Abstract
Cloning mammals by somatic cell nuclear transfer (SCNT) is highly inefficient. Most SCNT-generated embryos die after implantation because of unidentified, complex epigenetic errors in the process of postimplantation embryonic development. Here we identify the most upstream level of dysfunction leading to impaired development of clones by using RNAi against Xist, a gene responsible for X chromosome inactivation (XCI). A prior injection of Xist-specific siRNA into reconstructed oocytes efficiently corrected SCNT-specific aberrant Xist expression at the morula stage, but failed to do so thereafter at the blastocyst stage. However, we found that shortly after implantation, this aberrant XCI status in cloned embryos had been corrected autonomously in both embryonic and extraembryonic tissues, probably through a newly established XCI control for postimplantation embryos. Embryo transfer experiments revealed that siRNA-treated embryos showed 10 times higher survival than controls as early as embryonic day 5.5 and this high survival persisted until term, resulting in a remarkable improvement in cloning efficiency (12% vs. 1% in controls). Importantly, unlike control clones, these Xist-siRNA clones at birth showed only a limited dysregulation of their gene expression, indicating that correction of Xist expression in preimplantation embryos had a long-term effect on their postnatal normality. Thus, contrary to the general assumption, our results suggest that the fate of cloned embryos is determined almost exclusively before implantation by their XCI status. Furthermore, our strategy provides a promising breakthrough for mammalian SCNT cloning, because RNAi treatment of oocytes is readily applicable to most mammal species.
Keywords: somatic cell cloning, RNA FISH, DNA micro array, Tsix, trichostatin A
Somatic cell nuclear transfer (SCNT) is a unique technology for endowing the somatic cell genome with totipotency by genomic reprogramming (1). This technique has a distinct advantage over other similar biotechnologies, such as generating induced pluripotent stem cells, because a single somatic cell can give rise to a new individual with exactly the same genome as that of the donor cell (2, 3). Therefore, SCNT could become an invaluable tool with a broad range of applications including biological drug manufacturing, regenerative medicine, endangered species preservation, and commercial animal breeding. However, the success rate of cloning mammals by SCNT is low, predominantly because of the developmental arrest of cloned embryos after embryo transfer (1, 4).
Our recent in-depth gene expression analysis of cloned mouse blastocysts revealed ectopic expression of the noncoding RNA Xist—responsible for X chromosome inactivation (XCI) in female cells (5)—from the active X chromosome in both male and female clones (6). This ectopic expression of Xist had a global adverse effect on the gene expression of cloned embryos, because normalization of Xist expression by genetic knockout remarkably improved the expression patterns of not only X-linked genes but also a number of autosomal genes in the cloned embryos. As a result, many of the knockout-cloned embryos survived to term, resulting in an eight- to ninefold increase in overall cloning efficiency (6). These findings indicate that many, if not all, of the mechanisms that compromise the development of clones can be attributed to ectopic expression of the Xist gene in mice. The XIST gene is also known to be aberrantly expressed in bovine and pig SCNT-derived embryos and is implicated in their prenatal death (7–9). Thus, correction of X-linked gene expression is one of the most promising strategies now conceivable for improving the efficiency of mammalian SCNT.
Despite its potential efficacy, the Xist-knockout approach is impractical because it causes irreversible modification of the donor genome. Furthermore, gene targeting is still difficult for animal species other than mouse because of the difficulty in establishment of germ line-competent embryonic stem cells and the low cost-effectiveness of designed nucleases (10). In addition, it is still unclear from knockout studies how and when the ectopic expression of Xist affects the development of cloned embryos. The most probable solution to overcome these problems might be RNA interference (RNAi)-mediated gene knockdown, which works transiently and does not alter genomic DNA sequences (11–13). This strategy is easily applicable to many mammalian species and could provide us with clues to understanding the critical period of gene expression that might allow cloned embryos to survive to term. In this study, we examined whether RNAi-mediated knockdown against Xist could ameliorate the poor development of cloned mouse embryos by injecting a short interfering RNA (siRNA) into reconstructed embryos. As exact quantitative adjustment by conventional RNAi is technically difficult, we primarily focused on the silencing of Xist RNA in male cloned embryos with only a single X chromosome.
Results
Injected Xist-siRNA Repressed Ectopic Xist Expression at the Morula Stage but Not Later in Cloned Embryos.
