Abstract
Reverse transcriptases (RTs) polymerize DNA on RNA templates. They fall into several structurally related but distinct classes and form an assemblage of RT-like enzymes that, in addition to RTs, also includes certain viral RNA-dependent RNA polymerases (RdRP) synthesizing RNA on RNA templates. It is generally believed that most RT-like enzymes originate from retrotransposons or viruses and have no specific function in the host cell, with telomerases being the only notable exception. Here we report on the discovery and properties of a unique class of RT-related cellular genes collectively named rvt. We present evidence that rvts are not components of retrotransposons or viruses, but single-copy genes with a characteristic domain structure that may contain introns in evolutionarily conserved positions, occur in syntenic regions, and evolve under purifying selection. These genes can be found in all major taxonomic groups including protists, fungi, animals, plants, and even bacteria, although they exhibit patchy phylogenetic distribution in each kingdom. We also show that the RVT protein purified from one of its natural hosts, Neurospora crassa, exists in a multimeric form and has the ability to polymerize NTPs as well as dNTPs in vitro, with a strong preference for NTPs, using Mn2+ as a cofactor. The existence of a previously unknown class of single-copy RT-related genes calls for reevaluation of the current views on evolution and functional roles of RNA-dependent polymerases in living cells.
DNA-dependent polymerases are essential for cellular function, as they mediate the flow of genetic information from DNA to RNA to proteins (1). In contrast, RNA-dependent polymerases have long been associated with replication of selfish and parasitic genetic elements, such as viruses or transposons. Although the discovery of reverse transcriptase (RT) challenged the concept of unidirectionality of the flow of genetic information, this reverse direction has been reserved for retroviruses, pararetroviruses (hepadna- and caulimoviruses), and other RT-containing multicopy entities such as non-LTR and LTR retrotransposons, group II introns, retrons, and retroplasmids, as well as occasional retro(pseudo)genes (2–4). Similarly, viral RNA-dependent RNA polymerases (RdRPs), enzymes structurally related to RTs, serve to replicate the genomes of viruses that use RNA as genetic material (5). These and certain other polymerases are unified by the architecture known as “right hand,” composed of the three subdomains called fingers, palm, and thumb (6). Like all polymerases, they use two-metal-ion catalysis for phosphoryl transfer reactions resulting in nucleotide addition.
In 1997, this diverse superfamily of enzymes was joined by the telomerase reverse transcriptase (TERT), a specialized RT that maintains the ends of eukaryotic linear chromosomes by addition of short G-rich repeated DNA sequences that are copied multiple times via reverse transcription of a specific region of the associated RNA template constituting part of the holoenzyme (7). Telomerases are unique in being single-copy eukaryotic RT genes that do not represent a component of any mobile element or virus. It has been argued that, in early eukaryotic evolution, telomerases may have either descended from domesticated retrotransposons or given rise to them (8, 9). In fact, TERTs were shown to be most closely related to RTs from Penelope-like retroelements (PLEs) (10–12). However, TERT genes so far remain the only example of single-copy RT-related eukaryotic genes with a defined cellular function. In this study, we identify and characterize the second major group of RT-related cellular genes.
Results
Identification and Characterization of rvt Genes in Bdelloid Rotifers.
We discovered rvt genes in the course of cloning and sequencing of telomeric regions from rotifers of the class Bdelloidea, small freshwater invertebrates that are best known for having evolved for tens of millions of years apparently without males and meiosis, for their resistance to desiccation and ionizing radiation, and for their ability to acquire foreign genes from diverse sources (11, 13–16). During sequencing of fosmids from the genomic library of the bdelloid rotifer Adineta vaga (family Adinetidae), we found a member of a previously unrecognized group of RT-like genes, which we named rvt because it contains an identifiable rvt conserved domain (reverse transcriptase; pfam00078:RVT_1). The A. vaga rvt did not fall into any of the known RT categories, such as retrons, retroplasmids, group II introns, telomerases, non-LTR retrotransposons, LTR retrotransposons, retroviruses, and pararetroviruses. Its single-copy status was established by Southern blot hybridization of genomic DNA and by exhaustive screening of the A. vaga genomic library (17). In this library, we found a colinear pair of rvt-containing fosmids with 4% overall divergence, consistent with the genome structure of bdelloid rotifers in which chromosomes occur as colinear allelic pairs with overall divergence up to 6% (17, 18). The divergence between members of the rvt pair is <1%, either at the nucleotide level (13/2,490 nt substitutions) or at the protein level (6/829 aa substitutions). Sequencing of rvt-containing fosmids revealed that rvt genes are located in a subtelomeric region rich in telomeric repeats, telomere-associated Athena retrotransposons, and foreign genes of apparently bacterial or fungal origin (Fig. S1A).
