Abstract
Protein fate in higher eukaryotes is controlled by three complexes that share conserved architectural elements: the proteasome, COP9 signalosome, and eukaryotic translation initiation factor 3 (eIF3). Here we reconstitute the 13-subunit human eIF3 in Escherichia coli, revealing its structural core to be the eight subunits with conserved orthologues in the proteasome lid complex and COP9 signalosome. This structural core in eIF3 binds to the small (40S) ribosomal subunit, to translation initiation factors involved in mRNA cap-dependent initiation, and to the hepatitis C viral (HCV) internal ribosome entry site (IRES) RNA. Addition of the remaining eIF3 subunits enables reconstituted eIF3 to assemble intact initiation complexes with the HCV IRES. Negative-stain EM reconstructions of reconstituted eIF3 further reveal how the approximately 400 kDa molecular mass structural core organizes the highly flexible 800 kDa molecular mass eIF3 complex, and mediates translation initiation.
Keywords: protein synthesis, translation regulation, supramolecular complex assembly, electron microscopy
Three protein complexes with conserved composition and architectures regulate the synthesis and degradation of proteins in higher eukaryotes. These complexes, the proteasome, COP9 signalosome, and eukaryotic translation initiation factor 3 (eIF3), each contain eight conserved protein subunits, six of which bear proteasome, COP9, eIF3 (PCI) domains and two of which bear Mpr1-Pad1 N-terminal (MPN) domains (1), here grouped as PCI/MPN subunits. Although interaction maps have been proposed for each complex (2–5), their structures, assembly pathways, and the functional contributions of subunits within them remain poorly understood (1, 5–7). A major barrier to the study of these macromolecular protein regulators has been the inability to produce samples in genetically tractable recombinant systems in sufficient quantity for mechanistic studies.
Among these three complexes, eIF3 remains the least characterized, despite its central role in recruiting both mRNAs and the cellular translation machinery to form translation initiation complexes (8). In humans, eIF3 is an 800 kDa molecular mass assembly of 13 proteins (eIF3a-eIF3m) (3, 9), many of which have been implicated in cancers via misregulation of their expression (10). In addition to its involvement in regulation of cell proliferation, eIF3 also interacts directly with the hepatitis C viral (HCV) mRNA to promote the translation of viral proteins (11–13). The 5′-untranslated region (5′-UTR) of HCV contains an approximately 340-nt internal ribosome entry site (IRES) element that folds into defined secondary and tertiary structural elements (14) capable of specifically binding to eIF3 and the 40S ribosomal subunit (13, 15). These interactions trigger HCV IRES-mediated initiation complex formation with the GTPase eIF2 and methionyl-initiator tRNA (Met-tRNAi) (16, 17) or an alternate initiation complex with eIF5B (18).
A structural and functional understanding of eIF3’s roles in human translation initiation has relied on human eIF3 purified from human cell lines (19–21), expressed in part in insect cells (3, 21, 22), or by comparison to eIF3 purified from other mammalian sources (18, 23). Human eIF3 purified from human cell lines was used to derive a low-resolution structural model of the complex and its interactions with the small (40S) ribosomal subunit and HCV IRES (24). Attempts to reconstitute human eIF3 expressed in insect cells revealed parallels between the human and yeast eIF3 complexes, which share six conserved subunits (21). Additional reconstitution experiments in insect cells led to the model that the functional core of human eIF3 is composed of six subunits (eIF3a, eIF3b, eIF3c, eIF3e, eIF3f, and eIF3h) of which only eIF3a, eIF3b, and eIF3c are conserved in all eukaryotes (22). However, a full molecular understanding of eIF3’s roles in translation remains to be determined.
Here we have reconstituted human eIF3 in a stepwise manner using expression in Escherichia coli, resulting in a 13-subunit recombinant form of eIF3. We have tested the resulting subcomplexes and the 13-subunit recombinant eIF3 for their ability to bind components of the translational machinery, to the HCV IRES RNA, and to form initiation complexes. Finally, we compare the structures of the eIF3 complexes assembled using E. coli expression with that determined for natively purified human eIF3. Our results establish a unique paradigm for expression of intact human supramolecular assemblies in E. coli, and reveal the molecular basis for eIF3 assembly, structure, and function in translation initiation. This analysis also reveals architectural features likely to be shared between the proteasome, COP9 signalosome, and eIF3 and suggests a productive strategy for mechanistic and structural dissection of these and other large macromolecular assemblies that control the eukaryotic proteome.
Results
Stepwise Reconstitution of Human eIF3.