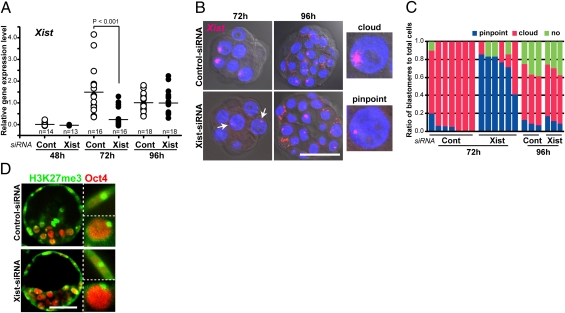
First, we examined the validity of an siRNA construct using parthenogenetic embryos, which express the Xist gene from the morula stage onward (14). We found that the siRNA treatment effectively decreased the Xist RNA level in the resulting blastocysts when injected into oocytes at 6 h postactivation (Fig. S1). As parthenogenetic embryos arrest their development at midgestation, we next injected the Xist-siRNA into in vitro fertilization (IVF)-derived embryos to determine whether the knockdown of Xist would affect normal postimplantation development. After embryo transfer, 44% of the siRNA-injected embryos reached term, showing a nearly 1:1 sex ratio, with no significant difference from that of control embryos (P > 0.05 by Fisher's exact test) (Table S1). This result indicates that Xist-siRNA did not affect the normal development of embryos of either sex. Therefore, in the next series of experiments, the same regimen was used for male SCNT embryos reconstructed using nuclei from neonatal Sertoli cells. Quantitative reverse transcription–PCR (RT-PCR) analyses revealed that Xist transcripts were first detected at the four-cell stage in cloned embryos and reached a steady level at the morula and blastocyst stages (Fig. 1A). When cloned embryos were pretreated with Xist-siRNA (hitherto termed Xist-siRNA embryos), the Xist level was reduced significantly compared with control siRNA embryos at the morula stage but not at the blastocyst stage (Fig. 1A). The effect of RNAi on ectopic Xist expression was further examined with Xist RNA fluorescent in situ hybridization (RNA FISH). At the morula stage, a clustered Xist RNA signal, referred to as a “cloud,” was found in the majority of nuclei in control siRNA-treated embryos (Fig. 1 B and C), as has been reported previously (6). By contrast, in Xist-siRNA–treated embryos, most blastomeres showed regionally restricted “pinpoint” signals, which presumably represented nascent Xist RNA on the X chromosome. This confirmed massive degradation of Xist induced by the injected Xist-siRNA (Fig. 1 B and C). At the blastocyst stage, the ectopic Xist expression spread as a cloud in some of the blastomeres of Xist-siRNA embryos, showing a pattern indistinguishable from that in control siRNA embryos (Fig. 1 B and C). We also investigated the XCI status of cloned blastocysts by staining for trimethylated histone H3 at lysine 27 (H3K27me3), a marker for repressed chromatin state in the inactive X chromosome (15). In blastocysts of both Xist-siRNA–treated and control embryos, the majority (>65%) of inner cell mass cells and trophectoderm cells were positive for punctate H3K27me3 staining (Fig. 1D), confirming the findings by RNA-FISH analysis (Fig. 1B). These results collectively indicate that the injected Xist-siRNA was effective up to about 72 h of embryo development but that the efficacy diminished thereafter.
Fig. 1.
Transient repression of ectopic Xist in SCNT-generated embryos by Xist-siRNA injection. (A) Quantitative RT-PCR of Xist in cloned embryos injected with control or Xist-siRNA and cultured for 48 h (four-cell), 72 h (morula), or 96 h (blastocyst). The expression levels of Xist were significantly decreased by Xist-siRNA at 72 h (P < 0.001 by Student's t test), whereas no significant difference was observed at 48 or 96 h. (B) RNA-FISH analyses of Xist in siRNA-injected cloned embryos. After 72 h in culture (morula), ectopic Xist expression was observed as a cloud pattern in most nuclei of the control cloned embryos, whereas it was detected as small pinpoint signals in Xist-siRNA–treated cloned embryos (arrows). At 96 h (blastocyst), the majority of nuclei showed cloud signals of Xist RNA in both groups. (Scale bar, 50 μm.) (C) The ratios of blastomeres classified according to the cloud or pinpoint expression patterns of Xist analyzed by RNA FISH. Each column represents a single embryo. It is apparent that the siRNA against Xist strongly repressed spreading of the Xist RNA over the X chromosome at 72 h but not at 96 h. (D) Immunostaining for H3K27me3 (green) and Oct4 (red) in control or Xist-siRNA–treated cloned embryos at 96 h. Strong punctate signals of H3K27me3 were observed in more than 65% of trophectoderm cells (Oct4-negative; Upper Inset) and inner cell mass cells (Oct4-positive; Lower Inset) in both siRNA-treated groups. (Scale bar, 50 μm.)
Injected Xist-siRNA Normalized Global Gene Expression of Cloned Embryos at the Morula Stage.
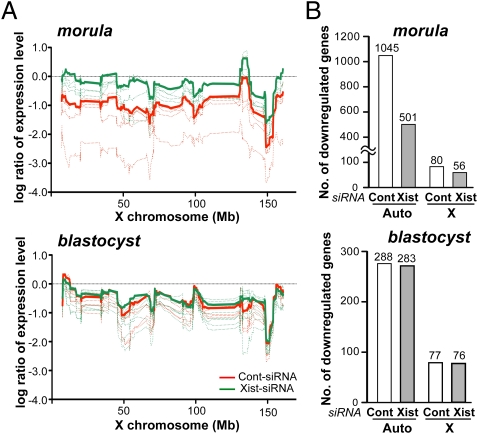
We then examined the effect of Xist-siRNA injection on the gene expression pattern of cloned embryos by DNA microarray analysis using single embryos (6). When relative expression levels of control cloned morulae were plotted on the positions of the X chromosome, X-linked genes were largely down-regulated over the entire chromosome (Fig. 2A) because of the high ectopic Xist expression at this stage, as shown in Fig. 1. By contrast, in cloned Xist-siRNA embryos, the relative expression levels were elevated considerably in a chromosome-wide manner, indicating that the repression of ectopic Xist had a profound effect on the correction of X-linked gene expression. Indeed, the number of down-regulated X-linked genes was reduced from 80 in control embryos to 56 in Xist-siRNA embryos (Fig. 2B). Interestingly, the number of down-regulated autosomal genes was also reduced by 52.1% (1,045 to 501) compared with control cloned embryos, indicating that the Xist-siRNA treatment had a genome-wide effect on the gene expression in cloned embryos (Fig. 2B).
