Abstract
Reverse transcriptases have shaped genomes in many ways. A remarkable example of this shaping is found on telomeres of the genus Drosophila, where retrotransposons have a vital role in chromosome structure. Drosophila lacks telomerase; instead, three telomere-specific retrotransposons maintain chromosome ends. Repeated transpositions to chromosome ends produce long head to tail arrays of these elements. In both form and function, these arrays are analogous to the arrays of repeats added by telomerase to chromosomes in other organisms. Distantly related Drosophila exhibit this variant mechanism of telomere maintenance, which was established before the separation of extant Drosophila species. Nevertheless, the telomere-specific elements still have the hallmarks that characterize non-long terminal repeat (non-LTR) retrotransposons; they have also acquired characteristics associated with their roles at telomeres. These telomeric retrotransposons have shaped the Drosophila genome, but they have also been shaped by the genome. Here, we discuss ways in which these three telomere-specific retrotransposons have been modified for their roles in Drosophila chromosomes.
Keywords: centromeres, chromosome evolution, heterochromatin, euchromatin, Y chromosome
Cells invest an unexpected amount of their resources in what might seem to be relatively insignificant parts of the chromosome, their telomeres. Multicellular eukaryotes tend to have 10 or more kb of telomere repeats on each chromosome end; even unicellular eukaryotes have a few hundred base pairs of telomere repeats per end. Telomere length is regulated in species- and cell type-specific ways; it is dynamic and can be influenced by diverse factors, including environment, genetic background, stress, and health (1–3). Telomere maintenance is complex. It is also essential for cell replication and genome integrity.
In this Perspective, we discuss a dramatic alternative to the almost universal use of telomerase to maintain telomeres. Drosophila lacks telomerase. Instead, specialized non-LTR retrotransposons (6–13 kb long) extend telomeres by transposing onto chromosome ends to form head to tail repeats (4, 5). Despite the apparent differences, Drosophila telomeres, like other telomeres, are extended by reverse transcription of an RNA template—a newly transcribed copy of a telomeric retrotransposon—and share many other characteristics of telomerase telomeres. For example, Drosophila telomeres are comparable in length with those of other metazoans and are much longer than telomeres of most single-celled eukaryotes. Accordingly, the Drosophila telomere-specific retrotransposons provide an unexpected link between chromosome structure and transposable elements, a link that raises questions about the evolution of both and specifically about mechanisms underlying telomere length homeostasis in Drosophila in which the retrotransposon repeats are more than three orders of magnitude longer than telomerase repeats.
HeT-A, TART, and TAHRE: A Ménage à Trois?
Like other complex repeat sequences, telomere database entries are prone to misassembly and require special verification. In fact, D. melanogaster is the only member of the genus whose genome has been sequenced thoroughly enough to allow reliable conclusions about the population distribution of telomere elements. Therefore, this section will be limited to this species, with brief comments on other species at the end.
HeT-A, TART, and TAHRE are the only telomere-specific retrotransposons found in D. melanogaster (Fig. 1). All are non-LTR retrotransposons belonging to the jockey clade (Fig. S1), an abundant group of non-LTR retrotransposons scattered over both euchromatic and heterochromatic regions of the Drosophila genome. These three retrotransposons are the only members of this clade found in telomeres. Potentially active copies are found nowhere except telomeres, although decayed fragments are present in other heterochromatin.
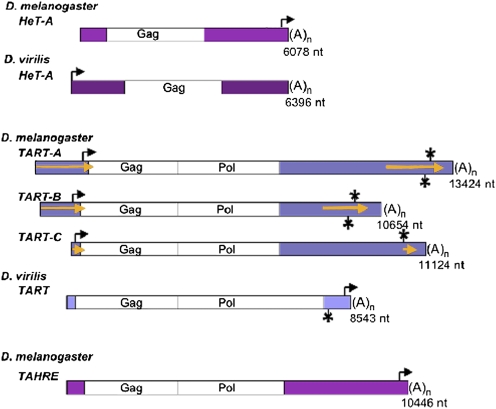
Fig. 1.
Telomeric retrotransposons from D. melanogaster and D. virilis (approximately to scale). Magenta, 5′ and 3′ UTRs of HeT-A and TAHRE; blue, 5′ and 3′ UTRs of TART; white, Gag and Pol ORFs; gold arrows in 5′ and 3′ UTRs of TARTmel elements (see Figs. 3B and 4 and accompanying text), PNTRs; (A)n, 3′ oligoA; bent arrows, transcription start sites for full-length sense-strand RNA (note that, for HeT-Amel and TARTvir, the element transcribed is immediately downstream of the element shown); asterisks above TART elements, start site for short sense-strand RNA; asterisks below TARTs, start site for nearly full-length antisense RNA (not determined for TART-C). The length of 5′ UTRs in TARTmel elements is extremely variable. Those lengths shown here representing the three subfamilies were the first to be sequenced; other members of these subfamilies have shorter or longer 5′ UTRs. 5′ PNTR extends to the 5′ end of the element; thus, the length of any element’s 3′ PNTR is defined by the length of its 5′ PNTR. Although the TARTvir 3′ UTR is much shorter than the 3′ UTR of the other elements, it is more than two times the length of the 3′ UTR of nontelomeric jockey clade elements that we have analyzed. TARTvir Pol ORF also has a 3′ extension of ∼1.2 kb, with no obvious motifs to indicate its function (14). This sequence is not seen in the other elements and might do double duty as 3′ UTR when the element forms telomere DNA. Elements shown are HeT-Amel, U06920, nucleotides 1,015–7,097; HeT-Avir, AY369259, nucleotides 7,211–13,612; TARTmel -A, AY561850; TARTmel -B, U14101; TARTmel -C, AY600955; TARTvir, AY219709, nucleotides 4,665–13,208; and TAHRE, AJ542581.
It Is Significant That HeT-A, TART, and TAHRE Are Non-LTR Retrotransposons: The Mechanism by Which This Set of Retroelements Transposes onto Chromosomes Is Basically Equivalent to That Used by Telomerase.
Non-LTR elements enter the nucleus as RNA, the 3′ end of this RNA associates with a nick in the chromosome, and its reverse transcription is primed off the 3′ OH of the nicked DNA, linking the new DNA to the chromosome (6–8). Although we lack iron-clad proof, there is plenty of evidence that telomeric retrotransposon RNA associates with the end of the DNA rather than with an internal nick like other elements. Thus, each transposition of a telomeric element adds a new end to the DNA, extending the chromosome. Successive transpositions produce long arrays of these elements, all oriented with their 5′ ends toward the end of the chromosome, and many showing some truncation of their 5′ end (Fig. 2 is an example). HeT-A, TART, and TAHRE probably have equivalent roles in telomere arrays, because all three retrotranposons seem to be distributed randomly in telomere arrays.
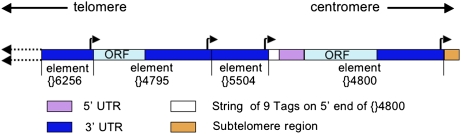
Fig. 2.