Moreover, we found rvt in four other species of bdelloid rotifers (Philodina roseola, Philodina acuticornis, and Macrotrachela quadricornifera from the family Philodinidae and Habrotrocha rosa from the family Habrotrochidae), using PCR and genomic library screens. On a contig from the P. roseola genomic library, rvt is located between two genes of apparently bacterial origin (Fig. S1B). There are two distinct lineages of rvt genes in bdelloids, A and B, which could originate from two independent acquisition events (Fig. S1C). However, although the A. vaga rvtA has a slightly higher GC content than neighboring genes, there is no detectable difference in GC content and codon usage between P. roseola rvtB and adjacent genes, indicating that if it was also acquired by lateral transfer, it took place a sufficiently long time ago for the differences to have ameliorated.
Structure and Distribution of rvt in Sequenced Genomes.
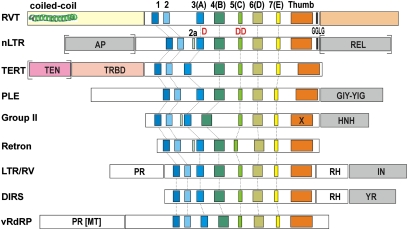
Comparison of rotifer rvt genes with their homologs in other kingdoms reveals their highly conserved overall structure, which deviates significantly from all presently known RT types (Fig. 1). A typical rvt ORF is 800–1,000 aa in length. The core RT domain, which contains RT motifs 1 through E (fingers and palm), plus the thumb subdomain, is framed at the N and C termini by well-conserved 300- and 200-aa extensions, respectively, which have no homology to known motifs other than the coiled-coil motif at the very N terminus. The two neighboring aspartates in the core motif C, which constitute part of the D, DD catalytic triad, are typically preceded by a noncanonical histidine residue. In agreement with the single-copy nature of rvt genes, their core RT domain is not associated with any domains resembling known endonucleases or integrases, which are usually responsible for intragenomic mobility. A distinctive structural feature of rvt genes is a large insertion loop separating motifs 1 and 2 from the rest of the core RT (70–100 aa and up to an additional 70 aa in the Mo lineage, see below; Fig. 1 and Fig. S2), which is enriched in acidic Asp and Glu residues and confers a net negative charge to the molecule, with an average isoelectric point of 5.5.
Fig. 1.
Domain structure of rvt and representative members of the RT-like sequence cluster (National Center for Biotechnology Information-Conserved Domains Database cl02808 superfamily). Shown are the conserved core RT motifs 1–7 and the adjacent N- and C-terminal domains. Associated endonuclease domains of various types (AP, REL, GIY-YIG, HNH, IN, and YR) are shown in gray. Other domains are abbreviated as follows: TEN, telomerase essential N-terminal; TRBD, telomerase RNA-binding domain; PR, protease; RH, RNase H; MT, methyltransferase; X, maturase. Domains indicated by brackets may or may not be present (e.g., non-LTR elements may contain either AP or REL endonuclease or both). Also shown are the positions of the catalytic D, DD triad and the GGLG motif shared between rvt and early-branching non-LTR retrotransposons. Not shown are RTs of pararetroviruses and copia-like LTR retrotransposons.
In blastp searches, rvt genes retrieve each other, but not other types of RTs (with occasional low-significance hits to non-LTR retrotransposons and group II introns). In a conserved domain (CD) search, many of them fit the profile RT_like_1 (cd01709: “an RT gene usually indicative of a mobile element such as a retrotransposon or retrovirus”) composed of 14 fungal sequences, although this profile lacks core RT motifs 1 and 2 as they are separated by the large loop.
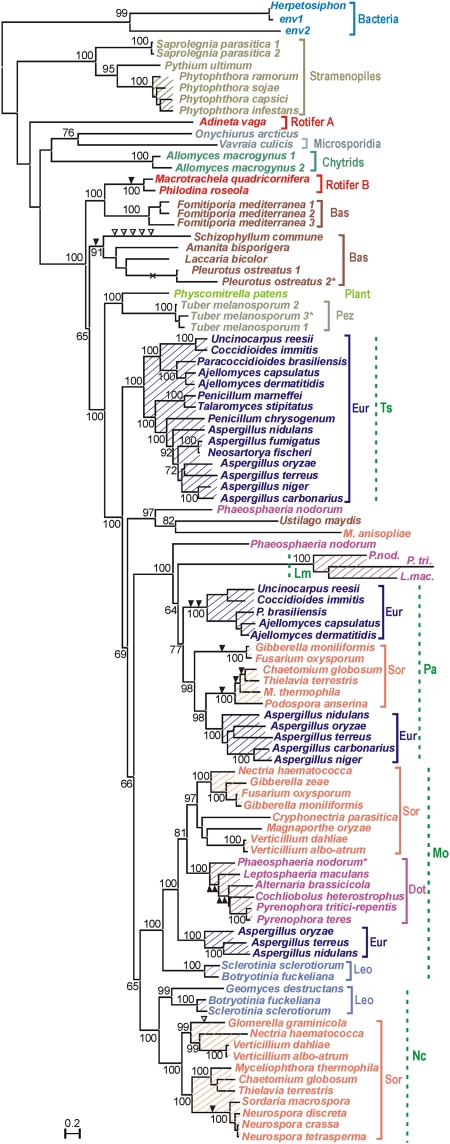
Although the distribution of rvt genes is rather patchy, they occur in all eukaryotic kingdoms: protists, fungi, animals, and plants. These genes are present in a highly diverse set of species with sequenced genomes: 60 fungi (not only euascomycetes and basidiomycetes, but also chytrids and microsporidia, the most basal fungal taxa); the moss Physcomitrella patens; six stramenopiles (heterokonts), including the genera Phytophthora, Saprolegnia, and Pythium; and a bacterium (Fig. 2 and Table S1). In EST databases, there are also two homologous fragments from an arthropod (Arctic springtail Onychiurus arcticus), which, however, exhibit some similarity to rvt from a microsporidian parasitizing on mosquitoes (Vavraia culicis). Of special interest is the existence of rvt genes in the sequenced genome of the filamentous gliding bacterium Herpetosiphon aurantiacus (Chloroflexi) and two uncultured environmental bacteria. Finding the same type of RT in both prokaryotes and eukaryotes is so far unprecedented. However, despite its basal position, this RT may have originated from a rare eukaryote-to prokaryote horizontal transfer, as rvt genes occasionally exhibit phylogenetic discordance (e.g., one of the bdelloid lineages groups with basidiomycetes; rvt from the moss Physcomitrella forms a clade with Tuber, a basal ascomycete; and rvt from the basidiomycete Ustilago is found within an ascomycete lineage) (Fig. 2).