Fig. 2.
Effects of Xist-siRNA injection on the global gene expression patterns of cloned embryos. (A) Relative expression levels of X-linked genes plotted on the X chromosome position in cloned embryos compared with IVF embryos at 72 h in culture (morula stage) and at 96 h (blastocyst stage) (n = 4 for each group). At the morula stage, X-linked genes were largely down-regulated over the entire chromosome in control cloned embryos (red), and the majority of them were increased in their expression levels by Xist-siRNA injection (green). By contrast, in blastocyst embryos, X-linked genes were down-regulated over the entire chromosome in both siRNA groups. Dotted lines represent a single embryo, and solid lines indicate their mean values. (B) Numbers of down-regulated genes (fold change >10) in cloned embryos compared with conventional IVF-generated embryos. At the morula stage, gene numbers were reduced by the injection of Xist-siRNA for genes not only on the X chromosome but also on autosomes, whereas they did not differ between the two groups at the blastocyst stage.
At the blastocyst stage, by contrast, cloned embryos from the two siRNA groups did not differ in their X-linked gene expression (Fig. 2A) or in the number of down-regulated genes (Fig. 2B), most likely because of the reappearance of ectopic Xist expression in Xist-siRNA embryos. However, despite such aberrant XCI status, there was a significant up-regulation of several developmentally important genes (e.g., Fras1, Car2, and Tet3) in Xist-siRNA blastocysts (Table S2). As implicated from the public database Mouse Genome Informatics (http://www.informatics.jax.org), these genes might play important roles in the cross-talk between the embryonic and extraembryonic parts of the conceptus that is a prerequisite for subsequent normal fetal development (16).
Xist-siRNA Increased the Birth Rates of Male Clones.
Next, we sought to explore whether the injection of reconstructed oocytes with Xist-siRNA could improve their subsequent development in vitro and in vivo. After a 96-h culture in vitro, 66% (53/80 embryos cleaved) of Xist-siRNA cloned embryos developed into blastocysts (Table S1). This efficiency was not statistically different from that of embryos injected with control siRNA (52%, 44/84; P > 0.05 by Fisher's exact test). The number of blastomeres per embryo did not differ between the groups either: 15 ± 2 (n = 7) and 16 ± 1 (n = 6) at the morula stage and 74 ± 4 (n = 3) and 73 ± 3 (n = 3) at the blastocyst stage, respectively. Thus, the Xist-siRNA injection seemed to have no effect on the development of cloned embryos during preimplantation stages.
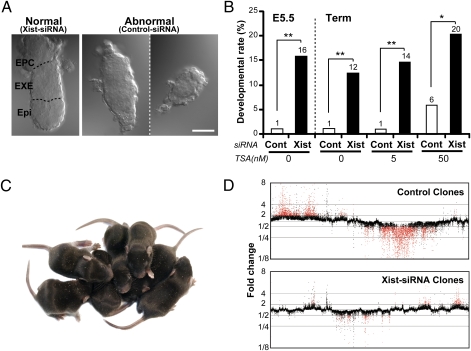
We then performed embryo transfer experiments to assess the postimplantation developmental ability of the siRNA-injected cloned embryos. At embryonic day (E)5.5, there was no effect of siRNA on the implantation rate, as determined by the endometrial decidual reaction and the embryo recovery rate (Table S1). However, the typical morphology of embryos retrieved from the implantation sites (Fig. 3A) was improved remarkably: 75% (9/12) of Xist-siRNA embryos showed normal morphology with distinct embryonic and extraembryonic compartments, whereas only 5% (1/20) of the control group showed normal morphology (Fig. 3B; P < 0.005 by Fisher's exact test). These rates corresponded to 16% and 1% of the embryos transferred in Xist-siRNA and control groups, respectively (Fig. 3B and Table S1). The remaining embryos showed some developmental retardation or various developmental defects in embryonic and/or extraembryonic regions (Fig. 3A), as reported for embryos cloned from embryonic stem cells (17). The distribution of Oct4-positive cells, which normally show exclusive localization to the epiblast, was also dysregulated (Fig. 4 A and C). We then allowed the recipient females to deliver young at term. The resultant birth rate was greatly improved: up to 12% of the Xist-knockdown embryos developed to normal-looking pups, attaining a more than 10-fold increase in birth rate compared with that of the control siRNA-treated group (1%; P < 0.005 by Fisher's exact test; Fig. 3B and Table S1). It is interesting to note that the rates of normal development examined at E5.5 and E19.5 (term) were not significantly different within the same group (16% vs. 12% in Xist-siRNA embryos and 1% vs. 1% in controls; P > 0.05 by Fisher's exact test). This clearly indicates that the developmental fate of cloned embryos is largely determined as early as E5.5, an early stage after implantation. Importantly, this birth rate could be further improved to about 20% by combining Xist-siRNA with 50 nM trichostatin A (TSA) treatment (Fig. 3 B and C and Table S1). TSA is a potent histone deacetylase inhibitor that is known to promote the in vitro and in vivo development of cloned embryos by relaxing the histone-related repression of the donor chromatin at the time of genomic reprogramming (18). This finding indicates that TSA treatment and Xist knockdown had synergistic effects on the development of cloned embryos. This is consistent with the finding that TSA had no corrective effect on the aberrant X chromosome inactivation in SCNT embryos (6). To our knowledge, this is the highest birth rate ever reported for mouse SCNT cloning since the first success by Wakayama et al. (19).