The four most proximal elements in the sequenced array from the XL telomere drawn approximately to scale. All elements are HeT-A, and each element is joined by its 3′ oligoA to its proximal neighbor. The most proximal element, HeT-A {}4,800, is complete, two elements are truncated in the 3′ UTR, and one element is truncated in the ORF. Elements are identified by FlyBase identifier number. Dark blue, 3′ UTR; light blue, ORF; magenta, 5′ UTR; white, string of Tags on the 5′ end of complete element {}4,800; gold, beginning of the subtelomere region; bent arrows, transcription start sites (each arrow indicates a cluster of three closely spaced sites at the 3' end of the element). The start site on {}5,504 initiates transcription of {}4,800, a transposition-competent element. Other starts will not produce productive transcripts. Physical mapping of BACs from this stock indicates that this telomere extends >100 kb further to the left (51), but no sequence is available.
The Three Telomere-Specific Elements Share Characteristics Not Seen in Other Jockey Clade Non-LTR Retrotransposons.
These characteristics include: (i) Telomere elements transpose only onto chromosome ends. There is no apparent DNA sequence specificity for the attachment site; they transpose onto the 5′ ends of other elements on the chromosome end whether those ends are intact or broken, including broken chromosome ends that have lost all telomere and sometimes, subtelomere sequences (7–10). (ii) Other transposons are not found in telomere arrays (11). (iii) The telomere-specific elements are not found in euchromatic DNA unless that DNA is the end of a broken chromosome. Fragments of the elements (mostly from the UTRs) have been found in nontelomere heterochromatic regions, but these fragments seem to have been passively moved to these locations rather than actively transposed (discussion of Y centromere below) (11). (iv) Telomere elements have atypical, very long UTRs.
Although They Apparently Share a Specific Role in Telomere Maintenance, HeT-A, TART, and TAHRE Are Surprisingly Different from Each Other.
For example, most, if not all, nontelomeric jockey clade elements encode a reverse transcriptase (RT). Both TART and TAHRE encode this enzyme, but HeT-A does not have an RT gene in any species studied. This lack of self-sufficiency might be thought to diminish the efficiency of HeT-A transposition, but in all the D. melanogaster stocks that we have studied, both HeT-A and TART are present, and HeT-A is always much more abundant than TART, no matter how much telomere DNA the stock contains (11). TAHRE is rare, although it seems to combine the best features of the other two elements because its UTR and gag sequences are very similar to those of HeT-A and its RT is similar to that of TART (12). Homologs of HeT-A and TART have been identified in D. virilis (>40 Myr separation from D. melanogaster), showing that these elements probably have been maintaining telomeres since before the separation of the extant Drosophila species (13, 14). TAHRE elements have been detected in several species of the melanogaster species subgroup (separation = 10–15 Myr) and may well be present in more distantly related species (15). This evolutionary conservation of the three elements suggests that each one makes a contribution to maintaining Drosophila telomeres.
As mentioned earlier, the apparently random distribution of HeT-A, TART, and TAHRE in telomere arrays argues that they are essentially equivalent in their ability to form telomere chromatin. However, there is strong evidence that the three elements collaborate in different ways in transposing to chromosome ends. In diploid somatic cells, the HeT-A Gag protein specifically localizes to chromosome ends in interphase nuclei. TART Gag moves into nuclei in these cells but moves to chromosome ends only if assisted by HeT-A Gag (16). TAHRE Gag is predominantly cytoplasmic but localizes adjacent to the nuclear membrane; it also requires HeT-A Gag for localization to telomeres (17). The colocalization of TART and TAHRE Gags with HeT-A Gag provides an opportunity for HeT-A to use the RT of either of the other two elements. It is possible that the choice of RT may depend on cell type: in oocytes, TAHRE RNA but not TART RNA is coexpressed with HeT-A (15).
Drosophila Cell Interactions with Telomere Elements Differ from Their Interactions with Nontelomeric Retrotransposons.
Although Gags of telomeric elements are efficiently localized to chromosome ends, other jockey clade Gags are almost entirely retained in the cytoplasm, even when HeT-A Gag is present (18). We suggest that cytoplasmic retention of the Gags of nontelomeric retrotransposons is one layer of cellular defense against parasitic elements, whereas telomeric Gags and their host cells have coevolved a nuclear localization mechanism to facilitate telomere maintenance.
The Rasi RNAi mechanism that protects cells from parasitic DNA is also involved in modulating the rate of transposition of telomeric elements in oocytes (19, 20), suggesting that this cellular defense has been modified for telomere regulation.
For Multicopy Elements, Phylogenetic Studies Provide Important Information About Evolutionary Origins and the Functional Importance of Conserved Features.
We began phylogenetic studies of Drosophila telomeres by selecting λ-phage clones of DNA from D. yakuba (5–15 Myr separation from D. melanogaster). Importantly, λ-phage clones carry enough contiguous DNA to contain entire elements and element junctions, eliminating the possibility of misassembly of smaller sequences. Low stringency hybridization with the most conserved part of TARTmel RT identified clones of D. yakuba TART (21), which also contained HeT-A in head to tail arrays. A similar strategy identified TART and HeT-A from D. virilis (40–60 Myr separation from D. melanogaster) (13, 14). Using the cloned sequences to characterize these elements in intact flies and cultured cells, we found strong conservation of many unusual features across the genus, e.g., the discussion of their 5′ end protection below. We conclude that retrotransposons constitute a robust mechanism of telomere maintenance that may have arisen well before the separation of the Drosophila genus.
Do other Drosophila species have telomeric elements not in the HeT-A, TART, and TAHRE families? One element (Uvir in our D. virilis telomere clones) has HeT-A 5′ and 3′ UTR sequences but lacks a Gag gene; instead, it has a complete and open RT gene (13). Neither molecular studies of our D. virilis stock nor analysis of Genome Project scaffolds reveals the multiple Uvir elements expected of a bona fide replication-competent element. Also, analysis of sequence from other Drosophila species has identified a number of putative telomere retrotransposons (22); functional studies of these sequences will be necessary to determine if they are bona fide telomeric elements and how they relate to HeT-A, TART, and TAHRE.
Telomere-Specific Retrotransposons Have Evolved Innovative Ways to Protect Essential Sequences at Their 5′ Ends
Because telomere elements are reverse-transcribed onto the chromosome end, each newly transposed element is oriented so that its 5′ end is also the end of the chromosome. Thus, the element is subject to terminal erosion until another element transposes to take over the terminal position. Established Drosophila telomeres consist of many kilobases of HeT-A, TART, and TAHRE; many of these elements are variably 5′ truncated, showing that these chromosome ends, like ends maintained by telomerase, are dynamic (11). We believe that the incomplete elements in Drosophila telomeres also fulfill the roles carried out by telomerase repeats in other organisms. However, retrotransposon telomeres have an added responsibility; they must preserve at least some intact retrotransposons to supply new transpositions to maintain chromosome ends, because we have found no other source of complete telomere elements. Analysis of sequence from telomere arrays reveals a statistically significant overabundance of intact elements, evidence for protection of transposition-competent elements (11, 23).
