Abstract
Although well defined in bacterial systems, the molecular mechanisms underlying ribosome recycling in eukaryotic cells have only begun to be explored. Recent studies have proposed a direct role for eukaryotic termination factors eRF1 and eRF3 (and the related factors Dom34 and Hbs1) in downstream recycling processes; however, our understanding of the connection between termination and recycling in eukaryotes is limited. Here, using an in vitro reconstituted yeast translation system, we identify a key role for the multifunctional ABC-family protein Rli1 in stimulating both eRF1-mediated termination and ribosome recycling in yeast. Through subsequent kinetic analysis, we uncover a network of regulatory features that provides mechanistic insight into how the events of termination and recycling are obligately ordered. These results establish a model in which eukaryotic termination and recycling are not clearly demarcated events, as they are in bacteria, but coupled stages of the same release-factor mediated process.
Keywords: peptide release, ABCE1, Pelota
The translation of messenger RNA into protein is traditionally thought to consist of four discrete steps: initiation, elongation, termination, and recycling (1, 2). Recycling, the final stage of translation, involves subunit dissociation and recovery of translational components (e.g., mRNA, tRNA, etc.) for reuse in subsequent rounds of translation. In bacteria, recycling depends on a specialized ribosome recycling factor (RRF) and elongation factor G, which together separate subunits in a GTP-dependent manner (3, 4). These factors selectively act on posttermination complexes following the facilitated removal of class 1 release factors (RF1 or RF2) by the class 2 release factor GTPase (RF3) (5). The departure of the termination factors ensures that ribosome recycling does not commence until after completion of peptide release. Separated subunits are subsequently bound by various initiation factors, including IF3, which prevent reassociation of subunits, allowing for the dissociation of mRNA and tRNA species and preparing subunits for the next round of initiation (3).
Neither RRF nor RF3 are conserved outside of bacteria, suggesting that this mode of recycling is also not conserved. Rather, a highly conserved ABC-family ATPase, ABCE1, was recently implicated in ribosome recycling in both eukaryotic and archaeal systems (6, 7). ABCE1 (or Rli1 in yeast) is a cytosolic ABC-family ATPase containing two NTP binding domains and a conserved N-terminal [4Fe-4S] domain that is required for its function (7–10). Characteristic of this family of ATPases, Rli1 is thought to convert the chemical energy of ATP hydrolysis into a mechanical tweezer-like motion (11). It is highly conserved throughout eukaryotes and archaea (12, 13) and is essential in all organisms tested (14–16). Consistent with this, Rli1 has been implicated in several essential, conserved cellular processes including ribosome maturation, translation initiation, and translation termination, with additional roles in RNAse L inhibition and HIV capsule assembly in mammals (16–24).
Biochemical studies have also suggested a direct role for eukaryotic release factors (eRF1 and eRF3) and related proteins (Dom34 and Hbs1) in recycling that is independent of peptide release (25, 26). Dom34 and Hbs1, originally identified in the No-Go Decay pathway (27), share significant structural similarity with the canonical eukaryotic release factors (28–31), but lack the residues necessary for both stop codon recognition and hydrolysis of peptidyl-tRNA (32). Like Rli1, eukaryotic release (eRF1, eRF3) and release-like (Dom34, Hbs1) factors are highly conserved from archaea to metazoans, albeit with the exception that the translational GTPases eRF3 and Hbs1 appear to be functionally replaced by the related GTPase aEF1α in archaea (33, 34).
Rli1 interacts with release factors both in vitro and in vivo (6, 20), consistent with the finding that eRF1 is necessary for Rli1-mediated recycling of posttermination ribosomes (6). Furthermore, nonsense suppression assays have shown that overexpression of Rli1 is able to compensate for the inefficient termination caused by certain release-factor mutations, although it is unknown whether this is due to the recycling activity of Rli1 or to a distinct activity (20). Although release factors act only in the context of a stop codon, several studies indicated that Pelota and ABCE1 (the human Dom34 and Rli1 orthologs, respectively) are involved in splitting empty 80S ribosomes or ribosomes stalled at the 3′ end of mRNAs (25, 33, 35). However, our own earlier work in yeast indicated that the Dom34∶Hbs1 complex is capable of splitting subunits without Rli1 (26). As such, there remain a number of important questions concerning the significance of a Dom34∶Hbs1 or eRF1∶eRF3 interaction with Rli1 in yeast.