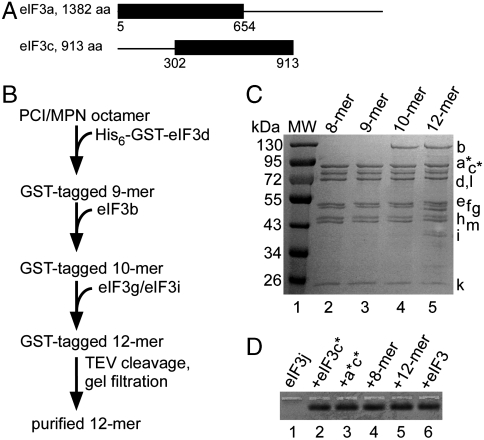
We tested whether subcomplexes identified during eIF3 disassembly (3), or assembled using baculovirus expression (21, 22) could be formed de novo in E. coli. Subunits eIF3a and eIF3c truncated to remove likely flexible regions of the proteins (Fig. 1A) (25) yielded a stable a*c* dimer (asterisks indicating the truncations, Fig. 1A) that further assembled with subunits containing PCI (subunits a, c, e, k, l, and m) and MPN (subunits f and h) domains (Fig. S1), as well as with subunits conserved in all eukaryotes (b, g, and i) (21, 26–28) (Fig. S1). These results indicate that the a*c* dimer is central to eIF3, as it interacts with each of the remaining PCI and MPN domain-containing subunits in human eIF3, and also assembles with the universally conserved bgi subcomplex. However, the resulting trimeric through heptameric complexes were not biochemically stable, and were highly prone to aggregation, as determined by gel filtration chromatography.
Fig. 1.
Subassemblies of eIF3 expressed in E. coli. (A) Truncations in subunits eIF3a and eIF3c, denoted a* and c*. All other subunits were full-length (9). (B) Purification scheme for 12-subunit eIF3, beginning with the purified PCI/MPN octamer. Purifications of untagged 9-mer and 10-mer complexes similarly used TEV cleavage of the GST tag and gel filtration, when needed. Details are in SI Materials and Methods. (C) Coomassie blue-stained SDS gel of the octameric PCI/MPN complex containing subunits a*c*efhklm, eIF3 nonamer (a*c*defhklm), eIF3 decamer (a*bc*defhklm), and eIF3 dodecamer (a*bc*defghiklm), affinity-purified sequentially starting from the PCI/MPN octameric core using GST-tagged eIF3d, and with added subunits (bold) expressed in E. coli. Molecular weight (MW) markers, in kilodaltons, are shown to the left, and subunit positions are marked to the right. (D) Native agarose gel of eIF3 complexes showing binding of eIF3j. Fluorescently labeled eIF3j (10 nM) was incubated with the other eIF3 subunits or complexes (30 nM) prior to loading the gel. In the absence of binding, eIF3j does not enter the gel as a discrete band (lane 1). Lane 1, eIF3j alone; lanes 2–6, eIF3j incubated with other recombinant eIF3 components (lanes 2–5), or native human eIF3 depleted of eIF3j (lane 6).
Coexpression of the eight subunits that contain PCI or MPN domains (1) resulted in a highly stable octameric a*c*efhklm, or PCI/MPN, complex (Fig. 1 B and C) that further assembled into larger recombinant human eIF3 complexes in vitro. Subunits d and b purified individually, and subunits g and i coexpressed with each other (3, 22, 26, 27), added serially to the PCI/MPN octamer formed 9–12 subunit complexes, respectively (Fig. 1 B and C). The final subunit eIF3j associates with the remainder of eIF3 in a salt-dependent manner (21), and bound well to recombinant eIF3c*, the a*c* dimer, and the PCI/MPN octameric complex (Fig. 1D), as well as to subunits f and h (Fig. S1). Addition of eIF3j to the recombinant dodecameric eIF3 formed a complete 13-subunit human eIF3 of approximately 700 kDa molecular mass, lacking only the truncated domains in subunits eIF3a and eIF3c (Fig. 1 A and D).
Function of Reconstituted eIF3 Assemblies Expressed in E. coli.
To test the function of reconstituted eIF3 complexes, we assessed each complex for its ability to bind components of the translation initiation machinery and the HCV IRES. The biochemically stable PCI/MPN complex bound tightly to the IIIabc RNA (apparent Kd of 20–40 nM) (Fig. 2A), similar to natively purified eIF3 and to the 13-subunit reconstituted eIF3 (Fig. S2). An a*c*efhl hexamer (Fig. S1B) also bound tightly to the IIIabc RNA (Fig. S2) (25), but was biochemically unstable and highly prone to aggregation. Addition of subunits k or m individually to the hexamer did not affect IIIabc domain binding (Fig. S2) or improve the biochemical stability of the subcomplexes.
Fig. 2.