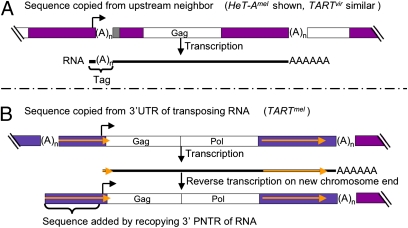
D. melanogaster and D. virilis HeT-A and TART have two unusual mechanisms to protect essential 5′ sequences from terminal erosion. D. melanogaster HeT-A (HeT-Amel) and D. virilis TART (TARTvir) add pilfered redundant sequences to the 5′ end of the transposing RNA to buffer sequence loss of the transposed element (Table 1 and Fig. 3A). D. melanogaster TART (TARTmel) also adds expendable sequence to its 5′ end (Table 1 and Fig. 3B); sequence copied from the element’s own 3′ UTR during reverse transcription (24, 25).
Table 1.
Mechanisms to add extra 5’ sequence
| Element | Mechanism |
| HeT-Amel and TARTvir | Start transcription in upstream element |
| HeT-Avir | Unknown |
| TARTmel | 2nd reverse transcription of 3’ PNTR |
Fig. 3.
Mechanisms for adding buffering 5′ sequence. (A) Sequence copied from upstream neighbor. Used by HeT-Amel and TARTvir. Telomere segment with a complete HeT-Amel flanked by other HeT-As. Transcription starts at the bent arrow in the upstream element and continues through the complete element. The resulting RNA (black line) has a Tag of the last nucleotides of the upstream element. On transposition, this Tag will become the 5′ end of the new element, undergo erosion, and if transposed again, be internalized into the string of variably eroded Tags indicated by the gray box at the 5′ end of the complete element. (B) Sequence copied from the 3′ UTR of transposing RNA. Used by TARTmel. Telomere segment with a complete TARTmel flanked by distal TART and proximal HeT-A. (A)n, 3′ oligoA in DNA; AAAAAA, polyA tail on RNA; gold arrows, PNTRs; other annotation as in Fig. 1. Transcription starts at the bent arrow and produces RNA with a very short 5′ UTR. When this is reverse-transcribed onto the chromosome end, the RT jumps back to the 3′ UTR and copies sequence to extend the 5′ UTR (Fig. 4).
D. virilis HeT-A (HeT-Avir) is an enigma (24). The HeT-Avir promoter, like the promoter of most non-LTR retrotransposons, is located in the 5′ UTR. Transcription starts upstream of the promoter, and the 5′-most sequence of the RNA is essential for transcription of the new element after transposition. There is no mechanism for adding buffering sequence; nevertheless, D. virilis telomere arrays contain a significant fraction of complete HeT-As. Thus, HeT-Avir must have another mechanism to protect its 5′ end.
On an evolutionary timescale, the different mechanisms in this small sample show an unexpected variation of end protection. However, only transposition-competent elements can give rise to lineages of new elements, which provides a very strong drive for evolving efficient 5′-end protection. The diversity of mechanisms seen in these studies seems to be a result of this drive.
HeT-Amel and TARTvir (and Maybe TAHRE) Share an Unusual Promoter Architecture That Adds Buffering Sequence to the 5′ End of the RNA Transcript.
Promoter sequences slightly upstream of the 3′ end of each of these two elements drive transcription, not of that element, but of its downstream neighbor (Figs. 2 and 3A). Transcription starts at sites within the 3′ end of the upstream element (26, 27). Thus, each new full-length sense-strand RNA has a very short copy of 3′ sequence, including an oligoA tail, added to its 5′ end as a Tag (23).
Although three of the four elements in Fig. 2 are 5′ truncated, all have intact 3′ ends carrying transcription start sites (indicated by bent arrows). Each appears capable of driving transcription of its downstream neighbor, even if the neighbor is truncated. However, our RNA studies show that most, if not all, detectible RNA is transcribed from full-length elements, suggesting some regulation of either transcription or turnover (28). Furthermore, statistical overabundance of complete elements in telomere DNA (11) and complete lack of Tags on partial elements support the idea that only complete elements are capable of transposing. In Fig. 2, only HeT-A {}4,800 is complete and expected to transpose.
A Tag on the RNA can be reverse-transcribed onto the chromosome, becoming an extension of the 5′ UTR of the new element (Fig. 3A). This Tag provides an expendable sequence to buffer loss of essential 5′ sequence from chromosome-end erosion. If a new transposition caps the chromosome end before the Tag has eroded completely, the truncated Tag remains as a 5′ extension of the element. A new Tag is added each time an element is transcribed, and this Tag will be attached to the existing string of truncated Tags. Thus, an element can have a 5′ string of variably truncated tags, providing evidence that it has transposed several times. For example, HeT-A {}4,800 (Fig. 2) has nine Tags of variable lengths on its end, showing that it has transposed at least nine times. When this element is next transcribed, the RNA will have a Tag of terminal sequence from HeT-A {}5,504 added to the existing Tags (in white). The presence of Tags confirms that the 5′ end of an element is functionally complete. Although some replicatively complete elements might lack Tags, we have not seen an element without Tags that has the complete 5′ UTR seen on elements with Tags.
The promoter used by HeT-Amel and TARTvir has not been found in other non-LTR elements but resembles the promoters of LTR retrotransposons and retroviruses. Promoters and transcription start sites of LTR elements are in the 5′ LTR (29). For two adjacent HeT-Amel (or TARTvir) elements in a telomere array, the 3′ end of the upstream element is essentially identical to the 3′ end of the downstream element; furthermore, the 3′ end of the upstream neighbor not only looks like but temporarily acts like a 5′ LTR for the downstream neighbor, because it contains the transcription start site. This pseudo-LTR promoter differs from bona fide LTR promoters in one important way: the Tag added to the new transcript does not contain the complete promoter sequence. Therefore, the newly transposed element does not possess its own promoter and remains a non-LTR element. Clearly, HeT-Amel and TARTvir pay a price for this arrangement; their transposition is possible only if two sister elements occur in tandem on the telomere. Strong selection for maintaining transposition-competent elements must balance the cost of this unusual arrangement.
TAHRE has not been identified in D. virilis, and there is not enough sequence information on TAHREmel to characterize a possible buffer mechanism. However, the strong similarity to HeT-A UTR sequences, supported by evidence that TAHREmel has a 3′ promoter similar to that of HeT-Amel, suggests that TAHREmel shares the HeT-Amel buffering mechanism (15).
TARTmel also Adds a Protective 5′ Sequence but Does so by Making a Second Copy from Its Own 3′ UTR When It Is Reverse-Transcribed onto the Chromosome.