Fig. 3.
Effects of Xist-siRNA on the postimplantation development of cloned embryos. (A) Representative photomicrographs of siRNA-treated cloned embryos recovered at E5.5. Most Xist-siRNA embryos showed normal morphology (Left) with a distinct embryonic epiblast region (Epi) and extraembryonic ectoderm (EXE) or an ectoplacental cone (EPC), whereas most control siRNA-treated embryos showed abnormal morphology such as developmental retardation or ambiguous embryonic and extraembryonic regions (Center and Right). (Scale bar, 50 μm.) (B) The developmental rate of embryos assessed at E5.5 and at term. In some experiments, TSA was added at 5 or 50 nM in the culture medium. The numbers at the top of the bars indicate the rates of normal-shaped embryos (E5.5) and full-term births per embryos transferred. *P < 0.05, **P < 0.001 by Fisher's exact test. (C) A litter of cloned pups produced by SCNT from Sertoli cell nuclei, obtained following treatment with Xist-siRNA and 50 nM TSA. They were born at the best efficiency we observed in this series: Seven pups were born from 23 embryos transferred (30%) to a single recipient mother. (D) Gene expression profiles of livers in neonatal mice generated by SCNT with or without Xist-siRNA injection. The values indicate the mean fold changes from the control IVF level (=1). Red dots represent genes of which expression levels exceeded a twofold change in all of the individual clones. Xist-siRNA clones showed much fewer dysregulated genes compared with control clones. For data on each individual clone, see Fig. S2.
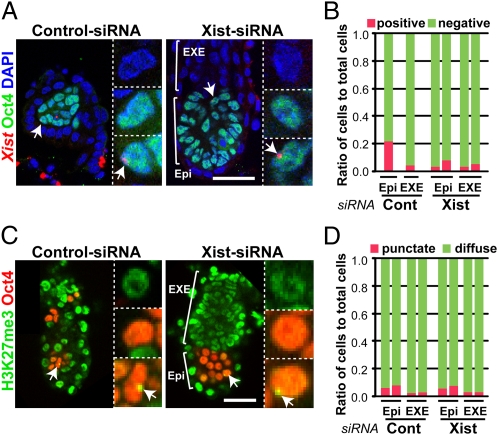
Fig. 4.
The XCI status of cloned embryos at E5.5. (A) RNA-FISH analyses of Xist (red) combined with immunostaining for Oct4 (green) in control or Xist-siRNA–injected cloned embryos. There were very few Xist signals in either Oct4-positive epiblast (Epi) (Middle Inset) or Oct4-negative extraembryonic ectoderm (EXE) (Top Inset) in either siRNA group. In these micrographs, only one Xist-positive cell was found within the epiblast (arrow; Bottom Inset, higher magnification). (Scale bar, 50 μm.) (B) Ratios of Xist-positive cells in the epiblast or extraembryonic ectoderm region analyzed by RNA FISH. Each column represents a single embryo. Only a few Xist-positive cells were observed in the regions irrespective of siRNA treatment. (C) Immunostaining for H3K27me3 (green), another marker for XCI, and Oct4 (red) in control or Xist-siRNA–treated cloned embryos. There were very few punctate H3K27me3 signals in either epiblast (Middle Inset) or extraembryonic ectoderm (Top Inset) in either siRNA group. The nuclei of some EXE cells showed relatively weak and diffuse staining for H3K27me3, which does not represent XCI. In these micrographs, only a single cell within the epiblast showed a punctate H3K27me3 signal (arrow; Bottom Inset). (Scale bar, 50 μm.) (D) Ratios of cells classified according to the punctate or diffuse patterns of H3K27me3 signals. Each column represents a single embryo. Consistent with the results of Xist RNA FISH, the punctate signals for H3K27me3 were rarely observed in cloned embryos in either group.
We also examined the gene expression patterns of the cloned neonates by microarray analysis, because we have previously shown that neonatal cloned mice had considerable diversity in their gene expression patterns despite their normal appearance and genetic identity (20). The number of transcripts commonly exhibiting a more than twofold up-regulation in the four Sertoli clone pups was 547, and the number exhibiting down-regulation was 1,752. In the siRNA-injected clone pups, the numbers of transcripts exhibiting more than twofold up- or down-regulation were 131 and 132, respectively. The gene expression profile was largely normalized, and the variation between clone individuals was also reduced in Xist-siRNA–injected clones (Fig. 3D and Fig. S2). These findings indicate that the SCNT-associated aberration in the gene expression patterns at birth can be corrected largely by injection of Xist-siRNA into early embryos. Thus, correction of XCI status in the preimplantation stage not only increased the cloning efficiency remarkably but also improved the epigenetic status of clones at birth.
Ectopic Xist Expression in Cloned Embryos Was Corrected Autonomously After Implantation in both Embryonic and Extraembryonic Regions.