Binding of eIF3 subassemblies to the HCV IRES IIIabc domain. (A) Native agarose gel showing binding of the PCI/MPN octamer to the HCV IRES IIIabc domain, schematically drawn to the right. The nanomolar concentrations of the octamer are listed. Lane 1 shows IIIabc RNA binding to natively purified human eIF3 as a control (lane 1). Fluorescent IIIabc RNA was used at 20 nM in concentration, and the reactions were carried out in the presence of 2 μM tRNA to prevent nonspecific binding. (B) Native agarose gel showing binding of the PCI/MPN octamer to the 40S ribosomal subunit, monitored by UV absorbance of the 40S subunit rRNA. The nanomolar concentrations of the PCI/MPN octamer are given, and the 40S ribosomal subunit was used at a concentration of 10 nM. Shown as a control, 40S subunit binding to natively purified human eIF3 (lane 1). (C) Coomassie blue-stained SDS gel showing PCI/MPN octamer affinity-purified using MBP-tagged eIF1A, or using MBP-tagged eIF1. MW markers are shown in kilodaltons. Arrows indicate the position of MBP-eIF1A (lanes 2, 4) and MBP-eIF1 (lanes 5, 7). Binding and wash conditions prevent nonspecific binding of the PCI/MPN octamer to the beads (lane 8). (D) Coomassie blue-stained SDS gel showing reconstituted eIF3 10-mer affinity-purified using the FLAG-tagged central domain of eIF4G. MW markers are shown to the left. Arrow indicates the position of subunit eIF3b. GroE copurified with eIF3d (asterisks), as determined by MS analysis. Antibody heavy (H) and light (L) chains are marked. The concentration of KCl used in the washes (200 mM) prevents nonspecific binding to the anti-FLAG beads (Fig. S4).
The PCI/MPN octamer also bound tightly to the 40S ribosomal subunit, with an apparent affinity two- to threefold weaker than that of natively purified human eIF3 (Fig. 2B, Fig. S3). The addition of subunits d, b, and the gi dimer to the octamer did not increase the apparent binding affinity of the recombinant eIF3 complexes for the 40S subunit (Fig. S3). Including eIF3j, which stabilizes eIF3 binding to the 40S subunit on sucrose gradients (21, 29), had little impact on the apparent affinity of recombinant and natively purified eIF3 complexes for the 40S subunit in native gels (Fig. S3). Initiation factors eIF1 and eIF1A, which bind to eIF3 (28, 30, 31) and aid the formation of stable preinitiation complexes with the 40S subunit (32), also bind to the PCI/MPN core in affinity pull-down assays (Fig. 2C).
Translation initiation factor eIF4G in humans binds tightly to eIF3 (33, 34), and is positioned near the mRNA exit site of the 40S subunit (24). Although the central domain of eIF4G did not bind to the PCI/MPN octamer (Fig. S4), it did bind to a 10-subunit complex that includes the octamer plus subunits b and d (Fig. S4). Notably, binding of eIF4G to the 10-subunit complex in some experiments resulted in loss of eIF3b during the affinity pull-down (Fig. 2D), due to eIF3b dissociation from the complex under the conditions used. Thus eIF4G likely interacts with the 9-subunit eIF3 complex containing the PCI/MPN core and subunit d, after a conformational change induced in the complex by subunit b.
To test whether reconstituted eIF3 forms intact HCV IRES-mediated initiation complexes, we programmed rabbit reticulocyte lysate (RRL) with an mRNA containing the HCV IRES. Affinity purification of reconstituted and GST-tagged eIF3 from RRL reactions that included 2 mM GMPPNP (35) showed that reconstituted 12-subunit eIF3 could form preinitiation complexes with the 40S subunit and eIF2/Met-tRNAi (Fig. S5) as well as HCV IRES-mediated initiation complexes (Fig. 3, Fig. S6) (15). The 9-subunit eIF3 containing the PCI/MPN octamer and GST-eIF3d was less efficient at forming HCV IRES-mediated initiation complexes (Fig. S7), indicating that eIF3 subunits b, g, and i are important for HCV IRES-driven translation initiation.
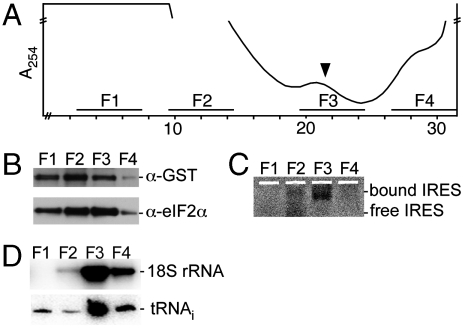
Fig. 3.
In vitro translation initiation complex formation in the presence of recombinant eIF3 dodecamer. (A) Sucrose gradient of RRL translation reaction stalled with GMPPNP and programmed with fluorescently labeled HCV IRES-containing mRNA. The top of the gradient is to the left, and the A254 absorbance is shown. The break marked on the absorbance axis corresponds to a fivefold decrease of sensitivity to account for heme absorbance. Fractions pooled for GST affinity purification and analysis are marked F1-F4, with F3 corresponding to HCV IRES-mediated initiation complexes (arrow). (B) Western blotting of GST-eIF3d and eIF2α, from samples GST-affinity-purified from pooled fractions F1-F4 in A. (C) Native agarose gel of fluorescently labeled HCV IRES copurified with GST-tagged eIF3 affinity-purified from pooled fractions F1-F4 in A. (D) Northern blotting of 18S rRNA and tRNAi present in GST-affinity-purified complexes from pooled fractions F1-F4 in A. In this experiment, recombinant eIF3 dodecamer must compete with endogenous eIF3 to form initiation complexes.