The direct involvement of eukaryotic release and release-like factors in recycling suggests immediate differences from the bacterial system. Our in vitro reconstituted yeast translation system and recombinant Rli1 protein provides us with a means to significantly advance our mechanistic understanding of eukaryotic recycling events through rigorous kinetic analysis. Moreover, by exploiting subtle differences in eRF1 and Dom34 function, we are better able to define the roles of these factors, and Rli1, in the final stages of translation. Surprisingly, we observe discrete contributions of Rli1 to both termination and ribosome recycling. This process initiates with either stop codon recognition and peptide release (for eRF1∶eRF3) or mRNA length selection (for Dom34∶Hbs1) and is followed by subunit dissociation catalyzed by either set of factors in concert with Rli1. We find that the sequence of these events is gated by sequential GTP (on eRF3 or Hbs1) and ATP (on Rli1) hydrolysis events. These results highlight a defining connection between eukaryotic termination and recycling that differs fundamentally from the independently evolved and distinct processes in bacteria.
Results
Rli1 Interacts with eRF1 and Dom34 in Vivo.
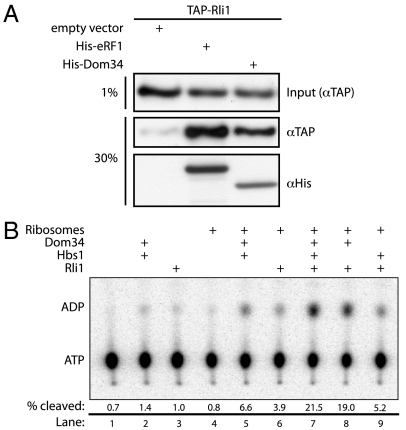
Recent studies in a mammalian system demonstrated that the multifunctional ATPase ABCE1 functionally interacts with Pelota (25). We asked whether yeast Rli1 similarly interacts with Dom34 using the previously documented eRF1∶Rli1 interaction as a positive control (20). This analysis was performed with an affinity pull-down in a TAP-tagged Rli1 yeast strain where eRF1-His6 and Dom34-His6 were overexpressed. TAP-tagged Rli1 associates with both eRF1 [positive control (6, 20)] and Dom34 at levels above those seen with an empty vector control (Fig. 1A). These preliminary results suggested that Rli1 interacts, either directly or indirectly, with both eRF1 and Dom34 in yeast.
Fig. 1.
Dom34 and Rli1 interact in vivo and in vitro. (A) Interaction of Dom34 and Rli1 in extract. TAP-tagged Rli1 selectively associates with His6-tagged eRF1 or Dom34 in an Ni-NTA resin pull-down from yeast lysate. Input lysates were derived from an Rli1-TAP strain expressing either eRF1-His6, Dom34-His6, or an empty vector (pYES2) control. Both input and pelleted protein were analyzed via Western blot analysis using anti-TAP and anti-His6 antibodies. (B) Rli1 interacts productively with Dom34 and ribosomes. Multiple turnover ATPase activity of Rli1 was monitored via thin-layer chromatographic analysis of 32P-ATP hydrolysis in the presence of various factors. The fraction of ATP hydrolyzed after 15 min was quantitated and is indicated under each representative lane.
Purification of Active Recombinant Rli1.
Interested in better defining the molecular interactions between Rli1, translational factors, and the ribosome, we purified recombinant Rli1-His6 from Saccharomyces cerevisiae for use in our in vitro reconstituted yeast translation system (Fig. S1). A UV-visible absorbance scan of purified Rli1 exhibits a pronounced shoulder at approximately 390 nm, characteristic of [4Fe-4S] cluster containing proteins (36) (Fig. S2). To evaluate activity, we conducted a multiple turnover ATPase assay in which Rli1 and excess nucleotide were incubated with Dom34, Hbs1, or empty 80S ribosomes. We observed high levels of ATPase activity only when Rli1 was incubated with Dom34 and 80S ribosomes; little additional activity was observed when Hbs1 was added to this mixture (Fig. 1B and Fig. S3). These data establish that the Rli1 preparation from S. cerevisiae yielded functional protein that appears to recognize features of the ribosome bound by specific translational factors.
Reconstitution of Rli1-Mediated Recycling in Yeast.
