity. A renilla luciferase expression plasmid, pRL-SV40, was used as an internal control for transfection efficiency. *P < 0.05. (G) Effects of overexpressing NS or GNL3L on the expression of c-MYC, OCT3/4, KLF4, and SOX2.
Cell surface markers are useful tools to identify cells that exhibit cancer stem cell activity but may not unequivocally mark this cell population. We also investigated whether NS expression alters pathways implicated in the maintenance of cancer stem cells. Specifically, we determined whether expression of NS or GNL3L affected WNT/β-catenin signaling and found that expression of NS or GNL3L correlated with increased nuclear localization of the active nonphosphorylated β-catenin (Fig. 3 D and E) and induced statistically significant increased β-catenin activity as measured by reporter assay (26, 27) (Fig. 3F). Expression of NS or GNL3L also induced higher steady-state levels of c-MYC, a direct β-catenin target gene (Fig. 3G). The introduction of OCT3/4, SOX2, c-MYC, and KLF4 into normal human and murine cells suffices to reprogram such cells into ES cells (28, 29). When we analyzed the expression of these genes in cells expressing either a control vector, NS, or GNL3L, we found that NS or GNL3L up-regulated c-MYC, OCT3/4, and KLF4 (Fig. 3G and Fig. S4). These observations confirmed that the expression of NS or GNL3L induces the expression of genes associated with ES cells and cancer stem cells.
NS or GNL3L Expression Activates the Epithelial Mesenchymal Transition and Induces Metastasis.
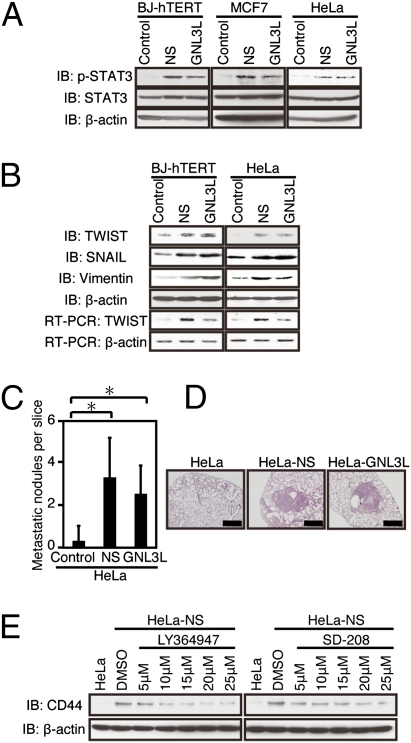
In addition to WNT/β-catenin signaling, several other pathways have been implicated in the maintenance of the cancer stem cell state. Specifically, recent evidence indicates that increased STAT3 signaling regulates the expression of the master regulator TWIST to induce epithelial mesenchymal transition (EMT) and metastasis (30). In consonance with these observations, we found that BJ-hTERT, MCF7, and HeLa cells expressing NS or GNL3L exhibited increased expression of the tyrosine phosphorylated form of STAT3 (Tyr705; Fig. 4A) and higher levels of TWIST, SNAIL, and vimentin (Fig. 4B). TGF-β signaling has been implicated in the regulation of both CD44 expression and TWIST (10). To investigate whether TGF-β signaling was required for EMT in cells expressing NS or GNL3L, we treated HeLa cells expressing NS with two different TGF-β inhibitors (LY364947 and SD-208) and found that the expression of CD44 was decreased in HeLa cells expressing NS in a dose-dependent manner (Fig. 4E). These observations suggest that the expression of NS or GNL3L induces an EMT.
Fig. 4.
Effects of NS and GNL3L on EMT and metastasis. (A) Effects of overexpression of NS or GNL3L on STAT3 and phospho-STAT3 (Tyr-705). IB, immunoblot. (B) Effects of overexpression of NS or GNL3L on TWIST, SNAIL, and vimentin expression. (C and D) Effects of overexpressing NS or GNL3L on metastasis. (C) Bar graph shows the number of metastatic foci found after tail vein injection. The mean ± SD for three independent experiments is shown. *P < 0.05. (D) Representative H&E images are shown (magnification: 100×). (Scale bar = 50 μm.) (E) Effects of TGF-β inhibitors on CD44 expression induced by NS/GNL3L. The indicated cells were treated with LY364947 or SD-208 for 24 h, followed by immunoblotting.