D. melanogaster TART apparently has evolved a mechanism for maintaining its 5′ end that differs, in all details but not in principle, from the one found for D. virilis TART. Surprisingly, TARTmel does not have the pseudo-LTR promoter used by both its D. virilis homolog and its HeT-A partner in D. melanogaster. (This mechanism might be unfavorable, because TARTmel is greatly outnumbered by HeT-Amel and is less likely to have another TART as an upstream neighbor.) Extensive searches have found only a single start site for transcription of full-length sense-strand TARTmel RNA. Maxwell et al. (27) used RACE analysis to determine the 5′ end of TARTmel RNAs, and identify transcription start sites, whereas we identified promoter activity with reporter constructs (24–26). Both methods gave the same result, identifying only one site for full-length sense RNA, a site conserved in all three subfamilies of TARTmel (Fig. 1). These results were supported by the presence at that site of a very good match to the downstream promoter element and initiator of the 5′ UTR promoters typical of many non-LTR retrotransposons. This result is paradoxical: the transcription start is ∼75 nt 5′ of the start codon of ORF 1 (Fig. 3B), but only one reported TARTmel 5′ UTR, 33 nt long, is short enough to have been transcribed from this start site. (The few available TARTmel 5′ UTRs range from 33 to 3,934 nt.) Whence came all other 5′ UTRs?
We have proposed an explanation using a model based on some of the other unusual features of TARTmel UTRs (27). There are three TARTmel subfamilies, A, B, and C (Fig. 1). As with other telomeric retrotransposons, coding sequences in these subfamilies are somewhat variable. In contrast to HeT-A subfamily UTRs, which anomalously are not much more variable than their coding regions (11), TARTmel UTRs differ markedly between subfamilies. Nevertheless, the UTRs of all TARTmel subfamilies share one unusual characteristic: each contains a pair of long repeat sequences (30), one in the 5 ′UTR and its match in the 3′ UTR (Fig. 1).
Within individual elements, these repeats are clearly evolving together (25), like the LTRs of retroviruses and LTR retrotransposons (29). We have proposed that, like LTRs, the two TARTmel repeats coevolve because both are reverse-transcribed from only one of two repeats in the RNA. However, in TARTmel (a non-LTR element), the coevolution mechanism differs in its details from those in LTR elements; most notably, the 3′ TARTmel repeat does not extend to the 3′ end of the element. Thus, the TARTmel repeats are not strictly terminal, and we refer to them as perfect nonterminal repeats (PNTRs).
In TARTmel, the transcription start site for the full-length sense strand lies in the 5′ PNTR, just upstream of ORF 1. Despite the marked sequence differences in the subfamily UTRs, all three subfamilies share the same site. Comparing subfamilies by BLAST (blastn), we find small islands of nucleotide similarity surrounding the sense-strand start sites, suggesting that these sequences have been conserved, while surrounding UTR sequences have diverged.
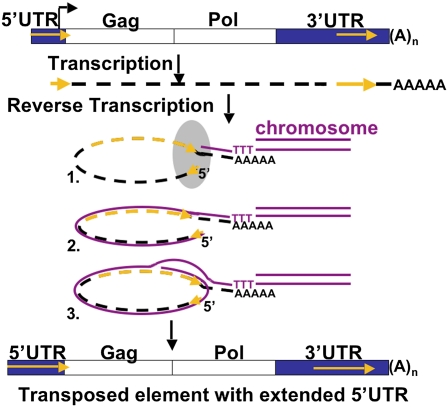
These sequence analyses suggested that all TARTmel RNA transcripts have a very short 5′ UTR, which is extended during transposition by repeating the reverse transcription of the 3′ PNTR (Figs. 3B and 4). Specifically, we postulated that when the reverse transcription of the RNA onto the chromosome reaches the 5′ end of the RNA, the RT makes a template jump to the identical sequence in the 3′ end of the 3′ PNTR (Fig. 4). It then continues to extend the 5′ UTR of the new element by making a second copy of the 3′ UTR, thus elongating the 5′ UTR of the transposed element and incorporating any changes that have occurred in the 3′ PNTR (25, 31). The proposed mechanism differs from that used by LTR elements to maintain the identity of their two ends. Specifically, the TARTmel RT must be capable of making a template jump from the 5′ end of the RNA back to the end of the 3′ PNTR. It must also dissociate the 3′ PNTR cDNA from its RNA template to make a second copy of the RNA. There is reason to expect the TARTmel enzyme can accomplish this dissociation because unlike the better known retroviral enzymes, RTs from at least two non-LTR elements (R2Bm and mouse L1) do make template jumps (31–33). The R2Bm enzyme is also able to separate the cDNA from its RNA template without destroying the template, whereas retroviral RNaseH degrades RNA bound to cDNA (34).
Fig. 4.
Proposed mechanism for extending the 5′ end of D. melanogaster TART. Transcription starts (bent arrow) near the ATG of ORF 1 (gag), producing a transposition intermediate RNA (dashed black line) lacking most of the 5′ UTR. This RNA has a small piece of the parent element’s 5′ PNTR (short gold arrow) and a complete 3′ PNTR (long gold arrow). Steps 1–3 show the RNA as it is reverse-transcribed into DNA on the chromosome end. (Step 1) The polyA tail associates with the chromosomal DNA (magenta), and RT begins to copy the RNA. The gray oval represents proteins proposed to hold the RNA in a conformation that brings the 5′ PNTR sequence into proximity to the 3′ end of the 3′ PNTR (omitted for clarity in later steps). (Step 2) When RT reaches the 5′ end of the transcript, it makes a template jump back to the matching 3′ end of the 3′ PNTR. (Step 3) RT dissociates the RNA–DNA complex and recopies some or all of the 3′ PNTR. As a result, the transposed element will have more 5′ UTR sequence than the RNA did and possibly more sequence and longer PNTRs than the element from which it was derived.
The second time around, the continuing reverse transcription of the TARTmel 3′ PNTR adds a sacrificial 5′ sequence that, like the Tags of TARTvir and HeT-Amel, can be lost to terminal erosion without affecting the transposition competence of the transposed element. The extreme variability in the length of the 5′ UTR in genomic TARTmel elements could be explained by variable termination of reverse transcription, terminal erosion, terminal deletions, or some combination thereof.
TARTmel Produces Two Other RNAs, a Small Sense-Strand RNA and a Nearly Full-Length Antisense RNA, Both of Unknown Function.
In addition to the transposition intermediate RNA discussed above, TARTmel produces two other abundant RNAs of unknown function. We do not believe that either RNA is involved in 5′ end buffering, but we mention them here to avoid confusion. The small sense strand is produced from the site in the 3′ PNTR that is identical to the start site in the 5′ PNTR. This 3′ site would produce RNA of a few hundred nucleotides depending on the TARTmel subfamily. There is a strong transcription termination site at the end of the element, which seems to act on both the large and small RNAs (27). A possible product of the 3′ start site has been seen on Northern blots, but no function is known (28).