Structures of the PCI/MPN Octamer and Reconstituted eIF3 Complexes.
Given the significant number of activities associated with the eIF3 PCI/MPN octamer, we wondered how its structure compares to that of natively purified eIF3. In an approximately 23 Å negative-stain reconstruction, the PCI/MPN octamer bears a striking resemblance to the previously determined cryoEM structure of native human eIF3 (24), as well as to the reconstituted 12-subunit eIF3 at a resolution of approximately 29 Å (Fig. 4A). This result is a surprise, as the PCI/MPN octamer is just over 400 kDa in molecular mass, compared to the 800 kDa molecular mass of native human eIF3 used in prior EM reconstructions (24), and the 700 kDa molecular mass of the reconstituted 12-mer. Quantitative difference maps between the reconstructions of the PCI/MPN octamer and reconstituted 12-subunit eIF3 show two major regions of additional density in the larger complex, at the end of the left arm and leg features (Fig. 4B). This difference density cannot account for the approximately 300 kDa in molecular mass missing in the PCI/MPN octamer, suggesting that a significant part of the added subunits (b, d, g, and i) exist in multiple conformations that average out during the reconstruction process.
Fig. 4.
Structural analysis of the PCI/MPN octamer and eIF3 dodecamer complexes. (A) Negative-stain EM reconstructions of reconstituted 12-subunit eIF3 and PCI/MPN octamer at resolutions of 29 and 23 Å, respectively. Both reconstructions are shown in the same orientations, and are compared to the cryoEM reconstruction of native human eIF3 (24), to the right. (B) Difference map comparing the 12-subunit complex to the PCI/MPN octamer, filtered to a resolution of 29 Å, and contoured at 5.4 sigma. Positive density is shown in gold, and negative density in gray. (C) Interactions of the eIF3 PCI/MPN octamer with the remaining eIF3 subunits, with identified binary interactions noted. No geometrical constraints on positioning are implied by the location of the connecting lines.
Discussion
Our approach of combining gene synthesis, multisubunit coexpression in E. coli, and stepwise assembly provides a framework for dissecting human eIF3 structure and function. In the assembly pathway for human eIF3 reconstituted here, a dimer of subunits eIF3a and eIF3c serves as a central scaffold to which most of the other subunits bind. Notably, the a*c* dimer directly interacts with all of the remaining PCI/MPN domain-containing subunits individually, as well as with eIF3b and eIF3j (Fig. S1). Although some of these interactions had been inferred from prior genetic, biochemical, and disassembly experiments (3, 36), the present results provide direct evidence for many of these interactions (Fig. 4C). Additionally, the core of eIF3 is only stable when all eight of these subunits are coexpressed in E. coli, forming a PCI/MPN octamer (Fig. 1C). This PCI/MPN octamer is distinct from the three modules identified by disassembly experiments using mass spectrometry (3), indicating that the spontaneous assembly of eIF3 follows a different pathway than salt-dependent disassembly. Many of the interactions observed here have not been modeled in the PCI/MPN cores of the proteasome lid or COP9 signalosome (1, 2, 4, 5), possibly due to the challenge of identifying direct orthologues among the subunits in each complex. The results obtained here may aid in identifying the phylogenetic relationship between corresponding subunits in these cores (1).
From the PCI/MPN octamer as a starting point, we were able to add subunits eIF3d, eIF3b, the eIF3g/eIF3i dimer, and eIF3j serially to form an intact 700 kDa molecular mass human eIF3, missing only two domains from subunits eIF3a and eIF3c (Fig. 1A). Attempts to assemble a preformed bgi trimer with the PCI/MPN octamer were not successful in our hands. Furthermore, binding of the eIF4G central domain to the 10-subunit eIF3 complex destabilized eIF3b association with the reconstituted eIF3 complex (Fig. 2D, Fig. S4), an effect not seen with natively purified eIF3. Whether b, g, and i, which are universally conserved in eukaryotes, bind the preassembled PCI/MPN octamer in a stepwise manner in vivo, or require chaperones for assembly into eIF3, remains to be determined. Notably, a functional 6-subunit complex assembled in insect cells lacked three of the PCI/MPN core proteins (22). This 6-subunit complex may have been stabilized by the presence of subunit eIF3b, indicating that interactions within the PCI/MPN core may be further stabilized by the remaining subunits (b, d, g, i, and j).