Fig. 2.
A maximum-likelihood phylogram of 100 representative rvt protein-coding sequences (Dataset S1). Bootstrap support values exceeding 60% are indicated at the nodes. Hatched areas indicate synteny in rvt genomic environments. Shared intron positions are denoted by solid triangles, unique positions by open triangles, and putative intron loss by ×. Asterisks denote copies with several frameshifts or in-frame stop codons. Color-coded taxonomic groups are as follows: Sor, Sordariomycetes; Eur, Eurotiomycetes; Dot, Dothideomycetes; Leo, Leotiomycetes; Pez, Pezizomycetes; Bas, Basidiomycetes. Nc, Mo, Lm, Pa, and Ts denote rvt lineages (see text).
Phylogenetic analysis of rvt genes reveals that they underwent duplications early in ascomycete evolution, as well as sporadic loss of members from each duplicated lineage. The phylogram in Fig. 2 shows an early duplication event leading to formation of lineages Mo and Nc and an even earlier duplication leading to formation of lineages Pa and Ts (each lineage is denoted after the species that carries only this lineage: Nc, Neurospora crassa; Mo, Magnaporthe oryzae; Ts, Talaromyces stipitatus; Pa, Podospora anserine; Lm, Leptosphaeria maculans). In some ascomycetes, such as Phaeosphaeria nodorum (Dothideomycetes) or Aspergillus spp. (Eurotiomycetes), representatives of three or four lineages are present simultaneously, which indicates early divergence and subsequent loss of different lineage members from certain species and may also indicate partial redundancy of rvt function in different lineages. Members of the minor lineage L are not likely to possess catalytic activity, because they lack one of the conserved aspartates in the D, DD triad, and hence form a very long branch. We also observed several recent rvt losses: For instance, in Epichloe festuca and Coprinopsis cinerea, only an ∼100-aa fragment can be recognized, and in Neosartorya fischeri, the Ts lineage appears intact, whereas the Pa lineage is represented by a fragment with a large internal deletion spanning nearly 700 aa. In addition, in six cases one of the lineages contains in-frame stop codons and/or frameshifts, whereas another appears intact. At the same time, rvt is absent from the genomes of 15 sequenced euascomycetes and is not found in any of the 35 sequenced yeast genomes. Although incomplete coverage may occasionally account for such absence, the lack of rvt in three Arthroderma spp., four Trichophyton spp., and three Trichoderma spp. is more likely to indicate secondary loss.
Synteny in rvt Genomic Environments.
In each host species, rvt is present either as a single-copy gene or as a two- to three-member gene family. If these genes are not mobile elements, and two or three rvt copies are found in related genomes, synteny in their genomic environment in related species would clearly indicate that such copies were not derived from recent retrotransposition, but from ancient duplication. We investigated whether rvt genes in sequenced fungal genomes are located in chromosomal regions exhibiting appreciable degrees of synteny. Analysis of genomic contigs carrying the most divergent members of the Ts lineage clearly shows that they are located in syntenic regions (Fig. S3A). Although several inversions occurred within syntenic blocks, the overall synteny can be traced before separation of the orders Eurotiales and Onygenales, dated between 150 and 400 Mya depending on molecular clock calibration (19). Members of other rvt lineages exhibit similar degrees of synteny (Fig. 2), which is typically observed within a class, although occasionally cannot be traced to that level due to insufficient contig length. Preservation of synteny for tens of millions of years is typical of nuclear genes and is not characteristic of mobile genetic elements.