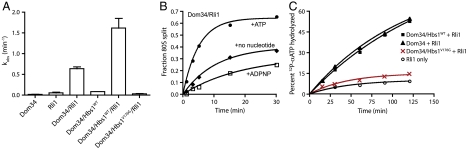
Dom34 and Hbs1 were previously shown to work together to split ribosomal subunits (and dissociate peptidyl-tRNA) in yeast, albeit at a modest rate (26). This splitting function is conserved in higher eukaryotes, although in this case efficient dissociation also depends on the presence of ABCE1 (25). We employed a reconstituted in vitro yeast translation system (37, 38) to ask whether yeast Rli1 similarly contributes to subunit splitting and thus to the early stages of ribosome recycling. In these experiments, stalled, elongated ribosomal complexes were prepared with a Met-Phe-tRNAPhe dipeptidyl-tRNA in the P site and a stop codon in the A site. Using a pre-steady-state kinetic approach, we find that Rli1 significantly accelerates (> 10-fold) the rate of subunit dissociation by Dom34 on these complexes in an ATP-dependent manner, and that the inclusion of Hbs1 further accelerates (approximately 2.5-fold) this rate (Fig. 2 A and B). Most strikingly, however, a GTPase-deficient derivative of Hbs1, Hbs1V176G (39), fully abolishes subunit-splitting activity, even in the presence of Rli1 (Fig. 2A). Similarly, although the ATPase activity level of Rli1 is independent of the presence of wild-type Hbs1, the addition of inactivated Hbs1V176G wholly inhibits ATPase activity (Fig. 2C). These data suggest that GTP turnover by Hbs1 precedes Rli1-dependent functions on the ribosome (including ATPase activity and subunit splitting), consistent with previous reports on the mammalian Rli1 homolog, ABCE1 (25).
Fig. 2.
Dom34∶Hbs1∶Rli1-facilitated subunit dissociation and its nucleotide dependence. (A) Observed rate constants for subunit dissociation in the presence of Dom34, Rli1, and/or Hbs1 and its derivatives. The reported rate constants were determined by monitoring the formation of free peptidyl-tRNA in a native gel system (n≥3, ± SEM). (B) Rli1-mediated subunit dissociation is dependent on ATP hydrolysis. Subunit splitting was monitored by peptidyl-tRNA formation in the presence of Dom34 and Rli1 with or without various nucleotides. (C) Multiple-turnover ATP hydrolysis by Rli1 on empty 80S ribosomes complexed with various factors, as indicated. The fraction of radiolabeled ATP that was hydrolyzed at each time point was monitored by thin-layer chromatography similar to Fig. 1B.
Mechanistic Insights on Dom34, Hbs1, and Rli1 in Recycling.
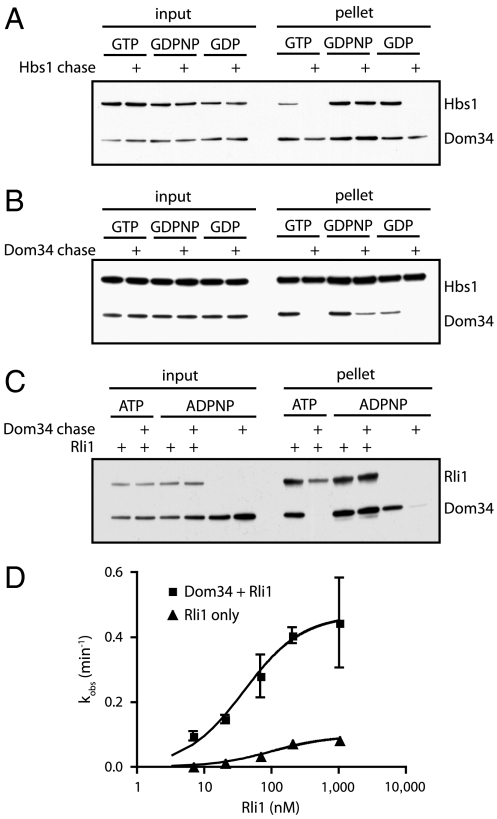
The apparent ordering of events suggested by the HbsV176G experiment led us to question whether GTP hydrolysis leads to the departure of Hbs1 from the ribosome or simply to conformational rearrangements in the complex, either of which could promote Rli1 function. To monitor Hbs1 dissociation, we incubated Dom34-His6, His6-Hbs1, and 80S ribosomes in the presence of various nucleotides (and their analogs). Relatively high levels of magnesium (10 mM) were included in these experiments to stabilize ribosome subunit interactions so that factor binding/dissociation could be monitored independently of subunit dissociation and recycling events. We then pelleted these reactions through a sucrose cushion to remove unbound protein and analyzed the resulting ribosome pellets for the presence or absence of each factor by Western blot analysis. To distinguish stable interactions from transient ones, we included a chase—an approximately 10-fold excess of an untagged version of the protein of interest—to prevent rebinding of the tagged factor following initial dissociation. Under these conditions, Hbs1 remained stably associated with 80S ribosomes only in the presence of the nonhydrolyzable GTP analog, GDPNP (Fig. 3A); incubation in the presence of either GTP or GDP allowed Hbs1 to fully dissociate from the ribosome. These data suggest that Hbs1 dissociates following nucleotide hydrolysis, setting the stage for subsequent Rli1-mediated events.
Fig. 3.