The striking resemblance of the PCI/MPN octamer to intact human eIF3 (Fig. 4) (24) suggests that eIF3 may envelope the 40S ribosomal subunit, using flexible regions emanating from the PCI/MPN core in the assembly of preinitiation and initiation complexes (24, 37–40). Genetic and biochemical experiments identified interactions between subunits eIF3a and eIF3c and the solvent side of the 40S subunit (37, 38). The flexible C terminus of eIF3a is located near the mRNA entry tunnel, whereas the N terminus of eIF3c is likely positioned near the 40S subunit platform where it interacts with eIF1 and eIF5 situated at the subunit interface (41). In addition, biochemical experiments place the C terminus of eIF3j on the 60S subunit interface side of the 40S subunit within the mRNA decoding site (39). Subunit eIF3j, which is involved in regulating mRNA binding and positioning on the 40S ribosomal subunit during translation initiation (39, 40), interacts with at least five subunits within human eIF3, including four of the PCI/MPN domain-containing subunits (c, f, h, and k) (Fig. 1, Fig. S1) (36). The only possible means for eIF3j to approach the decoding site in the 40S subunit is for eIF3 to nearly encircle the 40S subunit mRNA binding channel. Furthermore, models of the 40S subunit-eIF3 binary complex place one leg of eIF3 in the way of the 60S subunit binding surface of the 40S subunit platform domain (24). The evidence presented here shows that elements of the PCI/MPN core constitute this leg feature of eIF3. Finally, the PCI/MPN octamer interacts with eIF1 and eIF1A, both of which reside at the subunit interface during initiation (31, 42–44), indicating that even the PCI/MPN core identified here contains flexible domains (Fig. 5), in addition to subunits a and c (Fig. 1A), and the flexible attachment of subunits b, d, g, i, and j (Fig. 4B). Future efforts will be required to test the functional role of the PCI/MPN core in aiding a highly flexible eIF3 in the assembly of translation preinitiation and initiation complexes, and whether the proteasome lid and COP9 signalosome also exploit this architectural strategy for functional interactions with their cellular targets (45, 46).
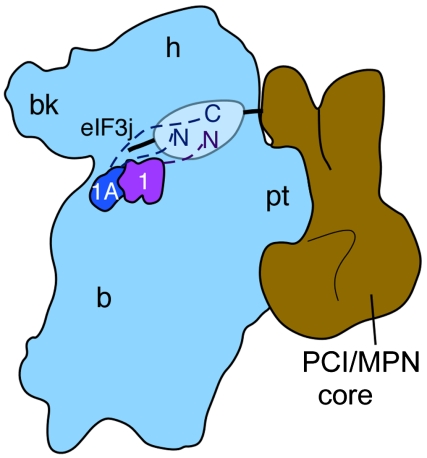
Fig. 5.
Model for interactions of the eIF3 PCI/MPN octamer with the 40S subunit, eIF3j, and initiation factors eIF1 and eIF1A. The position of the PCI/MPN octamer relative to the 40S subunit is based on that in ref. 24. The positions of eIF1 and eIF1A are based on the X-ray crystal structure of the eIF1/40S complex (44), and directed hydroxyl radical probing experiments of eIF1A bound to the 40S subunit (43), or eIF3j bound to the 40S subunit (39). Inferred flexible regions of the translation factors are indicated by dashed lines (eIF1 or eIF1A) or a solid line (eIF3j), as well as an oval for regions of the PCI/MPN octamer flexibly attached to the ordered core.
Materials and Methods
Purification of eIF3, 40S Ribosomal Subunits and HCV IRES RNAs.
Human eIF3 and 40S ribosomal subunits were purified from HeLa cell lysates as described in refs. 9 and 25. Subunit eIF3j was expressed and purified as described in ref. 39. HCV IRES RNA and the IRES IIIabc domain were transcribed and purified as described in ref. 47. The sequence of the IRES IIIabc domain is that used in ref. 25. The IRES sequence used in RRL initiation complex formation includes nucleotides 39–352 of the HCV subtype 1b genomic RNA.
In Vitro Reconstruction of eIF3 Subcomplexes.
All eIF3 subunits were PCR amplified from human cDNA (American Type Culture Collection) or were codon-optimized and inserted into transfer vectors 2AT, 2CT, 2GT, 2ST, and 2XT (SI Materials and Methods, Table S1, Table S2) using ligation-independent cloning (48, 49). The open reading frames for subunits eIF3b (N terminus), eIF3e, eIF3i, and eIF3j were codon-optimized by DNA 2.0 (SI Materials and Methods, Table S3). Vectors were developed by the MacroLab at the QB3 Institute, University of California, Berkeley, and are available from Addgene (www.addgene.org). The preparation of transfer vectors containing eIF3 subunits and of polycistronic expression vectors is described in SI Materials and Methods.
Expression and Purification of eIF3 Subcomplexes.