In addition, we found that tumorigenic cells expressing either NS or GNL3L exhibited increased capacity for metastasis as measured by the number of foci formed in the lungs of mice after tail vein injection of control, NS-expressing, or GNL3L-expressing cells (Fig. 4 C and D). Although this observed increase in the number of lung metastases in cells expressing NS may be attributable, in part, to the increased tumorigenicity of these cells, these observations show that NS-expressing cells exhibit several phenotypes associated with TICs (10, 11).
NS/GNL3L, BRG1, and TERT Form a Complex Necessary for TIC Function.
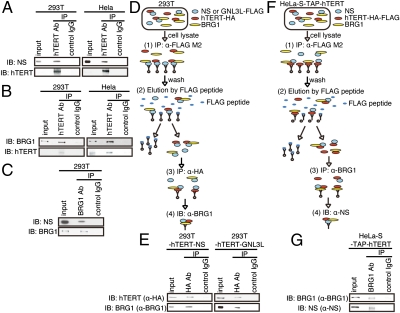
Because TERT has been implicated in the maintenance of stem cells by both telomere-dependent (31) and telomere-independent mechanisms (32–35) and had previously been reported to interact with GNL3L (36), we determined whether the interaction of NS/GNL3L with hTERT was involved in the induction of TICs. Specifically, we isolated hTERT immune complexes and found that endogenous NS interacts with endogenous hTERT in both HeLa and 293T cells (Fig. 5A). To characterize the interaction of hTERT with NS or GNL3L, we used a series of TERT truncation mutants and found that the amino-terminal end of hTERT (1–531) was necessary for hTERT to interact with both NS and GNL3L (Fig. S5A). We also found that the conserved consensus motifs G5 and G4 present in both NS and GNL3L (37–40) were required for the interaction with hTERT (Figs. S5 B and C, and S6). These observations identify a specific conserved region in NS and GNL3L responsible for the interaction with hTERT.
Fig. 5.
NS forms a complex with hTERT and BRG1. (A) hTERT interacts with endogenous NS. hTERT complexes from 293T or HeLa cells were purified with an anti-hTERT antibody (Rockland), and associated proteins were subjected to SDS/PAGE and immunoblotting with an anti-NS antibody. IB, immunoblot. (B) hTERT interacts with endogenous BRG1. hTERT immune complexes from 293T or HeLa cells were subjected to SDS/PAGE, followed by immunoblotting with an anti-BRG1 antibody. (C) BRG1 interacts with endogenous NS. BRG1 complexes from 293T cells were purified by immunoprecipitation with an anti-BRG1 antibody and subjected to SDS/PAGE, followed by immunoblotting with an anti-NS antibody. Schema (D) and results (E) of the sequential immunoprecipitation using cells expressing FLAG-tagged NS or GNL3L and hTERT-HA. NS or GNL3L immune complexes were isolated and then eluted with the FLAG M2 peptide. hTERT immune complexes were then purified, and endogenous BRG1 was detected by immunoblotting. Schema (F) and results (G) of the sequential immunoprecipitation using cells stably expressing TAP-hTERT. hTERT immune complexes were isolated and eluted with the FLAG M2 peptide. Endogenous BRG1 immune complexes were isolated, and endogenous NS was analyzed by immunoblotting.
As recently described (35), we confirmed that hTERT binds both overexpressed and endogenous BRG1 (Fig. 5 B and C). To determine whether hTERT, BRG1, and NS/GNL3L are found in the same complex, we performed two types of sequential immunoprecipitation experiments. First, we isolated FLAG–NS or FLAG–GNL3L immune complexes and confirmed that hTERT-HA was present. We then eluted these complexes with the M2 (FLAG) peptide, isolated hTERT immune complexes, and confirmed that endogenous BRG1 was present in these complexes (Fig. 5 D and E). Second, we isolated hTERT immune complexes and confirmed that BRG1 was present. We then eluted these complexes with the M2 (FLAG) peptide, isolated BRG1 immune complexes, and confirmed that endogenous NS was present (Fig. 5 F and G). These observations suggest that hTERT, BRG1, and NS or GNL3L are present in the same complex.