Selective Forces Acting on rvt Genes.
If rvt genes are not mobile elements, but have evolved to perform a certain function in the host, orthologous copies should exhibit evidence of selective pressure that acts to preserve that function. We asked whether rvt genes in related species evolve under purifying selection. To this end, we compared the rates of nonsynonymous and synonymous amino acid substitutions in pairs of orthologous rvt copies from fungal genomes. In each case, we observed a 4- to 10-fold excess of synonymous over nonsynonymous substitutions, which is strongly indicative of purifying selection (Fig. S3B). In bdelloid rotifers, rvt genes also evolve under purifying selection, as evidenced by comparison of rvtB in P. roseola and M. quadricornifera (Fig. S3B). The same pattern holds for all four species in the genus Phytophthora (Stramenopiles or Oomycetes). Interestingly, signatures of selection can be revealed even in comparisons between recent duplications: in the basidiomycete Fomitiporia mediterranea, three adjacent rvt copies display a fivefold excess of synonymous substitutions. Thus, rvt genes are under strong selective pressure in several unrelated groups of fungi, protists, and animals.
Intron Distribution.
Introns in nuclear genes are widespread, although in retroelements they are highly unusual, because the corresponding cDNA is synthesized on processed mRNA templates and is not expected to retain introns (but see ref. 10). Preservation of the exon–intron structure over evolutionarily long periods of time served as evidence for lack of retromobility of TERT genes, many of which contain introns (20). We therefore examined the patterns of intron occurrence in rvt genes. Whereas the coding regions of rvt from fungal lineages Lm and Ts do not contain introns, in lineages Pa, Mo, and Nc there are many copies that do (triangles in Fig. 2). Conserved introns can also be found in noncoding regions (see below), although these are more difficult to detect in the absence of adequate transcriptome coverage. Moreover, although a few introns appear to have arrived late, many intron positions are shared between different genera, indicating that these introns have been acquired relatively early in evolution (Fig. 2). Although alternative explanations, such as independent intron insertion into specific sites, cannot be ruled out, it appears likely that the presence of shared intron positions reflects common ancestry, as it is typically accompanied by synteny in rvt genomic environments.
Transcription Patterns.
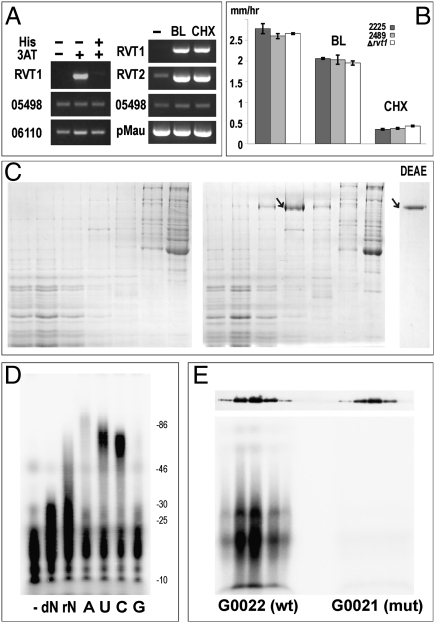
The first glimpse of rvt expression patterns may be obtained from BLAST searches of the available EST databases. EST analysis indicates that rvt genes are normally expressed at relatively low levels: Transcripts can be detected, on average, with a frequency of 1–2 per 10,000–15,000 sequenced EST tags, with a few exceptions (Table S1). Our RT-PCR experiments with N. crassa (mycelium) and A. vaga (whole animals) RNA show that in both species rvt is weakly transcribed, and 5′-RACE analysis demonstrates that the 5′-untranslated region (UTR) of the N. crassa rvt gene contains a 62-bp intron, which is conserved in Neurospora tetrasperma, Neurospora discreta, and Sordaria macrospora (Fig. S4A). Remarkably, inspection of transcriptional profiling data shows that in the N. crassa strain mutant for the gene encoding a global transcriptional regulator cross-pathway control-1 (CPC1), a yeast GCN4 ortholog, the level of rvt transcription undergoes a 47-fold increase under conditions of amino acid starvation induced by 3-aminotriazole (3-AT), an inhibitor of histidine biosynthesis (21). This increase is higher than that for any other gene of 10,526 N. crassa genes. We verified this result by RT-PCR and showed that addition of histidine restores normal expression levels (Fig. 3A). Thus, rvt expression in N. crassa appears to be under tight control, and under certain stress conditions its levels may rise dramatically.
Fig. 3.