The start site for the long TARTmel antisense RNA is similar in location to that of the antisense promoter for TARTvir, shown in Fig. 1 (25). Conservation of this promoter, despite the marked differences in the sense-strand promoter in the two species, suggests that the antisense RNA is important to the biology of the element. However, no role is known for the transcript. TARTmel is regulated by RasiRNA in the female germ line, but there is no evidence that the large antisense transcript is involved (19).
The Sequences of Telomeric Retrotransposon Arrays Provide a Chronological Record of Dynamic Activity at Chromosome Ends
Telomere arrays are elongated by successive transpositions onto chromosome ends. Thus, each element is older than the elements distal to it. The sequence information is rich in detail, because each element has several identifiers: (i) number of A residues copied when reverse transcription of that element was initiated, (ii) subfamily sequence of the element, (iii) extent of 5′ truncation, and (iv) amount of nonessential sequence remaining on the 5′ end of complete elements. In our analyses, these identifiers have allowed unique identification of elements. Furthermore, although these sequences are highly repetitive, there is enough microheterogeneity to identify products of recombination (except for exchanges within the most precisely aligned regions). We have seen no evidence for significant recombination. Thus, unlike telomerase telomeres with their myriads of identical repeats, each telomeric element bears a unique DNA fingerprint that allows determination of its history in the genome if its hierarchical position therein can be determined.
Even in D. melanogaster, difficulties in correctly assembling long sequences of highly repetitive DNA have largely precluded using whole-genome sequencing for analysis of the organization and possible roles of transposable elements in heterochromatic regions like centromeres and telomeres. Fortunately, some sequences derived from individual D. melanogaster BACs are available. We have analyzed sequences of a telomeric BAC from 4R and from directed finishing of a scaffold from the telomere of XL. Both the 4R and the XL sequences begin within their assembled chromosome and extend into the telomere, thus showing the precise relationship between these telomere arrays and the rest of the genome (11). Neither sequence extends to the distal end of the telomere, but together, they contain nearly 100 kb of telomeric arrays (76 kb from 4R and 20 kb from XL). Importantly, both include the most proximal and therefore, the oldest elements of the array and thus, present the most complete history available of D. melanogaster telomere maintenance.
The terminal arrays on both 4R and XL are composed entirely of head to tail telomeric retrotransposons. Each chromosome has a small transition zone at the proximal edge of the array where there are some fragments of nontelomeric elements mixed with fragments of telomeric elements. We do not include these transition zones in our discussion of telomere arrays. The most distal element in each array has been truncated by cloning and is also omitted. There is no available information on the organization of the most extreme end of Drosophila telomeres.
As explained below, the existing data justify positing three mechanisms for the maintenance of telomere-length homeostasis: small-scale end erosion averaging ∼20 nt between transpositions, large-scale terminal deletions that can encompass part or all of the telomere, and balancing sporadic transpositions that add large segments of DNA and renew the supply of transposition-competent elements.
HeT-A Elements in the Assembled Arrays Provide Sufficient Data to Make Quantitative Assessments of Telomere Dynamics in D. melanogaster.
The ∼100-kb sequence analyzed contains 4, possibly 5, intact and 10 or 11 5′-truncated HeT-A elements from six subfamilies as well as three 5′-truncated and two intact TART elements. There are no TAHRE elements. This distribution is consistent with the relative proportions of the three elements measured in different D. melanogaster stocks (11). The length of the arrays on XL and 4R suggests that these proximal elements should have been on the chromosome end long enough for significant sequence decay, because they are no longer under selection for function. However, this does not seem to be the case. Intact elements are distributed throughout the array, and all appear to be fully functional. Within the bounds of their natural variability, truncated elements have lost only 5′ sequence; remaining coding sequence is still open with no evidence of decay in the reading frame (23).
There is enough HeT-A sequence to allow statistically robust quantitative analysis of the overall dynamics of telomere turnover for these elements. There are few TART data, and we do not concatenate TART with HeT-A, because the two elements appear to be regulated differently. Most relevant to their turnover is the fact that, although we do not have enough TART 5′ UTR sequence to estimate the average amount of 5′ sequence added during TART transposition, it is clearly much longer than a HeT-A Tag; hence, details of TART turnover must be different (25).
HeT-A Sequence Provides Two Indicators of the Relative Rates of Sequence Loss from Telomeres.
These indicators are (i) the length and number of Tags on the 5′ end of each HeT-A and (ii) the distribution of complete and 5′ truncated elements in the telomere array. These observations, which describe the results of processes governing telomere maintenance and renewal, constrain models based on detailed numerical analysis. In turn, statistical analyses help in distinguishing and judging competing conclusions.
It is important to note that the sequence data analyzed here can be used only to determine relative rates for the different processes that we study. This finding is in contrast to measurements on the rate of loss from broken chromosome ends lacking telomere sequences. From broken chromosome studies, several groups (35–37) have determined that, unless healed by a telomere retrotransposon, the broken chromosome end recedes at about ∼70 nt per fly generation (i.e., between the measurement of its length in a male and its length in his son). It is interesting to think about the sequence losses seen in our telomere arrays in terms of the times measured for broken ends, but as discussed below, we conclude that sequence loss from telomere arrays is very different from the more or less regular, continuous erosion detected on broken ends. Instead, we conclude that maintenance of established telomeres involves at least three processes acting in concert to maintain relatively stable conditions: relatively small-scale terminal erosion, large-scale terminal deletion, and irregularly spaced transpositions (23).
HeT-A Tags Give Evidence for Relatively Small-Scale Terminal Erosion.
The initial length of a Tag is determined by its transcription start site (93, 62, or 31 nt upstream of the oligoA of the element providing the promoter) plus the oligoA of that element (mean OligoA = 8.3 nt, 95% confidence interval = 4.6–12.1 nt). Thus, the longest initial length would be ∼100 nt. This sequence is subject to chromosome-end erosion until another element transposes to cap the end of the chromosome. Because each new transposition adds 6–13 kb, depending on whether it is HeT-A, TART, or TAHRE, one would expect a Tag to be completely lost before the next transposition if simple erosion were the only telomere maintenance process. Instead, arrays have a good proportion of intact elements with truncated Tags on the 5′ end. Typically, the element has a string of several variably truncated Tags, indicating that it has transposed several times (23).
Analysis of Tag sequences allows us to measure the dynamics of erosion on established telomeres. On average, the Tags are surprisingly short. Their median length, including the oligoA tail, is 11 nt, their mean length is 14.0 nt, and the 95% confidence interval of the mean is 10.7–17.3 nt. Furthermore, the very shortest Tags are overrepresented (18% have the sequence TAAA), suggesting that the rate of sequence loss is reduced as Tags are eroded to their oligoA tail. There is also one very long Tag (68 nt) that is a distinct outlier; the next longest is 38 nt. The paucity of long Tags may be evidence that the two distant transcription starts, −93 and −62, are very rarely used. Alternatively, these longest Tags may be subject to more severe erosion, but Tags originating near −31 nt seem to be strongly protected until they loose several nucleotides (25). For shorter Tags, the median nucleotide loss is 25 nt, and the mean is 23.7 nt (SD = 6.3 nt).