To determine whether the interaction of hTERT with NS/GNL3L required the telomere elongation activities of TERT, we suppressed the expression of the telomerase RNA subunit hTERC and found that hTERT continued to associate with NS, indicating that this interaction occurs independent of telomerase activity (Fig. S7A). Moreover, overexpression or suppression of NS or GNL3L did not affect telomerase activity or telomere length as assessed by telomere repeat amplification protocol assays or telomere restriction fragment Southern blotting (Fig. S7 B–D). Together, these observations suggest that complexes containing hTERT, BRG1, and NS or GNL3L do not contribute directly to telomere maintenance but, instead, drive TIC formation by regulating the activity of BRG1.
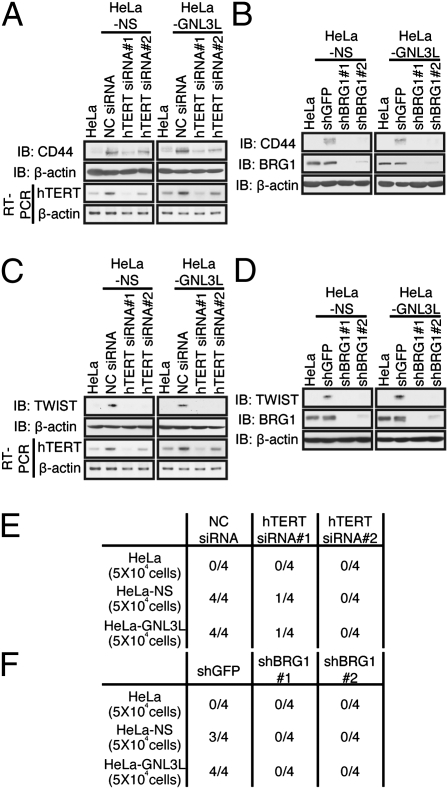
To confirm that the complex composed of hTERT, BRG1, and NS/GNL3L was involved in regulating TIC phenotypes, we assessed whether each of these components was necessary for NS-induced increase in CD44 levels. When we suppressed hTERT expression using hTERT-specific siRNAs, we failed to observe an increase in CD44 expression after expressing NS or GNL3L (Fig. 6A). Moreover, BRG1 suppression reversed the increase in CD44 expression induced by NS or GNL3L expression (Fig. 6B). In contrast, when we suppressed hTERC expression using hTERC-specific shRNAs, we found that the expression of hTERC was not required for the increase in CD44 expression induced by NS or GNL3L overexpression (Fig. S7E). Furthermore, when we suppressed hTERT or BRG1 expression, we failed to observe an increase in TWIST expression after expressing NS or GNL3L (Fig. 6 C and D). Suppression of hTERT or BRG1 in cells expressing NS or GNL3L, which are markedly enriched in TICs, ablated the ability of HeLa cells expressing NS or GNL3L to form tumors (Fig. 6 E and F). Together, these observations indicate that the expression of hTERT, NS/GNL3L, and BRG1 is each required to induce a TIC phenotype.
Fig. 6.
Complex of hTERT, BRG1, and NS/GNL3L is necessary for NS-induced TIC formation. (A) hTERT is required for NS/GNL3L-induced increase in CD44 expression. HeLa-NS cells or HeLa-GNL3L cells expressing either a control siRNA or two independent hTERT-specific siRNAs were immunoblotted for CD44. IB, immunoblot. (B) BRG1 is required for NS/GNL3L-induced increase in CD44 expression. CD44 expression was assessed by immunoblotting in HeLa-NS cells or HeLa-GNL3L cells stably expressing either a control vector or two independent BRG1-specific shRNAs. (C) hTERT is required for NS/GNL3L-induced increase in TWIST expression. HeLa-NS cells or HeLa-GNL3L cells expressing either a control siRNA or two independent hTERT-specific siRNAs were immunoblotted for TWIST. (D) BRG1 is required for NS/GNL3L-induced increase in TWIST expression. TWIST expression was assessed by immunoblotting in HeLa-NS cells or HeLa-GNL3L cells stably expressing either a control vector or two independent BRG1-specific shRNAs. (E) hTERT is required for tumor formation by HeLa cells. The number of tumors formed/number of injection sites in HeLa-NS cells or HeLa-GNL3L cells expressing either a control siRNA or two independent hTERT-specific siRNAs is shown. For E and F, 5 × 104 cells were injected per mouse. NC, negative control. (F) BRG1 is required for tumor formation by HeLa cells expressing NS or GNL3L. HeLa-NS cells or HeLa-GNL3L cells expressing either a control vector or two independent BRG1-specific shRNAs are shown.