Properties of N. crassa rvt. (A) Semiquantitative RT-PCR showing response of rvt to 3-AT in the cpc-1 mutant [Fungal Genetics Stock Center (FGSC) 4264], with and without histidine addition (Left), and response of rvt in the wild-type Mauriceville strain (FGSC 2225) to blasticidin (BL) and cycloheximide (CHX) (Right). Expression from genes NCU5498, NCU06110, and the Mauriceville plasmid (pMau) was monitored as a control. (B) Effect of blasticidin and cycloheximide on mycelial growth rates for Δrvt and two wild-type strains. (C) Sucrose gradient fractionation and DEAE chromatography purification of NcRVT from blasticidin-induced (Right) and noninduced (Left) Mauriceville strain. The position of the 102-kDa NcRVT protein in stained SDS/PAGE is indicated by an arrow. Odd-numbered fractions 17–29 are shown from left to right in each section of C; numbering begins from the bottom. The rightmost lane depicts the eluate from the DEAE column, which contains pure NcRVT protein. (D) Nucleotidyltransferase activity of NcRVT preincubated with [α-32P]dCTP in the presence of Mn2+. The reaction was chased with dNTP (dN), NTP (rN), ATP (A), UTP (U), CTP (C), or GTP (G). (E) Activity of His-tagged wild-type rvt (G0022) and the D529A mutant (G0021) in five consecutive peak fractions from the sucrose gradient. NcRVT was preincubated with [α-32P]dCTP in the presence of Mn2+, and the reactions were chased with CTP. (Upper) Comparison of the amount of wild-type and mutant protein by Western blotting of the same fractions probed with anti-His antibody.
Importantly, we found that rvt expression is strongly induced in the wild-type strain when protein synthesis is inhibited not only by the lack of histidine, but also by other means. Addition of antibiotics blasticidin S or cycloheximide to the exponentially growing N. crassa mycelium increases rvt expression by several orders of magnitude (Fig. 3A). These two antibiotics affect protein synthesis by different mechanisms, i.e., by blocking peptide bond formation and translocation steps, respectively (22, 23). Remarkably, the concentration of blasticidin needed for full induction (0.1 μg/mL) is at least an order of magnitude lower than that normally used to suppress protein synthesis (5–50 μg/mL) and does not strongly affect the growth rate, as does cycloheximide (Fig. 3B). Moreover, the increase in rvt transcription is paralleled by a similar increase in levels of the rvt-encoded protein (see below). We also find that several other genes, including an AAA+ ATPase, are strongly induced by addition of blasticidin at low concentration (Fig. S4B).
Protein Purification and in Vitro Activity Assays.
For initial characterization of RVT activity in vitro, we first sought to overexpress the full-length 890-aa NcRVT protein, tagged with an N-terminal 6xHis affinity tag, by introducing it into the rvt knockout N. crassa strain using homologous transformation (SI Materials and Methods). This strain displays no visible phenotype under laboratory conditions. Although the 6xHis tag did not bind to the Ni-affinity column as expected, it allowed us to track expression of the tagged protein in N. crassa extracts on SDS/polyacrylamide gels by Western blotting with the His-tag–specific antibody (Fig. S4E and Fig. 3E) and to adjust conditions under which it could be purified by ammonium sulfate fractionation and sucrose gradient centrifugation. Major improvement was achieved when we found that expression of the NcRVT protein is induced by several orders of magnitude under conditions that inhibit protein synthesis (Fig. 3A). Upon induction with 0.1 μg/mL blasticidin, untagged RVT protein could be purified to near homogeneity, as judged by SDS/PAGE that reveals a major 102-kDa band (Fig. 3C). In sucrose gradients, however, NcRVT sediments faster than all commercially available high molecular weight markers, and extrapolation of the calibration curve indicates that it likely forms a 1-MDa decameric complex (Fig. S5). The identity of untagged NcRVT protein was confirmed by mass spectrometry following tryptic digestion (Fig. S6).
We next sought to confirm the ability of NcRVT to act as a polymerase in vitro. The purified protein has the capacity to polymerize both NTPs and dNTPs, with a strong preference for NTPs, using Mn2+ as a cofactor. Purified NcRVT was first incubated with [α-32P]dCTP and then chased with an excess of cold NTPs or dNTPs (Fig. 3D). Addition of pyrimidine nucleotides typically results in longer extension products. Addition of Mg2+ instead of Mn2+ resulted in a significant decrease in length and intensity of extended products (Fig. S4C). Importantly, polymerization is completely abolished when one of the catalytic aspartates is replaced by alanine in the His-tagged version (Fig. 3E). Substitution of [α-32P]dCTP with [γ-32P]ATP does not result in appearance of visible extension products, indicating that de novo initiation is not occurring in vitro (Fig. S4C). All extension products have a minimum length of ∼10 nt, suggesting that [α-32P]CTP is being added to endogenous primers present in the purified NcRVT (Fig. S4 D and E). Addition of various exogenous primer/template combinations did not result in primer extension products, although endogenous RNA primers, represented by either natural 3′ termini or cleavage fragments of abundant cellular RNAs or short oligomers, were readily extended by the terminal nucleotidyltransferase activity upon incubation with an NTP and Mn2+, resulting in addition of up to 200-nt homopolymeric tails as verified by cloning and sequencing.
Relationship of rvt Genes to Other RT Sequences.
