Since its birth, systems biology has gained a great deal from the protocols devised to study phenomena at the level of single proteins and nucleic acids. Such protocols find broad markets and utility at higher levels of biological organization, from next-generation sequencing, which uses modified nucloeotides and fluorescent identifiers (1), to ChIP-seq analysis, which identifies histone modifications and binding sites in protein–DNA interactions (2). In PNAS, Sivaramakrishnan and Spudich introduce a system for interrogating interactions between pairs of proteins, domains, and peptides (3), and it is very possible that their invention will find applicability in the construction and analysis of large-scale protein–protein interaction networks (4).
The technology devised by Sivaramakrishnan and Spudich is based on Forster resonance energy transfer (FRET), which relies on the proximity-dependant excitation of an acceptor molecule by a donor (5). This excitation may be detected with spectral imaging, so FRET enables the investigator to directly probe interactions between candidate pairs of biomolecules, by using molecular cloning to tether the donor to one candidate and the acceptor to the second. For years this technology has enjoyed broad applicability (6–8), and biosensors using FRET have been applied in live cell imaging to investigate an array of biological processes, such as phosphorylation, protease activity, fluctuations in membrane potential, and changes in redox potentials, pH, and other environmental conditions (9, 10).
To engineer the effective concentrations of interacting species, Sivaramakrishnan and Spudich have recruited the ER/K single α-helix as an intradomain and intraprotein linker, in conjunction with FRET sensing, to quantitatively regulate and study interactions between proteins fused to the ends of the link. The effective concentrations are determined by the size of the linkage. Perhaps not surprisingly, the on-rate of the interaction decreases as the length of the linker increases. There is little effect on the dissociation rate. Importantly, the value of this system lies not only in the ability to modulate the interaction frequencies of two molecules without major perturbations to their structures, but also to obtain meaningful and accurate estimates of the natural binding affinities associated with interactions. Thus, this application has far-reaching implications for the large and diverse array proteins for which interactions with other cellular species are intrinsic to their basic functionality.
There are several advantages to this linker system. The linkage motif itself has been adopted from naturally occurring proteins, thereby precluding the need of designing the construct ab initio. Second, these biosensor constructs are designed with (Gly-Ser-Gly)4 bridges separating the interacting domains and FRET species, thereby allowing the elements of the construct to fully explore rotational degrees of freedom. The unique linker system has built-in modularity as well; protease sites separate its various domains, thereby lending the system separation and purification of its constituents, which are thus made available in stoichiometric amounts for bimolecular interactions and affinity calculations.
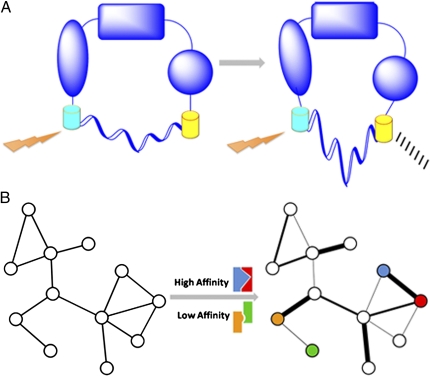
The interactions between proteins are inevitably tied to the motions and dynamics of the interaction participants, and a protein's conformation is an important factor in determining its interaction affinity. The linker system provides several avenues that may be of interest to the study of conformational changes. When fused to two distinct domains of a protein, one may be able to gain a crude knowledge of potential conformational changes in response to certain stimuli (Fig. 1A). The existence of a FRET signal only in the presence of a given stimulus would suggest that the protein undergoes a conformational change in a way that confers the needed proximity between the donor and acceptor FRET molecules. Of course, the absence of a FRET signal may, in fact, constitute a false negative, as much of the conformational space may not allow a favorable interaction between the two FRET molecules. In addition, the preparation of such a construct would not be the exact same as that described by Sivaramakrishnan and Spudich.
Fig. 1.
Applying the ER/K α-helix to studying conformational changes and network characterization. (A) A protein in two possible conformations. Detection of a FRET signal may be dependent on a change in conformation. The semicircle below each conformation represents the linker. (B) Two representations of a protein–protein interaction network. Left: This network is a conventional node-and-edge depiction. Quantifying the interaction affinities may enable the addition of interaction strength as a property of the network, yielding a network with weighted edges (Right). Colored interaction schematics above and below the central arrow correspond to examples of mapped high- and low-affinity interactions between similarly colored nodes in the affinity-based network.
Additionally, the linker may be applied to investigate the general freedom with which specific proteins explore conformational space. The authors note that this construct may be used to study the interaction affinities between distinct domains within a given protein. Careful analyses of intradomain affinities may provide valuable insights into the preferred conformations adopted by simple proteins. If two domains of a simple protein exhibit a high mutual affinity, the frequency of large-scale intramolecular motions may be low, with the closed state lying deep within an energy well. Conversely, two domains exhibiting a low mutual affinity may be one way to identify systems favoring more open configurations. Naturally, such interpretations would first entail knowledge of the protein's structure.
Questions regarding intramolecular motions and protein–protein binding affinities are relevant in the larger context of protein interaction networks (11, 12). The network representation has become an invaluable means of delineating interactions within cellular systems, and this more global view of interactions is generally provided in the form of nodes, which represent biomolecules, and edges, which represent physical interactions between those biomolecules (Fig. 1B, Left).