Association of Dom34 and Hbs1 with the ribosome. (A) Association of Hbs1 with ribosomes in the presence of Dom34 and various nucleotides was visualized by Western blot analysis following pelleting of ribosomes through a sucrose cushion. Unlabeled Hbs1 was used as a chase to distinguish between transient and stable interactions. (B) Similar to A, looking at the interaction of Dom34 with ribosomes under various conditions by using an unlabeled Dom34 chase. (C) Association of Dom34 with ribosomes in the presence or absence of Rli1 and various nucleotides. Visualized as above and using an unlabeled Dom34 chase. (D) kcat and K1/2 determinations for Rli1-mediated subunit dissociation. Subunit dissociation reactions were performed at various concentrations of Rli1 in the presence or absence of Dom34. Subunit dissociation was monitored by formation of peptidyl-tRNA via native gel electrophoresis (n = 3, ± SEM).
We next questioned whether Dom34 also dissociates from the ribosome following nucleotide hydrolysis by either Hbs1 or Rli1. When followed by a chase, we find that Dom34 spontaneously dissociates from the ribosome when incubated on its own or in the presence of Hbs1∶GTP/GDP or Rli∶ATP. However, the Dom34:ribosome interaction is partially or fully stabilized in the presence of Hbs1∶GDPNP or Rli1∶ADPNP, respectively (Fig. 3 B and C). Rli1 and Hbs1 are thus able to transiently “lock” Dom34 on the ribosome prior to NTP hydrolysis. Although the time resolution of these experiments is limited, longer chase times (30 min) eventually resulted in Dom34 dissociation even in the presence of Hbs1∶GDPNP, but not in the presence of Rli1∶ADPNP. These data suggest that the Dom34∶Rli1 interaction on the ribosome is particularly stable.
Given such a stable interaction, we sought to better define the nature of the relationship between Dom34, Rli1, and the ribosome. Namely, we asked whether Dom34 is involved in recruiting Rli1 more effectively to the ribosome complex (a K1/2 effect) or whether it directly contributes to the catalysis of recycling (kcat), two potential models that were proposed previously (25). To do this, we evaluated the rate of subunit splitting in the presence or absence of Dom34, titrating the concentration of Rli1 over two orders of magnitude. In these experiments, we observe only slight changes in K1/2 for Rli1, but significant effects on the rates of catalysis (kcat) (0.47 min-1 versus 0.09 min-1; Fig. 3D). These data provide clear evidence that Dom34 makes direct catalytic contributions to recycling.
mRNA Length Dependence of Dom34∶Hbs1-Mediated Recycling.
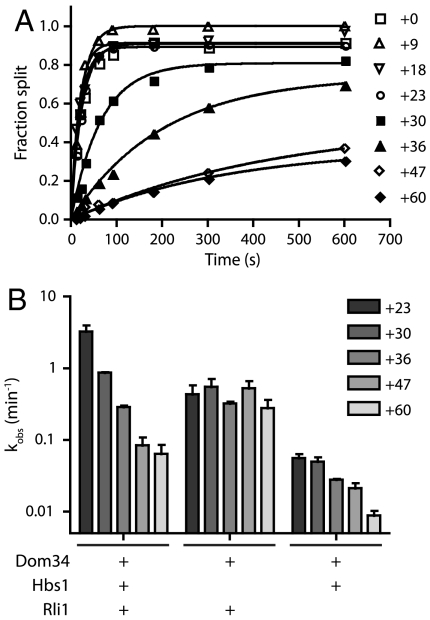
In the absence of codon dependence, we were interested in defining a mechanism for the regulation of Dom34-mediated recycling. A recent study in the mammalian system identified a strict mRNA length dependence for the human Pelota∶Hbs1∶ABCE1-mediated ribosomal recycling activity (25). In these studies, when the translated mRNA extended greater than 12 nucleotides past the P-site codon, subunit splitting was inhibited. Because we had not originally investigated such mRNA length dependence in the yeast system with Dom34 and Hbs1 (26), we took the opportunity to explore this parameter more fully here. Using a family of ribosomal complexes with increasingly long mRNAs (ranging from +0 to +60 nucleotides after the P-site codon), we observe a dependence of subunit-splitting activity on mRNA length (Fig. 4A); subunit splitting occurs unimpeded on mRNAs with up to 23 nucleotides extending beyond the P-site codon, whereas extensions longer than 30 nucleotides resulted in substantial decreases in rates. Although the precise limits of the length dependence in yeast varies from that observed for the mammalian system (where the cutoff occurred sharply at approximately 12 nucleotides), the data are broadly consistent with the previous report in suggesting that Dom34∶Hbs1∶Rli1-mediated recycling is principally targeted to empty ribosomes or those stalled near the 3′ end of messenger RNAs. However, differences in the observed length dependence suggest that the mechanism of monitoring may differ between organisms.