That Xist-siRNA embryos surviving at E5.5 showed a distinctively normal phenotype might suggest the importance of the short peri-implantation period for the survival of cloned mouse embryos. However, this seemed contradictory, because XCI aberration was suggested by the presence of the Xist RNA cloud and H3K27me3 signal in cloned blastocysts, which corresponded to around E4.5 (Fig. 1 B and D). Therefore, we investigated the XCI status of cloned embryos at E5.5 by localization of Xist RNA and H3K27me3. In both the embryonic (epiblast) and extraembryonic regions of cloned embryos, there were only a few cells with a Xist RNA signal, irrespective of siRNA treatment (Fig. 4 A and B). Consistent with this, the positive punctate signals for H3K27me3 were also rarely observed in cloned embryos in both regions (Fig. 4 C and D). These findings clearly suggest that the ectopic Xist expression found in cloned blastocysts was corrected autonomously shortly after implantation in both embryonic and extraembryonic tissues.
Discussion
The present study provides unequivocal evidence that the ectopic expression of Xist in SCNT-generated embryos could be mitigated by conventional siRNA, leading to more than a 10-fold increase in the birth rate of male clones. Although in our previous study we showed a remarkable improvement in SCNT by genetic deletion of Xist (6), we thought initially that conventional knockdown would not bring about such a high cloning efficiency. The results obtained in this study were beyond our expectations in three respects.
First, it is generally accepted that nuclear long noncoding RNA molecules such as Xist RNA are less amenable to conventional siRNA-knockdown techniques because RNAi-mediated RNA degradation occurs predominantly in the cytoplasm (11–13). This study reports successful knockdown of Xist using a specific siRNA, giving a remarkable improvement in cloning efficiency. Although we do not know the exact kinetics of the injected siRNA in the cloned embryos, it is possible that siRNA degraded Xist RNA molecules in embryonic cells at the metaphase stage, as has been reported in an RNAi experiment targeting meiotic proteins in metaphase II oocytes (21). In human cell lines, exogenous siRNA was reported to degrade nuclear RNA, as evidenced by the presence of nuclear RNA-induced silencing complexes (22). By contrast, the Xist-independent down-regulation of X-linked genes in cloned embryos reported previously (6, 23) was not corrected by RNAi treatment (SI Text, Fig. S3, and Table S3), indicating that RNAi strategies are not always effective for improving SCNT outcomes.
Second, unlike a genetic knockout approach, siRNA-mediated knockdown has inherent temporal and quantitative limitations. Indeed, under our experimental conditions, the ectopic Xist expression was down-regulated only at the morula stage, and it reappeared at the blastocyst stage. The repression itself was also incomplete, and there was still a modest Xist expression at this stage (Fig. 1 A and B). However, this temporal and incomplete repression of Xist resulted in considerable reactivation not only of X-linked genes but also of many autosomal genes. We could not exclude the possibility that Xist-siRNA might have also affected Tsix, an antisense repressor RNA of Xist, and thereby indirectly altered Xist expression levels. Thus, despite such inherent limitations and technical ambiguity in the use of siRNA, knockdown of Xist had a remarkable genome-wide effect on the expression of its downstream genes and could rescue severe impairment associated with clone development.
Third, we initially assumed that the ectopic Xist expression in early-stage cloned embryos would be transmitted, at least in part, to postimplantation cloned embryos because the imprinted XCI in female embryos is normally maintained in the trophectoderm lineage (24, 25) and the same was true for female SCNT embryos (26). However, contrary to our expectation, the eventual disappearance of the XCI markers in male cloned conceptuses suggested that the clone-specific ectopic Xist expression was corrected autonomously in both embryonic and extraembryonic tissues (Fig. 4). Although the exact molecular basis remains to be determined, this was likely caused by the initiation of another XCI mechanism that works specifically in postimplantation embryos. This mechanism primarily depends on Tsix, which is the antisense transcript from the Xist locus and ensures maintenance of the active X chromosome by repressing Xist in cis (27). In extraembryonic tissues, Tsix plays a particularly important role in both male and female embryos, as revealed by gene knockout studies (27, 28). The same mechanism might have worked in male cloned embryos and erased the aberrant Xist expression that had been imposed in earlier stages. Indeed, our RNA-FISH analysis for Tsix confirmed that Tsix was transcribed in most (>80%) cells of both embryonic and extraembryonic tissues in all E5.5 cloned embryos we examined (Fig. S4).
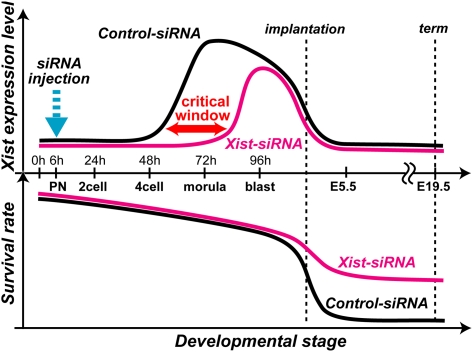
Taken together with the results we obtained, we can assume that the severely impaired development of SCNT mouse embryos might be largely attributed to an ectopic Xist expression from the active X chromosome that can be confined to a narrow stage before implantation. Thus, Xist knockdown (and knockout, too) probably exerted its greatest effect in rescuing cloned embryos during this very short period, leading to the remarkable improvements in cloning efficiency. A schematic representation of this assumption, together with the putative survival curve of clones, is shown in Fig. 5.
Fig. 5.