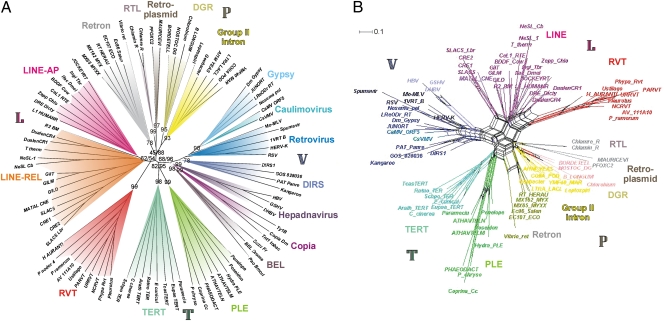
The ability of RVT to add NTPs and, to a lesser extent, dNTPs to 3′-OH termini places it apart from conventional RTs and closer to TERTs, which are known to exhibit RdRP and template-independent terminal deoxynucleotidyltransferase (TdT) activity in addition to RT activity (24, 25). The emergence of a unique type of RT-related genes raises questions about their relationship to other RT-like proteins. To clarify this relationship, we compiled an extended RT dataset (10) including representatives of every known group of RT-like proteins, including retrons, retroplasmids, group II introns, diversity-generating retroelements (DGR), retroviruses, pararetroviruses (hepadna- and caulimoviruses), LTR retrotransposons (gypsy, copia, BEL, and DIRS), non-LTR retrotransposons, PLEs, TERTs, and rvt genes. Using profile–profile searches and structure-based alignments, the core RT region was extended in both directions to span the entire ∼500-aa minimal functional RT, such as that found in retrons (Fig. S7). The rvt thumb domain aligns most readily with that of RT-derived Prp8 genes (26) and non-LTR retrotransposons (Fig. S2), ending with a highly conserved GGLG motif, which may serve as a flexible hinge connecting the thumb domain and a C-terminal extension. Similarities in secondary structure broadly subdivide RT-related sequences into four supergroups: A rather loose supergroup includes prokaryotic RTs (P); another unites virus-like entities such as LTR retrotransposons, retroviruses, and pararetroviruses (V); the third one consists of PLEs and TERTs (T); and the last one (L) shows affiliation between non-LTR retrotransposons and rvt genes, despite the lack of a conserved motif 2a in the latter.
Both traditional and phylogenetic network analyses (Fig. 4 A and B), which visualize uncertainties from conflicting phylogenetic signals, largely agree with these subdivisions, which can be further reinforced by additional synapomorphies, such as the presence of RNase H domain in virus-derived RTs or the presence of similarly structured N- or C-terminal extensions in other supergroups. Whereas all rvt genes are always grouped together with 100% support, other RT classes, such as non-LTR and LTR retrotransposons, are much more diverse. Overall, these findings demonstrate an ancient origin of rvt genes and point at independent evolutionary origins of different classes of retrotransposons, which were likely formed by fusion of ancestral RT domains with different types of endonucleases during early eukaryotic evolution.
Fig. 4.
Relation of rvt genes to other RT classes. (A) A phylogram indicating both minimum evolution (ME) and maximum-likelihood (ML) support values for the most basal branches and ME support for each colored clade in cases where it exceeds 70%. (B) A NeighborNet phylogenetic network built using protein ML distances under the WAG model with SplitsTree 4.10 (Materials and Methods).
Discussion
Forty years after the discovery of RTs in vertebrate retroviruses (2, 3), these enzymes have expanded into a large and diverse megafamily, members of which are usually assigned to various selfish genetic elements such as retrotransposons, retroviruses, or pararetroviruses. Also included in the RT-like sequence cluster are the RdRPs of dsRNA and positive-strand ssRNA viruses of the picorna-like superfamily (27). In all of the above cases, RNA-dependent synthesis serves the sole purpose of replication of selfish genetic elements of transposable or viral nature. A notable exception to this rule is telomerase, a specialized RT performing RNA-templated DNA synthesis at eukaryotic chromosome ends (28, 29).
The newest addition to the RT-like megafamily described herein, the rvt genes, bear a certain degree of resemblance to telomerases in being not multicopy, but single-copy genes, which evolve under purifying selection and do not exhibit varying localization in host genomes. It may therefore be argued that the role of RNA-dependent synthesis in eukaryotic cells is not restricted to maintenance of chromosome ends. Furthermore, rvt genes, like TERTs, have acquired additional domains that do not bear resemblance to endonuclease domains typically associated with core RT domains in retrotransposable elements and do not confer retromobility, but could be involved in primer, template, or protein–protein interactions.