Fig. 4.
Hbs1 preferentially targets recycling to ribosome complexes with short mRNAs. (A) Representative time courses of subunit splitting by Dom34∶Hbs1∶Rli1 on complexes containing mRNAs of increasing lengths. The length of mRNAs following the P-site codon is indicated. Subunit splitting was monitored by formation of peptidyl-tRNA using electrophoretic thin-layer chromatography. (B) Observed rate constants, as determined in A, for subunit splitting by the factors indicated within the sensitive window of mRNA lengths (n = 3, ± SEM). Length dependence is not observed in the absence of Hbs1.
We were subsequently interested in the mechanism by which length dependence is conferred. To evaluate the possible contributions of Dom34, Hbs1, and/or Rli1 to the length dependence observed above, we evaluated recycling with multiple combinations of these factors on complexes containing mRNAs of increasingly inhibitory lengths. All combinations that included Hbs1 displayed significant length dependence; strikingly, little length dependence was seen in its absence (Fig. 4B). These results indicate that Hbs1 plays a critical role in conferring length dependence upon Dom34:Rli1-mediated subunit dissociation in yeast.
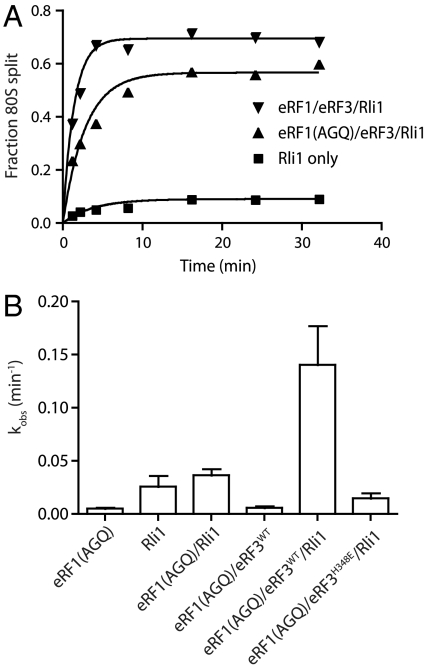
Rli1 Promotes Recycling by eRF1∶eRF3 Independent of Peptide Release.
Earlier studies showed that ABCE1 stimulates subunit dissociation on mammalian posttermination ribosome complexes when the peptide has been released by eRF1 (6). These results suggest that release of the nascent peptide is required for recycling in mammalian cells, reminiscent of recycling in bacteria. However, we have previously shown, in the context of yeast Dom34, that subunit dissociation can occur without peptide release (26). Indeed, we find that Rli1 promotes to a similar extent the recycling reaction with both wild-type eRF1 and a release-deficient mutant, eRF1G180A [eRF1(AGQ)] (Fig. 5A). With the short peptides used in our in vitro system, the peptidyl-tRNA is readily released. We suspect that with longer peptides the peptidyl-tRNA will partition with the 60S subunit and be resolved by peptidyl-tRNA hydrolase or other activities. As such, in the context of longer peptides, proper protein termination may accelerate downstream subunit dissociation processes by relieving interactions between the nascent peptide and the ribosome exit tunnel.
Fig. 5.
Rli1 accelerates eRF1∶eRF3 mediated recycling. (A) Representative time courses of the subunit-splitting activity of Rli1 and eRF1∶eRF3 on dipeptidyl-tRNA ribosome complexes containing 32P-labeled mRNA carrying a stop codon in the A site. Subunit splitting was monitored by native gel electrophoresis. Either eRF1(WT) or the eRF1(AGQ) variant was utilized. (B) Observed rate constants for subunit splitting mediated by eRF1(AGQ) in conjunction with other factors (n = 3, ± SEM).
We further find that ribosome splitting is greatly stimulated by eRF3 and is strictly codon dependent, as would be anticipated from an eRF1-stimulated reaction (Fig. 5B and Fig. S4). As with the Dom34-mediated recycling reactions explored in Fig. 2, we observe that the subunit dissociation reaction is inhibited when GTPase activity is inhibited in eRF3 (see eRF3H348E data in Fig. 5B) or when nonhydrolyzable ADPNP is included in the reaction to inhibit Rli1 (Fig. S5). As such, the biochemical features of Dom34-mediated and eRF1-mediated recycling as facilitated by Rli1 are remarkably similar, despite the specialized role of eRF1 in peptide release.