All eIF3 subcomplexes were expressed in Rosetta2(DE3)pLysS E. coli (EMD Biosciences), as described in the SI Materials and Methods and Table S3. Individual subunits of eIF3 expressed with N-terminal fusion tags (Table S1, Table S2) were purified by nickel affinity chromatography [His6-Saccharomyces cerevisiae small ubiquitin-like modifier (SUMO) and His6-γ-crystallin tags], glutathione affinity chromatography (His6-GST tag), or dextrin sepharose affinity chromatography [His6-E. coli maltose binding protein (MBP) tag], as described in SI Materials and Methods.
N-terminal tags were cleaved by tobacco etch protease (TEV) and the cleaved tag and TEV protease were removed from the subunits and complexes by passing the solution through the HisTrap HP column. The flow-through fractions were then passed through a HiTrap Q column (GE Healthcare) to remove nonspecifically bound RNA. Further details of the purification of different complexes are included in SI Materials and Methods.
EM Sample Preparation and Data Collection.
Samples diluted to a final concentration of 50 nM were placed onto continuous carbon grids, negatively stained with a 3% uranyl acetate solution and blotted dry. Data were acquired using a Tecnai T12 electron microscope equipped with a Tietz 4 × 4 K pixel CCD camera using low dose techniques. All the processing of two-dimensional data was performed using programs and utilities contained within the Appion processing environment (50). Further details of image acquisition, particle extraction, and generation of class averages is included in SI Materials and Methods. The particles included in the final set of class averages (21,472 and 12,452 particles for the PCI/MPN octamer and dodecamer stacks, respectively) were generated from phase-flipped micrographs using the estimated parameters from the CTFFind program (51). An initial model of human eIF3 (24) filtered at 120-Å resolution was used for three-dimensional refinement, performed using iterative projection matching in EMAN2 (52, 53). Further details of volume analysis and difference map calculation are included in SI Materials and Methods.
The 40S Ribosomal Subunit-eIF3 (Sub)complex Formation.
Binding reactions were carried out at 25 °C for 15 min in ribosome binding buffer (20 mM Hepes, pH = 7.5, 100 mM KCl, 2.5 mM MgCl2). Native agarose gel electrophoresis used to monitor binding was carried out at 4 °C in buffer containing 34 mM Tris, 66 mM Hepes, 0.1 mM EDTA, 2.5 mM MgCl2, 75 mM KCl, pH = 7.8, hereafter termed THEMK buffer, with frequent buffer exchanges.
eIF3 (Sub)complex Interactions with Labeled IIIabc HCV IRES and eif3j-Alexa488.
Binding reactions to the IIIabc HCV IRES domain contained reconstituted human eIF3, labeled HCV IRES IIIabc domain (SI Materials and Methods) at 20 nM, and THEMK buffer. The reactions were carried out at 25 °C for 15 min, then resolved by native 1% agarose gels containing THEMK buffer, at 4 °C for 45 min. Similarly, binding of eIF3 subcomplexes to labeled eIF3j (SI Materials and Methods) was assayed by native agarose gels containing THEMK buffer. The resulting agarose gels were analyzed by fluorescence imaging using a Typhoon Scanner (Amersham Biosciences) to detect the gel shifts. The excitation and emission wavelengths to detect the HCV IRES IIIabc domain and eIF3j were 494 and 518 nm, respectively.
eIF1, eIF1A, and eIF4G Pull-Down Assays.
For eIF1A or eIF1 pull-down assays, 50 μL of amylose resin (New England BioLabs) was washed three times with MBP column buffer A (20 mM Hepes, pH 1/4 7.5, 300 mM KCl, 1 mM EDTA, 1 mM DTT, 10% glycerol) and then incubated with 20 μg of purified His6-MBP-eIF1A or His6-MBP-eIF1 (SI Materials and Methods) for 1 h at 4 °C with gentle rotation. The beads were pelleted, to allow for removal of the supernatant. Following three washes with MBP column buffer A, 100 μg of purified PCI/MPN octamer was incubated with the beads for 2 h at 4 °C. After removing the supernatant and washing the beads three times, bound proteins were eluted using MBP column buffer B (20 mM Hepes, pH 1/4 7.5, 300 mM KCl, 10 mM maltose, 1 mM EDTA, 1 mM DTT, 10% glycerol).
A truncated version of human eIF4G (amino acids 1011–1104) with an N-terminal His6-FLAG tag (His6-FLAG-eIF4Gt) was used for eIF4G pull-down assays. Pull-down assays were carried out using resin linked to anti-FLAG antibodies (ANTIFLAG M2 affinity gel, Sigma). Briefly, 60 μL of resin slurry were washed three times with 500 μL of dilution buffer (20 mM Hepes pH = 7.5, 150 mM KCl, 10% glycerol), followed by addition of 10 μg of His6-FLAG-eIF4Gt to allow the FLAG-tagged truncated eIF4G to bind to the resin. The resin was then washed three times with 100 μL wash buffer (20 mM Hepes pH = 7.5, 200 mM KCl, 0.5% Triton X-100, 0.5 mM DTT, 5% glycerol). Natively purified eIF3 or reconstituted eIF3 complexes were then added to the resin and incubated with the resin for 30 min on ice. The resin was then washed three times with wash buffer to remove unbound protein. Any bound eIF3/eIF4G complex was then eluted by adding 7.5 μg FLAG peptide in dilution buffer.