One cannot help but wonder at a highly unusual pattern of rvt phylogenetic occurrence, which is not restricted to any specific domain of life, but nevertheless exhibits patchy distribution within each of the major kingdoms. So far, the most prominent rvt-carrying taxonomic group is the fungal kingdom, including 46 of 65 sequenced euasomycetes, 8 of 32 basidiomycetes, and 1 of 3 chytrids. This pattern may, to a certain extent, reflect the bias in genome sequencing: Ascomycete genomes are among the easiest to sequence and assemble, whereas basidiomycete genomes have not yet reached this level of coverage. The same logic may also be applied to other compact sequenced genomes, such as stramenopiles, half of which do carry rvt genes. However, several taxonomic groups exhibit clear-cut cases of rvt loss.
One of the most puzzling observations is the apparently universal occurrence of rvt genes in bdelloid rotifers: Of five bdelloid species investigated, at least one rvt lineage could be detected in each of them. Whereas rvt genes from lineage B are clearly related to rvt from basidiomycetes, those from lineage A appear more similar to oomycete rvt, pointing at independent introduction events. The fact that members of each lineage are found in genomic regions rich in genes of bacterial and fungal origin argues in favor of rvt introduction into bdelloids by horizontal transfers, which may have taken place before diversification of the major bdelloid families. So far, we have been unable to detect rvt in partially sequenced genomes of monogonont rotifers, which do not undergo frequent cycles of desiccation and rehydration and are apparently not subject to massive horizontal gene transfers. It is also unlikely that rvt genes exist in any of the chordate genomes, which have been extensively sampled (66 total, mostly mammalian). However, it is quite possible that additional rvt genes will be found in other invertebrate and plant genomes, which still remain undersampled and underassembled.
Preservation of synteny in the environment of fungal rvt lineages can be traced as far back as the taxonomic rank of a class (e.g., Eurotiomycetes), implying early divergence of rvt lineages by duplication. Intron distribution largely correlates with synteny. Whereas no intron position is conserved in all rvt lineages, several deep-branching lineages do share introns between all representatives. In bdelloid rotifers, intron insertion into lineage B occurred before divergence of the family Philodinidae, and both lineages were apparently introduced into the common bdelloid ancestor before divergence of the major bdelloid families, which took place tens of millions of years ago (13).
We also show that the purified RVT protein from N. crassa exerts the ability to polymerize NTPs, proving that it is a functional representative of the polymerase family. This activity depends on the presence of the highly conserved catalytic aspartate and could not be detected in the rvt knockout N. crassa strain, ruling out participation of an endogenous RdRP. Nevertheless, polymerization was observed only in the presence of Mn2+, which has less stringent coordination requirements than Mg2+ and allows use of suboptimal substrates and extra conformational flexibility (30). However, rvt sequence is much more similar to RTs than to viral RdRPs, and its conserved motifs A and B do not carry residues that are chemically similar and positionally equivalent to those responsible for choice of NTP over dNTP via formation of hydrogen bonds with the 2′- and 3′-ribose oxygens in RdRP (31). If rvt also exhibits preference for NTPs in vivo, the basis for such preference has to be different from that used by viral RdRPs, although other RTs can switch preferences rather easily (32). We hypothesize that the enzyme does not perform template-dependent synthesis indiscriminately and would likely require additional processing and/or interaction with cofactors for full activity. In particular, it may have to undergo dissociation from the multimeric state and/or conformational/structural changes leading to displacement or removal of the loop region. In vitro utilization of various endogenous RNA primers, including high-abundance host RNAs and shorter RNA oligomers, together with the inability to extend exogenously added primers and primer/template combinations, indicates that these RNAs could become captured by the enzyme either within the cell or during isolation, although it is not clear whether they represent natural primers and templates.
Although rvt is not an essential gene, its evolutionary conservation and strong signatures of purifying selection across a variety of species strongly argue in favor of its involvement in cellular processes, which are yet to be identified. Its expression appears to be tightly linked to protein metabolism: It is greatly enhanced upon inhibition of protein synthesis in a variety of ways, such as amino acid starvation and interference with peptide bond formation or ribosome translocation by antibiotic addition. Interestingly, we identified another N. crassa gene, AAA+ ATPase, which is strongly induced under the same conditions. These chaperone-like ATPases are associated with the assembly, operation, and disassembly of protein complexes. It remains to be seen whether these and other proteins functionally interact with rvt. Future studies will explore possible involvement of rvt genes in stress response, repair of radiation- and desiccation-induced DNA damage, and genome defense.
The unique phylogenetic position of rvt genes and their apparent relatedness to LINE-like RTs bear relevance to the long-standing question of whether cellular RTs, such as telomerases or rvt, could have originated from domesticated retrotransposons or represent originally cellular genes that could have given rise to RT-containing selfish genetic elements. In prokaryotes, single-copy DGR RTs can assist phages in tropism switching, conferring selective advantage in the arms race with a bacterial host (33); however, their function in bacterial genomes still remains a mystery. Telomerases are so far the only example of an essential RNA-dependent DNA polymerase gene in eukaryotes, as Prp8 genes have lost the catalytic aspartate responsible for polymerization. Our findings indicate that RT-related genes are a lot more common than previously thought and may have evolved different functions via acquisition of various N- and C-terminal extensions, and that retrotransposons could have originated several times in early evolution through association with different types of endonuclease domains that confer intragenomic mobility.