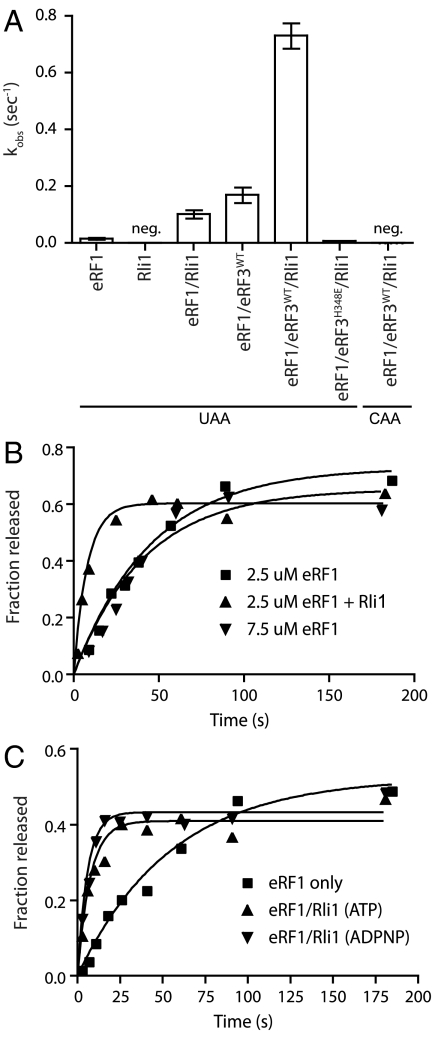
Rli1 Stimulation of eRF1-Mediated Peptide Release.
Given the dual roles for canonical termination factors in both peptide release and subunit splitting (6, 26), we asked whether Rli1 might also function in peptide release, and thus in both processes. In these experiments, elongated ribosome complexes were prepared with a dipeptidyl-tRNA in the P site and various codons in the A site, and the rate of peptide release was evaluated. We found that both Rli1 and eRF3 independently and substantially increased the rate of peptide release by eRF1 (Fig. 6A); the overall rate enhancement by the two together was approximately 48-fold. To ensure that the increased rate of peptide release did not result from increased binding of eRF1 mediated by Rli1 (i.e., a K1/2 effect), the eRF1 concentration was increased 3-fold, and we found that the rates of peptide release were unaffected (Fig. 6B). These data argue that eRF1 was present at a saturating concentration in these reactions, and thus that Rli1 increased the kcat of the release reaction (krelease). As anticipated, the release reaction is strictly dependent on the presence of a stop codon in the A site (Fig. 6A) and, when eRF3 is present, on GTP hydrolysis (Fig. S6). Surprisingly, the addition of ADPNP did not affect the ability of Rli1 to stimulate peptide release, as it did for the recycling reaction (Fig. 6C and Fig. S5). These latter data indicate that Rli1 binding is sufficient to mediate the release effect. Thus, the roles for Rli1 in peptide release and recycling are chemically distinct.
Fig. 6.
Rli1 accelerates eRF1-mediated release. (A) Observed rate constants for peptide release in the presence of eRF1, eRF3, and/or Rli1. Rates were determined using an electrophoretic TLC-based system to monitor peptide formation (n = 3, ± SEM; neg, negligible/nonmeasurable). (B) The observed effect of Rli1 on the rate of release is maintained in the presence of saturating levels of eRF1. (C) Rli1 accelerates peptide release in a step that is chemically distinct from its role in recycling. Time courses represent dipeptide formation as monitored by electrophoretic thin-layer chromatography. Both ATP and its nonhydrolyzable analog, ADPNP, allowed for Rli1-mediated stimulation of peptide release.
Discussion
Kinetic analysis of Rli1 function in an in vitro reconstituted S. cerevisiae translation system has revealed unique mechanistic insights into the regulation of translation termination and ribosome recycling in eukaryotes. We find that Rli1 stimulates ribosome subunit dissociation with both eRF1 and Dom34 in reactions dependent on ATP hydrolysis. Both recycling reactions are inhibited by GTPase-deficient variants of eRF3 (or Hbs1) or by the presence of GDPNP. Notably, subunit dissociation is not strictly dependent on peptide release, although it may play a role. This is in contrast to bacteria, where peptide release is essential for downstream recycling, and raises significant questions as to how termination and subunit dissociation are properly ordered in eukaryotes. Additionally, we show that Rli1 unexpectedly stimulates peptide release by eRF1 in a reaction that does not depend on ATP hydrolysis. These observations suggest a means by which the seemingly independent events of termination and recycling are directly coupled in yeast through a series of nucleotide hydrolysis events on the ribosome that ultimately result in subunit dissociation.