Schematic representation of siRNA-mediated Xist repression and its effect on the survival of male SCNT-generated embryos. (Upper) In control cloned embryos, ectopic Xist expression increased rapidly from 48 h through 72 h and maintained a high level until 96 h (black line; see also ref. 6). Injection of Xist-siRNA into pronuclear (PN)-stage cloned embryos at 6 h (dotted arrow) resulted in repression of the Xist level over 48–72 h, but this became ineffective at 96 h (magenta line). Thereafter, around implantation, ectopic Xist expression diminished spontaneously from all embryonic tissues in Xist-siRNA and control embryos. (Lower) Repression of Xist by siRNA had a remarkable effect on the survival of cloned embryos. As early as E5.5, more than 10 times as many Xist-siRNA embryos survived compared with control embryos, and this high survival persisted until term. These results suggest that the adverse effects of ectopic Xist expression in cloned embryos are confined to a short critical time window in the preimplantation period and that this can be reversed very efficiently by injecting Xist-siRNA into SCNT-derived embryos.
In addition to its scientific significance, our study also has an important implication for the practical applications of mammalian cloning. Because injecting siRNA into mammalian oocytes or embryos is technically feasible, our Xist-knockdown strategy should be readily applicable to other mammalian species such as bovines and pigs. Moreover, Xist knockdown had a synergistic effect with TSA, resulting in a birth rate of as high as 20% per embryo transferred. This combination would be a promising strategy for large-animal cloning, where histone deacetylase inhibitors have also proved effective for promoting the development of clones (29, 30). Unlike cloning mice, cloning domestic species by SCNT has often been associated with stillbirth or early neonatal death, which is also one of the major obstacles of SCNT that hamper its broad practical application (1). Although the underlying etiologies of these perinatal abnormalities are not yet understood, dysregulation of the XIST gene, in terms of its expression level and promoter hypomethylation, is associated with the neonatal death of clones in bovines and pigs (7–9). In mice, although most clones are alive and look normal at birth, there are also many aberrations in global gene expression patterns in their tissues (20). Interestingly, the present study with Xist-siRNA newborn clones revealed that they showed only a limited dysregulation of their gene expression (Fig. 3D), indicating that correction of Xist expression in preimplantation embryos might have long-term effects on their development and postnatal growth. If this is also the case with domestic species, we may expect that more clones would be born healthy at term following SCNT with a XIST-knockdown approach. This might realize the long-awaited breakthrough for SCNT technology in mammals, as it holds great potential in commercial animal breeding, producing gene-modified animals for medical and bioindustrial uses, and generating stem cells for regenerative medicine.
Methods
Animal Experiments.
All animal experiments described here were approved by the Animal Experimentation Committee at the RIKEN Tsukuba Institute and were performed in accordance with the committee's guiding principles.
Preparation of siRNAs and mRNAs.
Synthetic siRNA duplexes were designed by Stealth Designer (Life Technologies Japan) as follows. For Xist, 5′-AUAACAGUAAGUCUGAUAGAGGACA-3′ and 5′-UGUCCUCUAUCAGACUUACUGUUAU-3′; for negative control, 5′-UUACUCAUGUGUCAUAACACAGGUG-3′ and 5′-CACCUGUGUUAUGACACAUGAGUAA-3′. Primers MSS201293 and MSS293979 were used for G9a (Ehmt2) and Glp (Ehmt1), respectively. siRNA duplex mixtures were prepared as 200 μM stock solutions and stored at –80 °C until use. mRNAs for Jhdm2a were synthesized by a T7 mMESSAGE mMACHINE Kit (Ambion) and dissolved in water to a final concentration of 100 pg/mL.
Preparation of Donor Sertoli Cells.
Testicular masses of 1- to 9-d-old (C57BL/6 × DBA/2) F1 (BDF1) male mice were treated with 0.1 mg/mL collagenase (Sigma-Aldrich) for 30 min at 37 °C, followed by 0.2 mg/mL trypsin (Sigma-Aldrich) for 5 min at 37 °C (31). The cell suspension was washed with PBS containing 4 mg/mL BSA and used for nuclear transfer.
Nuclear Transfer.
Nuclear transfer was carried out as described (19, 31). Briefly, recipient oocytes were collected from BDF1 female mice by superovulation and enucleated in Hepes-buffered potassium modified simplex optimization medium (KSOM) containing 7.5 μg/mL cytochalasin B. Thereafter, the donor Sertoli cell nuclei were injected into enucleated oocytes using a Piezo-driven micromanipulator (PMM-150FU; Prime Tech). After 1 h of culture, the SCNT-treated oocytes were activated with 2.5 mM SrCl2 for 1 h. The reconstructed embryos were cultured in KSOM containing 5 μg/mL cytochalasin B for 5 h, followed by further culture in KSOM. In some experiments, trichostatin A (Sigma-Aldrich) was added to each medium (5 and 50 nM final concentrations) from the beginning of oocyte activation for 6 and 8 h in total, respectively.
siRNA and mRNA Injection.
Microinjection of siRNA or mRNA was carried out by using a Piezo-driven micropipette (Prime Tech). To examine the time schedule of siRNA injection, siRNAs (5 mM final concentration) were injected into BDF1 oocytes before and after parthenogenetic activation with SrCl2 (Fig. S1). SCNT-generated or IVF embryos were injected with siRNA 6–7 h after activation, corresponding to the pronuclear (one-cell) stage (Fig. 3). In some experiments, injections of mRNA (1–100 pg/mL) into oocytes were performed just before SCNT.