In Vitro Translation Assays of Reconstituted eIF3.
The purification of the 12-subunit eIF3-containing GST-tagged eIF3d was as described in SI Materials and Methods, but with the GST tag retained prior to the final gel filtration step. In vitro translation assays were carried out using nuclease-treated RRL (Promega), essentially as described in refs. 54 and 55. Details of the translation assays are given in SI Materials and Methods. Gradients of the RRL reactions were fractionated using an ISCO UV detector and 0.33 mL fractions were collected for subsequent native agarose gel and affinity purification experiments.
To track the HCV IRES, 10 μL of each fraction was resolved by native agarose gel electrophoresis in THEMK buffer, as described above. Fractions from the sucrose gradients were pooled into four larger fractions (F1-F4), to which additional RNasin Plus RNase Inhibitor and Complete Protease Inhibitor Cocktail were added. The four fractions were then dialyzed into THEMK buffer for 2 h at 4 °C. To affinity purify complexes containing GST-tagged eIF3, the pooled fractions were mixed gently with 0.1 mL reduced Glutathione-Sepharose 4B beads (GE Healthcare) for 2 hr at 4 °C. The beads were collected by brief centrifugation and washed three times with THEMK buffer. Proteins bound to the beads were eluted with THEMK buffer containing 10 mM reduced glutathione.
The affinity-purified complexes were then analyzed by Western and Northern blotting, as follows. The presence of GST-eIF3d was probed with an anti-GST antibody (Abcam), as described in ref. 56. Similarly, the presence of eIF2 was probed with an anti-eIF2α antibody (Cell Signaling), and subunit eIF3a or its truncated version (a*) was probed using an anti-eIF3a antibody (Santa Cruz). To probe for the 40S ribosomal subunit and tRNAi, an 18S rRNA-specific probe (5′-ACGGTATCTGATCGTCTTCGAACC-3′) (57) and a tRNAi-specific probe (5′-TGGTAGCAGAGGATGGTTTCGAT-3′) were used. The probes were labeled on the 5′ end with [γ-32P] ATP (Perkin Elmer) using T4 polynucleotide kinase (New England BioLabs). Details of the Northern blotting analyses are given in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Robert Tjian for HeLa cell cytoplasmic extracts, Anthony Iavarone for help with mass spectrometry data acquisition and interpretation, Masaaki Sokabe for help with protocols for Northern analyses, and John Hershey for helpful comments on the manuscript. This work was funded by the National Institutes of Health Grant P01GM073732 (to J.H.D.C. and J.A.D.), R01GM092927 (to C.S.F.), and 1S10RR022393-01 to the QB3/Department of Chemistry Mass Spectrometry facility. J.A.D. and E.N. are Howard Hughes Medical Institute Investigators.
Footnotes
The authors declare no conflict of interest.
Data deposition: The negative-stain EM maps of the eIF3 PCI/MPN octamer and eIF3 dodecameric complex have been deposited in the Electron Microscopy Data Bank, www.ebi.ac.uk/pdbe/emdb/, (EMDB accession codes EMD-1975 and EMD-1976).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116821108/-/DCSupplemental.