In establishing a generalized kinetic model for both eRF1∶eRF3∶Rli1 and Dom34∶Hbs1∶Rli1 in termination and recycling, we were struck by the extensive literature on the structural mimicry of tRNA by eRF1 and Dom34 (28, 29, 31, 33, 34, 40). These studies call to mind the well-defined model for tRNA selection in bacteria as facilitated by EFTu (for review, see ref. 41), a bacterial translational GTPase homologous to the eukaryotic GTPases eRF3 and Hbs1 (42). This tRNA selection process has been extensively characterized biochemically and, in conjunction with our biochemical data, provides an appealing framework for deciphering the activity of translational GTPases in eukaryotes.
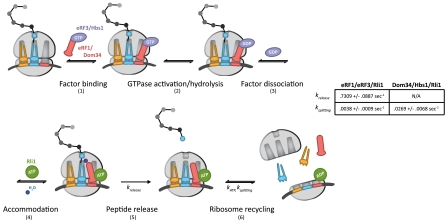
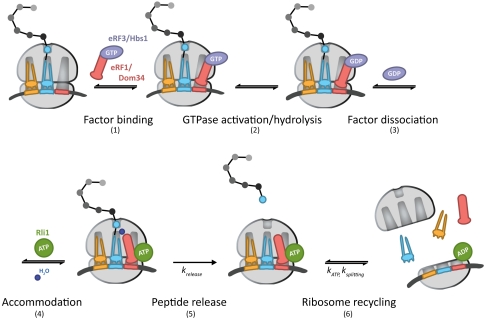
In this model (Fig. 7, steps 1–6), eRF1∶eRF3 or Dom34∶Hbs1 initially bind to the ribosome, likely as a heterodimer (step 1) (43). Analogous to GTPase hydrolysis by EFTu upon recognition of cognate codon:anticodon interactions, GTPase hydrolysis of eRF3 or Hbs1 occurs as a consequence of stop codon recognition, in the case of eRF3, or another signal, in the case of Hbs1 (step 2). Only complexes that have successfully undergone GTP hydrolysis by eRF3 or Hbs1 will ultimately be competent for rapid recycling by Rli1 at later steps (Fig. 2A and Fig. 5B). These data suggest the existence of a key eRF3/Hbs1-triggered conformational rearrangement in either eRF1/Dom34 or the ribosome that facilitates recycling. Following GTP hydrolysis, eRF3 and Hbs1 in the GDP bound state dissociate from the complex (Fig. 3A) (step 3). Accommodation of eRF1 or Dom34 into the active site can then occur, analogous to accommodation of aminoacyl-tRNA (step 4). In the case of eRF1, peptide release occurs after accommodation (krelease, step 5) and is promoted by Rli1 in an ATP-independent process (Fig. 6). The acceleration of release by Rli1 may actually derive from an acceleration of the preceding accommodation step (step 4). Specifically, we favor a model in which the binding of Rli1 locks eRF1 [or Dom34 (Fig. 3C)] on the ribosome in an accommodated, release-competent state prior to ATP hydrolysis. In the case of Dom34, however, peptide release does not occur. Postaccommodation complexes then proceed to the final step, in which Rli1 (in concert with either eRF1 or Dom34) initiates recycling (ksplitting, step 6) in an ATP-dependent manner (Fig. 2 and Fig. 5). The dual activities of Rli1 in peptide release and ribosome recycling are thereby effectively bridged through ATP hydrolysis.
Fig. 7.
A kinetic model for the integrated events of translation termination and ribosome recycling in yeast (modeled after ref. 41). The targeted ribosome complex either contains a stop codon in the A site, in the case of eRF1∶eRF3, or is stalled near the 3′ end of an mRNA, in the case of Dom34∶Hbs1. The determined rate constants relevant to this model are indicated. A full walk-through of steps 1–6 can be found in Discussion.
The strength of this model is in its ability to rationalize how termination and recycling are ordered in the cell, through sequential nucleotide hydrolysis steps, though both processes depend on a common set of factors. The acceleration of peptide release in the presence of Rli1 increases the likelihood that release will occur prior to subunit dissociation. Indeed, kinetic partitioning based on the relative rates of peptide release and subunit dissociation that we report in our minimal in vitro system (Fig. 7, table) suggests that subunit dissociation would aberrantly precede peptide release no more often than 1 in approximately 200 events. Additional mechanisms likely exist in vivo to make the fidelity of these processes even higher (44–46).
In our previous work, Dom34-mediated subunit dissociation did not require Rli1 (26). Subsequent reports, however, indicated a role for Rli1 in Dom34-mediated subunit dissociation in a mammalian system (25). The kinetic analysis described herein both explains and resolves the distinct behavior of the two experimental systems. Although Dom34∶Hbs1 is capable of splitting subunits alone, Rli1 binds the Dom34:ribosome complex (Fig. 1A) and significantly accelerates (approximately 19-fold) the process (Fig. 2B) (step 6). These observations are consistent with the mammalian study and suggest a broad conservation of these processes.