Embryo Transfer and Recovery.
Embryos at the two-cell stage were transferred into the oviducts of ICR strain recipient mice at day 1 of pseudopregnancy. On day 20, the recipient females were examined for the presence of term fetuses. Some recipients were killed on day 6, corresponding to E5.5, and the implanted embryos were recovered carefully from the uteri.
RNA Amplification and Microarray Analyses.
Total RNA was extracted with TRIzol (Invitrogen) from single embryos generated by nuclear transfer or IVF (32). They were subjected to linear amplification using TargetAmp Two-Round Aminoallyl-aRNA Amplification Kits (Epicentre Biotechnologies). Amplified RNA was labeled with Cy3 dye (GE Healthcare) and hybridized to a whole mouse genome oligo DNA microarray (4 × 44 K; Agilent Technologies) for 17 h at 65 °C. The scanned images of microarray slides were processed using Feature Extraction software (Agilent Technologies). All raw data were loaded into Gene Spring GX 11 (Agilent Technologies) and transformed by quantile normalization. For analyzing gene expression patterns on the X chromosome, signal intensities with mean values of more than 50 units in IVF embryos were chosen, and the mean values of 20 genes were plotted in accordance with their chromosomal locations.
For the examination of gene expression in neonatal mice, total RNA was also prepared using TRIzol (Invitrogen) and further purified using an RNeasy Mini Kit (Qiagen). The probe for hybridization was synthesized according to the manufacturer's protocol (Agilent Technologies). Data were analyzed using the R package (http://www.r-project.org/) with Bioconductor (http://www.bioconductor.org/) and Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm).
Quantitative RT-PCR.
cDNAs of single embryos were synthesized with Cell to cDNAII Kits (Ambion). Quantitative PCR was performed using QuantiTect SYBR Green PCR Kits (Qiagen) and the Prism 7900HT System (Applied Biosystems). All PCR runs were performed at an annealing temperature of 60 °C for 50 cycles. The primer sequences were as follows. For Xist, 5′-GTCAGCAAGAGCCTTGAATTG-3′ and 5′-TTTGCTGAGTCTTGAGGAGAATC-3′; for Gapdh, 5′-CAACAGCAACTCCCACTCTTC-3′ and 5′-CCTGTTGCTGTAGCCGTATTC-3′.
Immunofluorescence.
Embryos were fixed with 4% paraformaldehyde overnight at 4 °C. After permeabilization with PBST (0.5% Triton X-100 in PBS), they were incubated in a mixture of rabbit anti-H3K27me3 antibody (1:100 dilution; Millipore) and goat anti-Oct4 antibody (1:100 dilution; Santa Cruz Biotechnology) overnight at 4 °C. Immunostaining was revealed with Alexa Fluor-488- and -546-conjugated secondary antibodies (Invitrogen) and observed using a confocal scanning laser microscope (Digital Eclipse C1; Nikon).
RNA FISH.
A probe to detect Xist RNA was prepared by nick translation with Cy3-dCTP (GE Healthcare) from a Xist genomic clone encompassing a 7.5-kb fragment of exon 1. An antisense single-strand DNA probe for detecting Tsix RNA was labeled with Cy3-dCTP by random-primed reverse transcription from in vitro transcribed Tsix RNA. A plasmid containing a 9.2-kb Tsix genomic fragment encompassing exons 2 and 3 of Tsix was used as a template for synthesizing sense-strand Tsix RNA. Embryos were incubated in PBST for 10 s on ice and fixed with 4% paraformaldehyde for 10 min at room temperature. Hybridization was carried out at 37 °C overnight. The nuclei of embryos were stained with TO-PRO-3 (Invitrogen). In some experiments, immunostaining against Oct4, as described above, was followed by Xist RNA FISH.
Statistical Analysis.
Developmental rates of embryos were compared between groups using Fisher's exact test. The relative transcription levels determined by quantitative RT-PCR were analyzed by Student's t test for comparing group means. The microarray datasets were analyzed using fold-change analyses (cutoff >10) or one-way ANOVA followed by Tukey's post hoc test, and P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This research was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology (Japan) (to S.M., K.I., and A.O.), Joint Usage/Research Program of Medical Research Institute Tokyo Medical and Dental University (T.K. and A.O.), and the Novartis Foundation (Japan) for the Promotion of Science (to K.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE33208).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112664108/-/DCSupplemental.