Sivaramakrishnan and Spudich introduce a system for interrogating interactions between pairs of proteins, domains, and peptides.
Although valuable, these simple 2D renderings alone fail to relay many important biological phenomena. Thus, more recent efforts have been aimed at incorporating additional forms of data into simple node-and-edge pictures, including structural data of the constituent biomolecules (13, 14), evolutionary features (15), and expression patterns (16).
Proteins’ varying conformations also constitute essential attributes in understanding networks, and how the magnitude and general nature of conformational changes influence interactions is an area of active research. Recent work has highlighted the relevance of conformational flexibility. For instance, proteins with many interaction interfaces and binding partners have been found to undergo larger conformational changes (17). In this regard, should the helical linker be implemented on a large scale, its potential ability to elucidate conformational changes may equip investigators with easier means of further studying the role of conformational flexibility in large-scale networks.
Currently, most network edges are represented as simple binary entities, wherein the presence or absence of an edge denotes an interacting or noninteracting pair, respectively, without including information on the strength of the interaction itself (18). Notably, an additional potential benefit of recruiting this linker for network studies may be characterizing the confidence and strength with which two proteins interact. For example, it may be possible to weigh interactions on the basis of experimentally determined affinities (Fig. 1B), which may ultimately lead to new insights into network topology and organization. Such insights may include, for instance, the identification of highly stable or highly dynamic modules within the network. It should not be neglected that a limitation intrinsic to this line of thinking is the binary nature of the linker system itself. Many interactions within the network occur between many proteins simultaneously (16), and simultaneous interactions with a given protein are likely to influence that protein's properties in ways that may not surface in calculations on simple binary interactions.
Despite this, the genetically encoded construct to modulate interactions may very well have promise at the level of entire networks, even if only to assign varying degrees of confidence to network edges. One of the major future challenges to more fully realizing this promise will be deploying this type of system to investigate large combinatorial numbers of potential protein pairs. Although development of such high-throughput systems will constitute challenges, such a scaling up would bring huge rewards and greater dimension to network-based analyses.
Footnotes
The authors declare no conflict of interest.
See companion article on page 20467.
References
- 1.Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 2.Park PJ. ChIP-seq: Advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sivaramakrishnan S, Spudich JA. Systematic control of protein interaction using a modular ER/K α-helix linker. Proc Natl Acad Sci USA. 2011;108:20467–20472. doi: 10.1073/pnas.1116066108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert R. Scale-free networks in cell biology. J Cell Sci. 2005;118:4947–4957. doi: 10.1242/jcs.02714. [DOI] [PubMed] [Google Scholar]
- 5.Jares-Erijman EA, Jovin TM. FRET imaging. Nat Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 6.Aoki K, Kiyokawa E, Nakamura T, Matsuda M. Visualization of growth signal transduction cascades in living cells with genetically encoded probes based on Förster resonance energy transfer. Philos Trans R Soc Lond B Biol Sci. 2008;363:2143–2151. doi: 10.1098/rstb.2008.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aye-Han NN, Ni Q, Zhang J. Fluorescent biosensors for real-time tracking of post-translational modification dynamics. Curr Opin Chem Biol. 2009;13:392–397. doi: 10.1016/j.cbpa.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Allen MD. FRET-based biosensors for protein kinases: Illuminating the kinome. Mol Biosyst. 2007;3:759–765. doi: 10.1039/b706628g. [DOI] [PubMed] [Google Scholar]
- 9.Gao X, Zhang J. FRET-based activity biosensors to probe compartmentalized signaling. ChemBioChem. 2010;11:147–151. doi: 10.1002/cbic.200900594. [DOI] [PubMed] [Google Scholar]
- 10.Lu S, Wang Y. Fluorescence resonance energy transfer biosensors for cancer detection and evaluation of drug efficacy. Clin Cancer Res. 2010;16:3822–3824. doi: 10.1158/1078-0432.CCR-10-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gursoy A, Keskin O, Nussinov R. Topological properties of protein interaction networks from a structural perspective. Biochem Soc Trans. 2008;36:1398–1403. doi: 10.1042/BST0361398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiel C, Beltrao P, Serrano L. Analyzing protein interaction networks using structural information. Annu Rev Biochem. 2008;77:415–441. doi: 10.1146/annurev.biochem.77.062706.133317. [DOI] [PubMed] [Google Scholar]
- 13.Aloy P, Russell RB. Interrogating protein interaction networks through structural biology. Proc Natl Acad Sci USA. 2002;99:5896–5901. doi: 10.1073/pnas.092147999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aloy P, Russell RB. The third dimension for protein interactions and complexes. Trends Biochem Sci. 2002;27:633–638. doi: 10.1016/s0968-0004(02)02204-1. [DOI] [PubMed] [Google Scholar]
- 15.Kim PM, Korbel JO, Gerstein MB. Positive selection at the protein network periphery: Evaluation in terms of structural constraints and cellular context. Proc Natl Acad Sci USA. 2007;104:20274–20279. doi: 10.1073/pnas.0710183104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han JD, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 17.Bhardwaj N, Abyzov A, Clarke D, Shou C, Gerstein MB. Integration of protein motions with molecular networks reveals different mechanisms for permanent and transient interactions. Protein Sci. 2011;20:1745–1754. doi: 10.1002/pro.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]