Discussion
Here, we show that expression of NS or GNL3L in genetically engineered human cancer cells confers a TIC phenotype. Tumorigenic human cells of defined genetic constitution expressing NS or GNL3L exhibited the full range of phenotypes associated with cancer stem cells, including the expression of cell surface markers such as CD44 and CD133, genes that induce induced pluripotant stem (iPS) cells, the ability to form tumors when implanted at limiting numbers, activation of an EMT program, and increased metastatic potential. Because genetically engineered human cancer cells (BJELR and HA1ER) and even established human cancer lines (HeLa and MCF7) have both TIC and non-TIC populations (Fig. S3), expressing NS or GNL3L enhances the TIC fraction.
NS was initially found to be expressed at high levels in highly proliferative multipotential cells (14, 19) as well as a subset of aggressive brain tumors (16). We found that NS and GNL3L contribute directly to the maintenance of the TIC phenotype. Prior work demonstrated that NS physically interacts with p53 and that this interaction regulates cell proliferation (14). However, it is clear that NS also contributes to stem cell function independent of p53 (19). Although we have confirmed that depletion of NS is associated with cell cycle arrest accompanied by p53 up-regulation (18), the tumorigenic cells in which we expressed NS or GNL3L lack p53 function, suggesting that NS contributes to the generation of TIC independent of its regulation of p53.
TERT was recently shown to interact with GNL3L in a manner that regulated telomere length (36). We have confirmed that GNL3L and NS interact with TERT through a highly conserved region present in both of these nucleolar GTP binding proteins. However, in this setting, we failed to find evidence that the interaction of NS or GNL3L affected telomere length or telomerase activity. Instead, we found that NS or GNL3L forms a ternary complex with TERT and BRG1 in a manner that does not depend on hTERC. However, it remains possible that TERT–GNL3L complexes regulate telomere dynamics in other cells or conditions. These observations suggest that the NS/GNL3L–TERT–BRG1 complex operates in a telomere-independent manner to drive transcriptional programs essential to the maintenance of the TIC state. Thus, in addition to facilitating immortalization, hTERT may contribute to tumorigenicity through the NS/GNL3L–TERT–BRG1 complex by influencing the balance of cancer cells that exhibit TIC phenotypes.
Recent work indicates that TERT regulates stem cell homeostasis independent of its function at telomeres (32) and that TERT, together with BRG1, modulates stem cell homeostasis by modulating the WNT/β-catenin signaling pathway (35). We confirmed that the NS/GNL3L–TERT–BRG1 complex is essential for the maintenance of TICs. Further work will be necessary to determine whether this complex regulates normal stem cells by a similar mechanism.
In prior work, we identified a limited number of introduced genes that sufficed to convert normal human cells into tumorigenic cells (22). We and others have subsequently found specific combinations of genes mutating in spontaneously arising cancers that permitted the creation of tumorigenic human cells of a defined genetic composition (41–43). These experimental models have proven useful to decipher the cooperation between mutated genes and the discovery of new cancer genes (44, 45). However, like most established cancer cell lines, such cells formed tumors only after the implantation of a large number of cells, suggesting that additional perturbations were necessary to drive the formation of TICs. The observation that expression of NS or GNL3L in this setting leads to cells that exhibit all the phenotypes associated with cancer stem cells establishes an experimental model to define the pathways required to program the cancer stem cell state. Moreover, because the acquisition of stem cell characteristics correlates with treatment resistance and metastatic disease, these experimental models may provide the means to identify new targets to inhibit these phenotypes associated with lethal disease.
Materials and Methods
Cell Culture and Stable Expression of FLAG-NS/GNL3L and hTERT.