References
- 1.Pick E, Hofmann K, Glickman MH. PCI complexes: Beyond the proteasome, CSN, and eIF3 Troika. Mol Cell. 2009;35:260–264. doi: 10.1016/j.molcel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Sharon M, Taverner T, Ambroggio XI, Deshaies RJ, Robinson CV. Structural organization of the 19S proteasome lid: Insights from MS of intact complexes. PLoS Biol. 2006;4:e267. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou M, et al. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci USA. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharon M, et al. Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure. 2009;17:31–40. doi: 10.1016/j.str.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Enchev RI, Schreiber A, Beuron F, Morris EP. Structural insights into the COP9 signalosome and its common architecture with the 26S proteasome lid and eIF3. Structure. 2010;18:518–527. doi: 10.1016/j.str.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Gallastegui N, Groll M. The 26S proteasome: Assembly and function of a destructive machine. Trends Biochem Sci. 2010;35:634–642. doi: 10.1016/j.tibs.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Moretti J, et al. The translation initiation factor 3f (eIF3f) exhibits a deubiquitinase activity regulating notch activation. PLoS Biol. 2010;8:e1000545. doi: 10.1371/journal.pbio.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damoc E, et al. Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol Cell Proteomics. 2007;6:1135–1146. doi: 10.1074/mcp.M600399-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 11.Honda M, et al. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 12.Kamoshita N, Tsukiyama-Kohara K, Kohara M, Nomoto A. Genetic analysis of internal ribosomal entry site on hepatitis C virus RNA: Implication for involvement of the highly ordered structure and cell type-specific transacting factors. Virology. 1997;233:9–18. doi: 10.1006/viro.1997.8600. [DOI] [PubMed] [Google Scholar]
- 13.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieft JS, et al. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 15.Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 16.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraser CS, Hershey JW, Doudna JA. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nat Struct Mol Biol. 2009;16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- 19.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraser CS, et al. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40S ribosomal subunits in vitro. J Biol Chem. 2004;279:8946–8956. doi: 10.1074/jbc.M312745200. [DOI] [PubMed] [Google Scholar]
- 22.Masutani M, Sonenberg N, Yokoyama S, Imataka H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 2007;26:3373–3383. doi: 10.1038/sj.emboj.7601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 24.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 25.Cai Q, et al. Distinct regions of human eIF3 are sufficient for binding to the HCV IRES and the 40S ribosomal subunit. J Mol Biol. 2010;403:185–196. doi: 10.1016/j.jmb.2010.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verlhac MH, Chen RH, Hanachi P, Hershey JW, Derynck R. Identification of partners of TIF34, a component of the yeast eIF3 complex, required for cell proliferation and translation initiation. EMBO J. 1997;16:6812–6822. doi: 10.1093/emboj/16.22.6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asano K, Phan L, Anderson J, Hinnebusch AG. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:18573–18585. doi: 10.1074/jbc.273.29.18573. [DOI] [PubMed] [Google Scholar]
- 28.Phan L, Schoenfeld LW, Valasek L, Nielsen KH, Hinnebusch AG. A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNA(i)-Met. EMBO J. 2001;20:2954–2965. doi: 10.1093/emboj/20.11.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fletcher CM, Pestova TV, Hellen CU, Wagner G. Structure and interactions of the translation initiation factor eIF1. EMBO J. 1999;18:2631–2637. doi: 10.1093/emboj/18.9.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsen DS, et al. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 2003;22:193–204. doi: 10.1093/emboj/cdg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majumdar R, Bandyopadhyay A, Maitra U. Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40 S preinitiation complex. J Biol Chem. 2003;278:6580–6587. doi: 10.1074/jbc.M210357200. [DOI] [PubMed] [Google Scholar]
- 33.Korneeva NL, Lamphear BJ, Hennigan FL, Rhoads RE. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J Biol Chem. 2000;275:41369–41376. doi: 10.1074/jbc.M007525200. [DOI] [PubMed] [Google Scholar]
- 34.LeFebvre AK, et al. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem. 2006;281:22917–22932. doi: 10.1074/jbc.M605418200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 36.Mayeur GL, Fraser CS, Peiretti F, Block KL, Hershey JW. Characterization of eIF3k: A newly discovered subunit of mammalian translation initiation factor elF3. Eur J Biochem. 2003;270:4133–4139. doi: 10.1046/j.1432-1033.2003.03807.x. [DOI] [PubMed] [Google Scholar]
- 37.Valasek L, et al. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev. 2003;17:786–799. doi: 10.1101/gad.1065403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiu WL, et al. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol Cell Biol. 2010;30:4415–4434. doi: 10.1128/MCB.00280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fraser CS, Berry KE, Hershey JW, Doudna JA. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 40.Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinnebusch AG. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 42.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Y, et al. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res. 2009;37:5167–5182. doi: 10.1093/nar/gkp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 45.Kato JY, Yoneda-Kato N. Mammalian COP9 signalosome. Genes Cells. 2009;14:1209–1225. doi: 10.1111/j.1365-2443.2009.01349.x. [DOI] [PubMed] [Google Scholar]
- 46.Stadtmueller BM, Hill CP. Proteasome activators. Mol Cell. 2011;41:8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stols L, et al. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expression Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 50.Lander GC, et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 52.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 53.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Kozak M. Evaluation of the fidelity of initiation of translation in reticulocyte lysates from commercial sources. Nucleic Acids Res. 1990;18:2828. doi: 10.1093/nar/18.9.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berry KE, Waghray S, Doudna JA. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA. 2010;16:1559–1569. doi: 10.1261/rna.2197210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun C, Pager CT, Luo G, Sarnow P, Cate JHD. Hepatitis C virus core-derived peptides inhibit genotype 1b viral genome replication via interaction with DDX3X. PLoS One. 2010;5:e12826. doi: 10.1371/journal.pone.0012826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ikeda K, Arao Y, Otsuka H, Kikuchi A, Kayama F. Estrogen and phytoestrogen regulate the mRNA expression of adrenomedullin and adrenomedullin receptor components in the rat uterus. Mol Cell Endocrinol. 2004;223:27–34. doi: 10.1016/j.mce.2004.06.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.