Analysis of the strings of Tags on individual elements yields information on the relative rate of transposition onto those elements. The narrow limit on Tags per string (5–9) and Tag string length (69–161 nt) indicates that erosion is under some sort of control; we find neither intact HeT-As without Tags nor Tag strings that have grown without limit, as they would have if not effectively pruned (23).
These analyses show that the erosion process at the telomere end is more complex than the relatively regular loss described by studies of broken chromosomes, which may be the simple result of end replication losses. They also show that many, perhaps all, new transpositions occur before the terminal Tag has been completely eroded.
The two stochastic processes described here (relatively regular erosion of Tags and a tendency to protect the very shortest ones) cannot be the whole story. Given that one Tag is added per transposition, terminal erosion between transpositions, measured from individual Tag lengths, is very slow compared with sequence addition by new transposition (by a factor of several hundred); also, because only complete elements seem to be transposition-competent, the existence of multiple tags on complete elements implies that sequence addition is two to three orders of magnitude more rapid than gradual erosion (23). The result of transposition of elements that are much longer than the sequence eroded between transpositions should be extensive growth of telomeres, but telomere length remains relatively stable within each line studied (11). Analyses of the more truncated elements in the telomere (below) help explain how length balance is achieved.
Analyses of 5′-Truncated Elements Suggest Sporadic Terminal Deletions.
In contrast to Tag erosion, sequence loss from the 5′-truncated HeT-As is on a much larger scale and clearly contributes to telomere-length homeostasis. Two elements are truncated in the 5′ UTR, three elements are truncated in the ORF, and six in the 3′ UTR. All have enough 3′ UTR to provide promoter activity for a downstream neighbor, although the shortest element would provide only weak activity.
Lengths of the truncated elements scale from 5,892 to 241 bp and have no obvious correlation with position in the array. The relation of sequence loss to length for these elements is very different from that seen by analyzing Tag strings, suggesting that these truncations are the result of a different process. We suggest that at least some of this truncation results from terminal deletions that may occur anywhere within the array, leading to occasional rebuilding of all or part of the array (23).
There is no a priori reason to expect that some terminal deletions will not remove the entire telomere array and possibly, extend farther into the chromosome; furthermore, there is evidence of such deletions from studies of subtelomeric regions in natural populations. Subtelomeric regions have high levels of gene presence/absence polymorphism not seen in the adjacent euchromatin. At least some of this structural polymorphism is due to terminal deletions that were subsequently healed by transposition of HeT-A, as shown by early studies of lethal giant larvae(2) near the 2L tip (38) and a recent, more extensive study of the tip of 3L (39).
The loss of long segments of telomeres has been shown to be part of the regulation of telomere length in other organisms. The first evidence for such regulation came from studies of terminal rapid deletion in budding yeast (40). More recently, mammalian telomeres have been shown to use a similar mechanism (41, 42). Although the mechanism for generating terminal deletions may be different in Drosophila, the result, rapid regulation of telomere length is the same. For Drosophila, these deletions have a second important consequence; deletions remove decayed elements, allowing replacement by transposition-competent elements when the deleted telomere is regenerated by new transpositions. Deletion and rapid replacement would explain the lack of decayed elements found deep in telomere arrays. Replacement by new transpositions might also select against any nontelomeric retrotransposons that had managed to sneak into the telomere array.
HeT-A Arrays That Now Reside in Centromeric Heterochromatin Have Become Structurally Modified to Be Very Different from Het-A Arrays in Telomere Regions
Although the telomeric retrotransposons transpose only onto chromosome ends, in situ hybridization identified a large cluster of HeT-A DNA in the centromere region of the D. melanogaster Y chromosome (43). A similar cluster binding antibody to centromere-specific histone was found on the Y chromosomes of other members of the melanogaster species subgroup (44). Thus, telomeric HeT-A sequences appear to have been moved into the Y centromere before this species subgroup split >13 Mya. The Y chromosome is now metacentric in some of these species and telocentric in others, but despite this structural reorganization, the HeT-A sequence remains in the centromere region of every species. This conserved localization suggests that the HeT-A cluster has acquired some role at the centromere, possibly forming the kinetochore, affecting sister chromatid cohesion, or maintaining the heterochromatic environment. Mendez-Lago et al. (45) recently sequenced a D. melanogaster BAC that allowed them to characterize the molecular structure of this centromeric HeT-A cluster. That structure reveals dramatic changes from the structure of telomere arrays.
The Y chromosome BAC contained 159 kb of HeT-A DNA. Mendez-Lago et al. (45) concluded from their sequence that this DNA arose from a founder sequence that initially consisted of nine telomere retrotransposons in a typical telomeric head to tail array (Fig. 5). This founder sequence could have been either a Y chromosome telomere moved to the interior by an inversion or a segment of telomere from the Y or another chromosome that was inserted into the Y, which has a record of accepting sequence from other chromosomes (46). In either case, the founder was a typical telomere array of about 30 kb. Thus, the sequence of this BAC provides an unusual opportunity to compare a telomere array that has resided in centromeric heterochromatin for significant evolutionary time with telomere arrays that have remained on chromosome ends. We find that there are striking differences in the ways that the sequences have been maintained in the two regions.
Fig. 5.
Evolution of HeT-A sequences in the centromere region of the Y chromosome, deduced by Mendez-Lago et al. (45) (not to scale). The bottom diagram shows telomere sequence transposed into the Y chromosome: eight HeT-A elements (orange arrows) and one partial TART (#4; yellow arrow). Elements 1, 2, 3, and 5 are truncated HeT-As, and elements 6–9 are complete HeT-As. The top diagram shows 159 kb cloned in the sequenced BAC. The partial elements underwent complex amplifications to make up the 18HT satellite, which is partially represented by pentagons and black arrows on the left and is not further considered here. The end result of the several amplifications of the initially complete elements is shown on the right (numbering retained from ref. 45 to indicate origin of different parts of the sequence). Elements with two numbers result from amplifications of parts of two elements. Triangles, nontelomeric retrotransposons (copia, mdg1, diver, F, and 1731) that inserted at various times during the sequential amplifications of this DNA; green boxes, segment of autosomal region 42A transposed into element 8 and later duplicated. [Based on figure 7 in the work by Mendez-Lago et al. (45) and reproduced with permission from Oxford University Press.]
The Centromeric HeT-A Cluster Has Grown Extensively by Amplifications of Various Parts of the Sequence.