The human cell lines 293T, MCF7, HeLa-S, HA1ER (22), and HeLa were maintained in DMEM supplemented with 10% (vol/vol) heat-inactivated FBS. BJ fibroblasts and BJELR cells were cultured as described (22). Amphotropic retroviruses were created as described (22) using the expression vector pWZL-neo, pBABE-Hygro, pWZL-neo-FLAG-NS, pWZL-neo-FLAG-GNL3L, or pBABE-Hygro-hTERT. After infection, cells were selected with neomycin (G418; 2 mg/mL) for 7 d or with hygromycin (50 mg/mL) for 3 d.
Stable Expression of shRNA.
The pLKO.1-puro vector was used to express shRNAs targeting NS, GNL3L, hTERC, BRG1, and GFP. These vectors were used to make amphotropic lentiviruses, and polyclonal cell populations were purified by selection with puromycin (2 mg/mL).
Immunoblotting and Immunoprecipitation.
Cells were lysed using a Nonidet P-40/SDS–based lysis buffer. Details concerning the conditions and antibodies used are found in SI Materials and Methods.
Anchorage-Independent Growth and Tumorigenicity Assays.
Growth in soft agar was performed as described (22) and scored at 4 wk. For tumor experiments, cells were mixed with BD Matrigel Matrix (BD Bioscience) at 4 °C and injected s.c. in BALB/c-nu/nu mice.
Neurosphere Formation Assay.
GBM-derived 0308 tumor stem cells were a gift from H. Fine (US National Institutes of Health, Bethesda, MD) and were cultured in neurobasal media (NBE; Invitrogen), 0.5× each N2 and B27 supplements (Invitrogen), and 50 ng/mL each human recombinant basic FGF and EGF (R&D Systems). BT145 and BT112 cells were cultured in NeuroCult NS-A Basal Medium (StemCell Technologies) supplemented with NeuroCult NS-A Proliferation Supplement (StemCell Technologies) and 20 ng/mL human recombinant bFGF and EGF. For shRNA experiments, cells were infected with viral supernatant diluted 1:5, spun at 930 × g for 30 min, and incubated at 37 °C for 1.5 h before changing to fresh media. Cells were selected with 0.4 mg/mL puromycin 24 h after infection. At 72 h after infection, the cells were trypsinized and plated to assay for neurosphere formation; after 72 h, imaging with a light microscope was performed and cell lysates were made for immunoblot analysis. To determine average sphere size, images were analyzed using ImageJ software (National Institutes of Health).
Metastasis Assay.
A total of 5 × 106 cells were injected into the tail vein of BALB/c-nu/nu mice. After 4 wk, lungs were dissected to evaluate tissue morphology and to detect metastases.
Supplementary Material
Acknowledgments
We thank A. Miyajima, S. Saito, S. Inanobe, S. Takahashi, T. Ochiya, and R. Takahashi for their technical assistance. We thank H. Fine for the gift of 308 cells. This work was supported, in part, by Grant-in-Aid for Young Scientists (A) 21689012 (to K.M.) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology; by Third-Term Comprehensive Control Research for Cancer (K.M.) from the Japanese Ministry of Health, Labor, and Welfare; by the NOVARTIS Foundation (Japan) for the Promotion of Science (K.M.); by the Funding Program for Next Generation World-Leading Researchers (K.M.); and by Grant R01 AG23145 from the US National Institutes of Health (to W.C.H.). N.O. was supported by a Research Fellow of the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Telomerase and Retrotransposons: Reverse Transcriptases That Shaped Genomes,” held September 29–30, 2010, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at www.nasonline.org/telomerase_and_retrotransposons.