Materials and Methods
Library screening, subcloning, and sequencing were done as described in refs. 11 and 15. Detailed procedures for protein purification, PCR, activity assays, and bioinformatic analyses are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank M. Meselson for providing access to A. vaga and P. roseola genomic libraries and for stimulating discussions, D. Mark Welch for sharing unpublished rotifer sequences, and three anonymous reviewers for constructive comments. This work was supported by National Science Foundation Grant MCB-0821956 (to I.R.A.).
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Telomerase and Retrotransposons: Reverse Transcriptases that Shaped Genomes” held September 29–30, 2010, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at www.nasonline.org/telomerase_and_retrotransposons.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JN235987–JN235989).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100266108/-/DCSupplemental.
References
- 1.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 2.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 3.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 4.Eickbush TH, Malik H. Origins and evolution of retrotransposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM; 2002. pp. 1111–1144. [Google Scholar]
- 5.Ng KK, Arnold JJ, Cameron CE. Structure-function relationships among RNA-dependent RNA polymerases. Curr Top Microbiol Immunol. 2008;320:137–156. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steitz TA. DNA polymerases: Structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 7.Lingner J, et al. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 8.Eickbush TH. Telomerase and retrotransposons: Which came first? Science. 1997;277:911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura TM, Cech TR. Reversing time: Origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- 10.Arkhipova IR, Pyatkov KI, Meselson M, Evgen'ev MB. Retroelements containing introns in diverse invertebrate taxa. Nat Genet. 2003;33:123–124. doi: 10.1038/ng1074. [DOI] [PubMed] [Google Scholar]
- 11.Gladyshev EA, Arkhipova IR. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci USA. 2007;104:9352–9357. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang GS, et al. Phylogenetic profiles reveal evolutionary relationships within the “twilight zone” of sequence similarity. Proc Natl Acad Sci USA. 2008;105:13474–13479. doi: 10.1073/pnas.0803860105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mark Welch D, Meselson M. Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science. 2000;288:1211–1215. doi: 10.1126/science.288.5469.1211. [DOI] [PubMed] [Google Scholar]
- 14.Gladyshev E, Meselson M. Extreme resistance of bdelloid rotifers to ionizing radiation. Proc Natl Acad Sci USA. 2008;105:5139–5144. doi: 10.1073/pnas.0800966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320:1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- 16.Gladyshev EA, Arkhipova IR. Genome structure of bdelloid rotifers: Shaped by asexuality or desiccation? J Hered. 2010;101(Suppl 1):S85–S93. doi: 10.1093/jhered/esq008. [DOI] [PubMed] [Google Scholar]
- 17.Hur JH, Van Doninck K, Mandigo ML, Meselson M. Degenerate tetraploidy was established before bdelloid rotifer families diverged. Mol Biol Evol. 2009;26:375–383. doi: 10.1093/molbev/msn260. [DOI] [PubMed] [Google Scholar]
- 18.Mark Welch DB, Mark Welch JL, Meselson M. Evidence for degenerate tetraploidy in bdelloid rotifers. Proc Natl Acad Sci USA. 2008;105:5145–5149. doi: 10.1073/pnas.0800972105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor JW, Berbee ML. Dating divergences in the Fungal Tree of Life: review and new analyses. Mycologia. 2006;98:838–849. doi: 10.3852/mycologia.98.6.838. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura TM, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 21.Tian C, Kasuga T, Sachs MS, Glass NL. Transcriptional profiling of cross pathway control in Neurospora crassa and comparative analysis of the Gcn4 and CPC1 regulons. Eukaryot Cell. 2007;6:1018–1029. doi: 10.1128/EC.00078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen JL, Moore PB, Steitz TA. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- 23.Schneider-Poetsch T, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lue NF, et al. Telomerase can act as a template- and RNA-independent terminal transferase. Proc Natl Acad Sci USA. 2005;102:9778–9783. doi: 10.1073/pnas.0502252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maida Y, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dlakić M, Mushegian A. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA. 2011;17:799–808. doi: 10.1261/rna.2396011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koonin EV, Wolf YI, Nagasaki K, Dolja VV. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat Rev Microbiol. 2008;6:925–939. doi: 10.1038/nrmicro2030. [DOI] [PubMed] [Google Scholar]
- 28.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 29.Wyatt HD, West SC, Beattie TL. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010;38:5609–5622. doi: 10.1093/nar/gkq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: Two-Mg2+-ion catalysis and substrate specificity. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 31.Gong P, Peersen OB. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 2010;107:22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao G, Orlova M, Georgiadis MM, Hendrickson WA, Goff SP. Conferring RNA polymerase activity to a DNA polymerase: A single residue in reverse transcriptase controls substrate selection. Proc Natl Acad Sci USA. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medhekar B, Miller JF. Diversity-generating retroelements. Curr Opin Microbiol. 2007;10:388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.