The founder sequence consisted of nine elements. Five of these, four HeT-As and one TART, were extremely 5′-truncated. These elements formed a 3.1 kb repeat that has been amplified to make up more than 100 kb of relatively homogeneous simple sequence repeats typical of the satellite DNAs that are abundant in pericentric heterochromatin. The other segment of the founder contained four complete HeT-As. This segment has undergone a series of head to tail amplifications of different regions of the array to yield 10 elements and another element truncated by cloning. The centromeric cluster has also grown by insertion of members of seven families of nontelomeric transposable elements (45). In contrast, neither amplifications nor insertion of nontelomeric transposable elements is seen in telomeric regions, which grow entirely by transpositions onto the chromosome ends. (There is one qualification to this statement; subfamilies of telomeric elements can differ by small insertions/deletions in both coding and untranslated regions. Some indels are repeats of adjacent sequence; the origin of others is not obvious. In coding regions, these indels do not introduce stop codons or alter the reading frame and they do not affect the A + C strand bias conserved throughout these elements. In all cases, they are found in multiple elements and therefore, do not compromise transposition of elements.)
Full-Length HeT-A Elements in the Centromeric Array Have Undergone Extensive Internal Deletion.
The centromeric sequences differ from telomeric sequences not only in their mechanism of sequence addition but also in their mechanism of sequence loss. Loss from elements in telomere arrays is exclusively from their 5′ ends, except for the small indels noted above. In contrast, each centromere element has several large internal deletions scattered through its sequence. There has been little rearrangement of the remaining sequence, most of which is collinear with the canonical HeT-A, with relatively few inversions and rearrangements. Surprisingly, the only regions that are conserved in every centromere element are the extreme 5′ and 3′ ends (23).
Many deletions in the centromere elements are shared with siblings derived from the same amplification. Thus, these 10 elements and the partial element have become a complex array of repeats. Because some amplification events apparently involved more than a single unit, higher order repeats arise.
As a result of both sequence loss and nucleotide changes, the centromeric HeT-A elements have lost much of their protein coding capacity. The longest ORFs in these elements range from 246 to 558 nt; in comparison, the shortest complete HeT-A gag gene is 2,766 nt (23). Whether any of these short ORFs in the centromeric DNA are expressed is an open question.
Telomere Elements Now in the Centromere Cluster Have Been Shaped into Complex Repeats Similar to Those That Characterize the Heterochromatic Centromere Regions in Multicellular Organisms.
Centromeres in multicellular organisms are determined epigenetically (47, 48); thus, it is not possible to identify centromeres by sequence alone. Nevertheless, this Y cluster is similar (in size, repeated sequence structure, and presence of transposable elements) to the only functionally characterized centromere in Drosophila, the X chromosome centromere (49), supporting the cytological evidence that this cluster is in some way involved in centromeric activity. It is not surprising that the Y chromosome cluster does not share sequences with the centromere of the X chromosome: these two chromosomes do not pair normally, and a difference in centromere sequences could well be either a cause or a result of this lack of meiotic pairing. Y-specific centromere sequences also have been documented for the mouse Y chromosome (50).
Conclusion
Drosophila telomeres provide a detailed picture of the interactions between a metazoan genome and retrotransposons in coevolving a robust mechanism to maintain dynamic chromosome ends. These interactions are not a one-way street. The retrotransposons have maintained their identity as retrotransposons, while acquiring other characteristics important for their roles at telomeres. The end result of this coevolution is that Drosophila telomeres share many, if not most, operational characteristics with telomeres maintained by telomerase in other organisms. This picture of the coevolution of telomeric retrotransposons with the genome is strengthened by the fate of those retrotransposons that, after being moved into the centromere region, produced a repetitive sequence that was shaped, probably passively, into the complex DNA repeats that typify metazoan centromere regions.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM50315 (to M.-L.P.).
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Telomerase and Retrotransposons: Reverse Transcriptases That Shaped Genomes” held September 29–30, 2010, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at www.nasonline.org/telomerase_and_retrotransposons.
This article is a PNAS Direct Submission. J.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100278108/-/DCSupplemental.
References
- 1.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Greider CW. Telomerase activity, cell proliferation, and cancer. Proc Natl Acad Sci USA. 1998;95:90–92. doi: 10.1073/pnas.95.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epel ES, et al. Accelerated telomere shortening in response to life stress. Proc Natl Acad Sci USA. 2004;101:17312–17315. doi: 10.1073/pnas.0407162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardue ML, DeBaryshe PG. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu Rev Genet. 2003;37:485–511. doi: 10.1146/annurev.genet.38.072902.093115. [DOI] [PubMed] [Google Scholar]
- 5.Pardue ML, DeBaryshe PG. Adapting to life at the end of the line. How Drosophila telomeric retrotransposons cope with their job. Mobile Genetic Elements. 2011;1:128–134. doi: 10.4161/mge.1.2.16914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 7.Biessmann H, et al. Addition of telomere-associated HeT DNA sequences “heals” broken chromosome ends in Drosophila. Cell. 1990;61:663–673. doi: 10.1016/0092-8674(90)90478-w. [DOI] [PubMed] [Google Scholar]
- 8.Biessmann H, et al. Frequent transpositions of Drosophila melanogaster HeT-A transposable elements to receding chromosome ends. EMBO J. 1992;11:4459–4469. doi: 10.1002/j.1460-2075.1992.tb05547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- 10.Kahn T, Savitsky M, Georgiev P. Attachment of HeT-A sequences to chromosomal termini in Drosophila melanogaster may occur by different mechanisms. Mol Cell Biol. 2000;20:7634–7642. doi: 10.1128/mcb.20.20.7634-7642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George JA, DeBaryshe PG, Traverse KL, Celniker SE, Pardue ML. Genomic organization of the Drosophila telomere retrotransposable elements. Genome Res. 2006;16:1231–1240. doi: 10.1101/gr.5348806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abad JP, et al. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol Biol Evol. 2004;21:1620–1624. doi: 10.1093/molbev/msh180. [DOI] [PubMed] [Google Scholar]
- 13.Casacuberta E, Pardue ML. HeT-A elements in Drosophila virilis: Retrotransposon telomeres are conserved across the Drosophila genus. Proc Natl Acad Sci USA. 2003;100:14091–14096. doi: 10.1073/pnas.1936193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casacuberta E, Pardue ML. Transposon telomeres are widely distributed in the Drosophila genus: TART elements in the virilis group. Proc Natl Acad Sci USA. 2003;100:3363–3368. doi: 10.1073/pnas.0230353100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shpiz S, et al. Characterization of Drosophila telomeric retroelement TAHRE: Transcription, transpositions, and RNAi-based regulation of expression. Mol Biol Evol. 2007;24:2535–2545. doi: 10.1093/molbev/msm205. [DOI] [PubMed] [Google Scholar]