This article is a PNAS Direct Submission. N.F.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015171108/-/DCSupplemental.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Dalerba P, Cho RW, Clarke MF. Cancer stem cells: Models and concepts. Annu Rev Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 3.Dick JE. Looking ahead in cancer stem cell research. Nat Biotechnol. 2009;27:44–46. doi: 10.1038/nbt0109-44. [DOI] [PubMed] [Google Scholar]
- 4.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 6.Rich JN, Bao S. Chemotherapy and cancer stem cells. Cell Stem Cell. 2007;1:353–355. doi: 10.1016/j.stem.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Todaro M, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Dalerba P, Clarke MF. Cancer stem cells and tumor metastasis: First steps into uncharted territory. Cell Stem Cell. 2007;1:241–242. doi: 10.1016/j.stem.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Hermann PC, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagahama Y, et al. PSF1, a DNA replication factor expressed widely in stem and progenitor cells, drives tumorigenic and metastatic properties. Cancer Res. 2010;70:1215–1224. doi: 10.1158/0008-5472.CAN-09-3662. [DOI] [PubMed] [Google Scholar]
- 12.Quintana E, et al. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: Cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baddoo M, et al. Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J Cell Biochem. 2003;89:1235–1249. doi: 10.1002/jcb.10594. [DOI] [PubMed] [Google Scholar]
- 16.Tamase A, et al. Identification of tumor-initiating cells in a highly aggressive brain tumor using promoter activity of nucleostemin. Proc Natl Acad Sci USA. 2009;106:17163–17168. doi: 10.1073/pnas.0905016106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Q, Yasumoto H, Tsai RY. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol Cell Biol. 2006;26:9279–9290. doi: 10.1128/MCB.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma H, Pederson T. Depletion of the nucleolar protein nucleostemin causes G1 cell cycle arrest via the p53 pathway. Mol Biol Cell. 2007;18:2630–2635. doi: 10.1091/mbc.E07-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beekman C, et al. Evolutionarily conserved role of nucleostemin: Controlling proliferation of stem/progenitor cells during early vertebrate development. Mol Cell Biol. 2006;26:9291–9301. doi: 10.1128/MCB.01183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 21.Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6:425–436. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- 22.Hahn WC, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 23.Shipitsin M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Singh SK, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 25.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller JR, Hocking AM, Brown JD, Moon RT. Mechanism and function of signal transduction by the Wnt/beta-catenin and Wnt/Ca2+ pathways. Oncogene. 1999;18:7860–7872. doi: 10.1038/sj.onc.1203245. [DOI] [PubMed] [Google Scholar]
- 27.Firestein R, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 30.Cheng GZ, et al. Twist is transcriptionally induced by activation of STAT3 and mediates STAT3 oncogenic function. J Biol Chem. 2008;283:14665–14673. doi: 10.1074/jbc.M707429200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flores I, Cayuela ML, Blasco MA. Effects of telomerase and telomere length on epidermal stem cell behavior. Science. 2005;309:1253–1256. doi: 10.1126/science.1115025. [DOI] [PubMed] [Google Scholar]
- 32.Sarin KY, et al. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi J, et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;4:e10. doi: 10.1371/journal.pgen.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imamura S, et al. A non-canonical function of zebrafish telomerase reverse transcriptase is required for developmental hematopoiesis. PLoS ONE. 2008;3:e3364. doi: 10.1371/journal.pone.0003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park JI, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell. 2007;28:773–785. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daigle DM, et al. YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with circularly permuted G-motifs and marked burst kinetics. Biochemistry. 2002;41:11109–11117. doi: 10.1021/bi020355q. [DOI] [PubMed] [Google Scholar]
- 38.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 39.Meng L, Yasumoto H, Tsai RY. Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm. J Cell Sci. 2006;119:5124–5136. doi: 10.1242/jcs.03292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yasumoto H, Meng L, Lin T, Zhu Q, Tsai RY. GNL3L inhibits activity of estrogen-related receptor gamma by competing for coactivator binding. J Cell Sci. 2007;120:2532–2543. doi: 10.1242/jcs.009878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao JJ, et al. Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase. Cancer Cell. 2003;3:483–495. doi: 10.1016/s1535-6108(03)00088-6. [DOI] [PubMed] [Google Scholar]
- 42.Boehm JS, Hession MT, Bulmer SE, Hahn WC. Transformation of human and murine fibroblasts without viral oncoproteins. Mol Cell Biol. 2005;25:6464–6474. doi: 10.1128/MCB.25.15.6464-6474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao JJ, et al. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci USA. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolfschoten IG, et al. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005;121:849–858. doi: 10.1016/j.cell.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 45.Boehm JS, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.