References
- 1.Meissner A, Jaenisch R. Mammalian nuclear transfer. Dev Dyn. 2006;235:2460–2469. doi: 10.1002/dvdy.20915. [DOI] [PubMed] [Google Scholar]
- 2.Gurdon J, Murdoch A. Nuclear transfer and iPS may work best together. Cell Stem Cell. 2008;2(2):135–138. doi: 10.1016/j.stem.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thuan NV, Kishigami S, Wakayama T. How to improve the success rate of mouse cloning technology. J Reprod Dev. 2010;56(1):20–30. doi: 10.1262/jrd.09-221a. [DOI] [PubMed] [Google Scholar]
- 5.Okamoto I, Heard E. Lessons from comparative analysis of X-chromosome inactivation in mammals. Chromosome Res. 2009;17:659–669. doi: 10.1007/s10577-009-9057-7. [DOI] [PubMed] [Google Scholar]
- 6.Inoue K, et al. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science. 2010;330:496–499. doi: 10.1126/science.1194174. [DOI] [PubMed] [Google Scholar]
- 7.Xue F, et al. Aberrant patterns of X chromosome inactivation in bovine clones. Nat Genet. 2002;31:216–220. doi: 10.1038/ng900. [DOI] [PubMed] [Google Scholar]
- 8.Jiang L, et al. Expression of X-linked genes in deceased neonates and surviving cloned female piglets. Mol Reprod Dev. 2008;75:265–273. doi: 10.1002/mrd.20758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su JM, et al. Expression and methylation status of imprinted genes in placentas of deceased and live cloned transgenic calves. Theriogenology. 2011;75:1346–1359. doi: 10.1016/j.theriogenology.2010.11.045. [DOI] [PubMed] [Google Scholar]
- 10.Gama Sosa MA, De Gasperi R, Elder GA. Animal transgenesis: An overview. Brain Struct Funct. 2010;214(2-3):91–109. doi: 10.1007/s00429-009-0230-8. [DOI] [PubMed] [Google Scholar]
- 11.Scherr M, Eder M. Gene silencing by small regulatory RNAs in mammalian cells. Cell Cycle. 2007;6:444–449. doi: 10.4161/cc.6.4.3807. [DOI] [PubMed] [Google Scholar]
- 12.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 13.Ketting RF. The many faces of RNAi. Dev Cell. 2011;20(2):148–161. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Nesterova TB, Barton SC, Surani MA, Brockdorff N. Loss of Xist imprinting in diploid parthenogenetic preimplantation embryos. Dev Biol. 2001;235:343–350. doi: 10.1006/dbio.2001.0295. [DOI] [PubMed] [Google Scholar]
- 15.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300(5616):131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 16.Ang SL, Constam DB. A gene network establishing polarity in the early mouse embryo. Semin Cell Dev Biol. 2004;15:555–561. doi: 10.1016/j.semcdb.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Jouneau A, et al. Developmental abnormalities of NT mouse embryos appear early after implantation. Development. 2006;133:1597–1607. doi: 10.1242/dev.02317. [DOI] [PubMed] [Google Scholar]
- 18.Kishigami S, et al. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem Biophys Res Commun. 2006;340(1):183–189. doi: 10.1016/j.bbrc.2005.11.164. [DOI] [PubMed] [Google Scholar]
- 19.Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 20.Kohda T, et al. Variation in gene expression and aberrantly regulated chromosome regions in cloned mice. Biol Reprod. 2005;73:1302–1311. doi: 10.1095/biolreprod.105.044958. [DOI] [PubMed] [Google Scholar]
- 21.Amanai M, Shoji S, Yoshida N, Brahmajosyula M, Perry AC. Injection of mammalian metaphase II oocytes with short interfering RNAs to dissect meiotic and early mitotic events. Biol Reprod. 2006;75:891–898. doi: 10.1095/biolreprod.106.054213. [DOI] [PubMed] [Google Scholar]
- 22.Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat Struct Mol Biol. 2005;12(2):133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 23.Fukuda A, et al. Identification of inappropriately reprogrammed genes by large-scale transcriptome analysis of individual cloned mouse blastocysts. PLoS One. 2010;5:e11274. doi: 10.1371/journal.pone.0011274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huynh KD, Lee JT. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature. 2003;426:857–862. doi: 10.1038/nature02222. [DOI] [PubMed] [Google Scholar]
- 25.Sado T, Ferguson-Smith AC. Imprinted X inactivation and reprogramming in the preimplantation mouse embryo. Hum Mol Genet. 2005;14(Spec No 1):R59–R64. doi: 10.1093/hmg/ddi117. [DOI] [PubMed] [Google Scholar]
- 26.Eggan K, et al. X-chromosome inactivation in cloned mouse embryos. Science. 2000;290:1578–1581. doi: 10.1126/science.290.5496.1578. [DOI] [PubMed] [Google Scholar]
- 27.Sado T, Wang Z, Sasaki H, Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128:1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- 28.Ohhata T, Hoki Y, Sasaki H, Sado T. Tsix-deficient X chromosome does not undergo inactivation in the embryonic lineage in males: Implications for Tsix-independent silencing of Xist. Cytogenet Genome Res. 2006;113:345–349. doi: 10.1159/000090851. [DOI] [PubMed] [Google Scholar]
- 29.Wang YS, et al. Production of cloned calves by combination treatment of both donor cells and early cloned embryos with 5-aza-2/-deoxycytidine and trichostatin A. Theriogenology. 2011;75:819–825. doi: 10.1016/j.theriogenology.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 30.Zhao J, et al. Significant improvement in cloning efficiency of an inbred miniature pig by histone deacetylase inhibitor treatment after somatic cell nuclear transfer. Biol Reprod. 2009;81:525–530. doi: 10.1095/biolreprod.109.077016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogura A, et al. Production of male cloned mice from fresh, cultured, and cryopreserved immature Sertoli cells. Biol Reprod. 2000;62:1579–1584. doi: 10.1095/biolreprod62.6.1579. [DOI] [PubMed] [Google Scholar]
- 32.Mochida K, et al. Birth of mice after in vitro fertilization using C57BL/6 sperm transported within epididymides at refrigerated temperatures. Theriogenology. 2005;64(1):135–143. doi: 10.1016/j.theriogenology.2004.11.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.