- 16.Rashkova S, Karam SE, Kellum R, Pardue ML. Gag proteins of the two Drosophila telomeric retrotransposons are targeted to chromosome ends. J Cell Biol. 2002;159:397–402. doi: 10.1083/jcb.200205039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuller AM, Cook EG, Kelley KJ, Pardue M-L. Gag proteins of Drosophila telomeric retrotransposons: Collaborative targeting to chromosome ends. Genetics. 2010;184:629–636. doi: 10.1534/genetics.109.109744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rashkova S, Karam SE, Pardue ML. Element-specific localization of Drosophila retrotransposon Gag proteins occurs in both nucleus and cytoplasm. Proc Natl Acad Sci USA. 2002;99:3621–3626. doi: 10.1073/pnas.032071999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shpiz S, Kwon D, Rozovsky Y, Kalmykova A. rasiRNA pathway controls antisense expression of Drosophila telomeric retrotransposons in the nucleus. Nucleic Acids Res. 2009;37:268–278. doi: 10.1093/nar/gkn960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casacuberta E, Pardue ML. Coevolution of the telomeric retrotransposons across Drosophila species. Genetics. 2002;161:1113–1124. doi: 10.1093/genetics/161.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villasante A, et al. Drosophila telomeric retrotransposons derived from an ancestral element that was recruited to replace telomerase. Genome Res. 2007;17:1909–1918. doi: 10.1101/gr.6365107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeBaryshe PG, Pardue ML. Differential maintenance of DNA sequences in telomeric and centromeric heterochromatin. Genetics. 2011;187:51–60. doi: 10.1534/genetics.110.122994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traverse KL, George JA, Debaryshe PG, Pardue ML. Evolution of species-specific promoter-associated mechanisms for protecting chromosome ends by Drosophila Het-A telomeric transposons. Proc Natl Acad Sci USA. 2010;107:5064–5069. doi: 10.1073/pnas.1000612107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.George JA, Traverse KL, DeBaryshe PG, Kelley KJ, Pardue ML. Evolution of diverse mechanisms for protecting chromosome ends by Drosophila TART telomere retrotransposons. Proc Natl Acad Sci USA. 2010;107:21052–21057. doi: 10.1073/pnas.1015926107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danilevskaya ON, Arkhipova IR, Traverse KL, Pardue ML. Promoting in tandem: The promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell. 1997;88:647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- 27.Maxwell PH, Belote JM, Levis RW. Identification of multiple transcription initiation, polyadenylation, and splice sites in the Drosophila melanogaster TART family of telomeric retrotransposons. Nucleic Acids Res. 2006;34:5498–5507. doi: 10.1093/nar/gkl709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danilevskaya ON, Traverse KL, Hogan NC, DeBaryshe PG, Pardue ML. The two Drosophila telomeric transposable elements have very different patterns of transcription. Mol Cell Biol. 1999;19:873–881. doi: 10.1128/mcb.19.1.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voytas DF, Boeke JD. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, DC: American Society for Microbiology; 2002. pp. 631–662. [Google Scholar]
- 30.Sheen FM, Levis RW. Transposition of the LINE-like retrotransposon TART to Drosophila chromosome termini. Proc Natl Acad Sci USA. 1994;91:12510–12514. doi: 10.1073/pnas.91.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bibiłło A, Eickbush TH. The reverse transcriptase of the R2 non-LTR retrotransposon: Continuous synthesis of cDNA on non-continuous RNA templates. J Mol Biol. 2002;316:459–473. doi: 10.1006/jmbi.2001.5369. [DOI] [PubMed] [Google Scholar]
- 32.Bibillo A, Eickbush TH. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J Biol Chem. 2004;279:14945–14953. doi: 10.1074/jbc.M310450200. [DOI] [PubMed] [Google Scholar]
- 33.Babushok DV, Ostertag EM, Courtney CE, Choi JM, Kazazian HH., Jr L1 integration in a transgenic mouse model. Genome Res. 2006;16:240–250. doi: 10.1101/gr.4571606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurzynska-Kokorniak A, Jamburuthugoda VK, Bibillo A, Eickbush TH. DNA-directed DNA polymerase and strand displacement activity of the reverse transcriptase encoded by the R2 retrotransposon. J Mol Biol. 2007;374:322–333. doi: 10.1016/j.jmb.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biessmann H, Carter SB, Mason JM. Chromosome ends in Drosophila without telomeric DNA sequences. Proc Natl Acad Sci USA. 1990;87:1758–1761. doi: 10.1073/pnas.87.5.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levis RW. Viable deletions of a telomere from a Drosophila chromosome. Cell. 1989;58:791–801. doi: 10.1016/0092-8674(89)90112-8. [DOI] [PubMed] [Google Scholar]
- 37.Mikhailovsky S, Belenkaya T, Georgiev P. Broken chromosomal ends can be elongated by conversion in Drosophila melanogaster. Chromosoma. 1999;108:114–120. doi: 10.1007/s004120050358. [DOI] [PubMed] [Google Scholar]
- 38.Walter MF, et al. DNA organization and polymorphism of a wild-type Drosophila telomere region. Chromosoma. 1995;104:229–241. doi: 10.1007/BF00352254. [DOI] [PubMed] [Google Scholar]
- 39.Kern AD, Begun DJ. Recurrent deletion and gene presence/absence polymorphism: Telomere dynamics dominate evolution at the tip of 3L in Drosophila melanogaster and D. simulans. Genetics. 2008;179:1021–1027. doi: 10.1534/genetics.107.078345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lustig AJ. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat Rev Genet. 2003;4:916–923. doi: 10.1038/nrg1207. [DOI] [PubMed] [Google Scholar]
- 41.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 42.Pickett HA, Cesare AJ, Johnston RL, Neumann AA, Reddel RR. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 2009;28:799–809. doi: 10.1038/emboj.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agudo M, et al. Centromeres from telomeres? The centromeric region of the Y chromosome of Drosophila melanogaster contains a tandem array of telomeric HeT-A- and TART-related sequences. Nucleic Acids Res. 1999;27:3318–3324. doi: 10.1093/nar/27.16.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berloco M, Fanti L, Sheen F, Levis RW, Pimpinelli S. Heterochromatic distribution of HeT-A- and TART-like sequences in several Drosophila species. Cytogenet Genome Res. 2005;110:124–133. doi: 10.1159/000084944. [DOI] [PubMed] [Google Scholar]
- 45.Méndez-Lago M, et al. Novel sequencing strategy for repetitive DNA in a Drosophila BAC clone reveals that the centromeric region of the Y chromosome evolved from a telomere. Nucleic Acids Res. 2009;37:2264–2273. doi: 10.1093/nar/gkp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koerich LB, Wang X, Clark AG, Carvalho AB. Low conservation of gene content in the Drosophila Y chromosome. Nature. 2008;456:949–951. doi: 10.1038/nature07463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan BA, Blower MD, Karpen GH. Determining centromere identity: Cyclical stories and forking paths. Nat Rev Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- 48.Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 49.Sun X, Le HD, Wahlstrom JM, Karpen GH. Sequence analysis of a functional Drosophila centromere. Genome Res. 2003;13:182–194. doi: 10.1101/gr.681703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pertile MD, Graham AN, Choo KH, Kalitsis P. Rapid evolution of mouse Y centromere repeat DNA belies recent sequence stability. Genome Res. 2009;19:2202–2213. doi: 10.1101/gr.092080.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abad JP, et al. Genomic analysis of Drosophila melanogaster telomeres: Full-length copies of HeT-A and TART elements at telomeres. Mol Biol Evol. 2004;21:1613–1619. doi: 10.1093/molbev/msh174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.