A further distinction of the mammalian Dom34∶Hbs1∶Rli1 recycling activity was its marked dependence on 3′ mRNA length, a parameter not originally explored in the yeast system (25). Here, upon additional analysis, we do observe mRNA length dependence in yeast, although it appears to be more broadly permissive than its mammalian counterpart (Fig. 4A). Despite these differences, it seems likely that Dom34∶Hbs1 operates primarily on ribosome complexes containing short or no mRNA. We extend these observations with the finding that this mRNA length dependence is conferred upon Dom34-mediated recycling events by Hbs1, as recycling events occurring in the presence of Dom34 and Rli1 alone are insensitive to mRNA length (Fig. 4B). Interestingly, cryoEM structures of Dom34∶Hbs1 on the yeast ribosome indicate that the N-terminus of Hbs1 extends away from the body of the protein and contacts the mRNA entry site (43). These results suggest that GTP hydrolysis by Hbs1 is dependent on proper surveillance of 3′ mRNA length by the N-terminus of Hbs1.
By extension, we speculate that eRF3 activity may be similarly modulated by its N-terminus, which both binds to polyA-binding protein (Pab1) and is required for [PSI+] prion formation (47–49). The regulation of eRF3 through its N-terminus by Pab1 or other factors could have broad implications for distinguishing between authentic stop codons and, for example, nonsense-mediated decay targets in yeast and other organisms. However, these parameters remain outside the purview of the data presented here.
Our results suggest that the strict designation of factors as belonging to “release” or “recycling” is misplaced in the eukaryotic system. These processes appear to operate in a coupled manner requiring the tight regulation of a common set of factors. Thus, significant mechanistic differences exist between bacterial and eukaryotic termination processes. Following dissociation, subunits are subsequently trapped by initiation factors (e.g., eIF3/1/1A) (25, 50), which facilitate the recycling of remaining translation components and prepare ribosome subunits for the next round of initiation. Thus, it seems likely that reinitiation during the much discussed “circle of translation” in eukaryotic systems depends heavily on the tight regulation of the recycling process. Consistently, Rli1 has also been characterized as having a role in subsequent rounds of initiation (16, 19). If confirmed, such a role would further blur the lines that mark the defined phases of the translation cycle.
Materials and Methods
Pull-Down and Western Blot Analysis.
eRF1-His6, Dom34-His6, or empty vector (pYES2, Invitrogen) were expressed for 6 h at 30 °C in an Rli1 TAP-tagged yeast strain with 2% galactose/1% raffinose. Cells were collected and lysed via bead beating in lysis/wash buffer [10 mM Tris-Cl pH 7.5, 150 mM NaCl, 0.1% Tween-20, 10 mM imidazole, 5 mM beta-mercaptoethanol, 0.5 mM ADP, COMPLETE protease inhibitor (Roche)]. Lysate was clarified and purified over Ni-NTA beads blocked in 50 μg/mL BSA. Protein was eluted in lysis buffer +300 mM imidazole. Eluate was separated on an SDS-polyacrylamide gel and analyzed via Western blot using anti-TAP (GenScript) and anti-His (Qiagen) HRP-conjugated antibodies.
Rli1-His6 purification.
RLI1 was cloned from yeast genomic DNA into pYES2 using primers adding a C-terminal His6 tag (primers, see Table S1) and induced in INVSc1 cells (Invitrogen) at 30 °C for 16 h. Cells were resuspended in Ni-NTA lysis buffer (75 mM HEPES pH 8.0, 300 mM NaCl, 5 mM beta-mercaptoethanol, 1% Tween, 20 mM imidazole, 10% glycerol), frozen in pellets and lysed in a liquid nitrogen Freezer/Mill (SPEX SamplePrep, LLC). Lysate was clarified and purified over a HisTrap FF column (GE Healthcare) on an ÄTKA FPLC (GE Healthcare). Additional purification was conducted over an S100 size exclusion column (GE Healthcare) preequilibrated in Buffer SE (20 mM Tris-Cl pH 7.5, 200 mM NaCl, 5 mM beta-mercaptoethanol, 5% glycerol). Purified protein was observed to contain a brown/yellow color.
Unlabeled Dom34 Purification.
DOM34 was cloned into pTYB2 (New England Biolabs) using primers listed in Table S1. Dom34 was expressed in RIPL cells (Agilent) overnight at 16 °C. Cells were lysed by French press and bound to chitin resin (lysis/wash buffer: 20 mM HEPES-KOH pH 7.4, 1 M KCl, 10% glycerol, 1 mM EDTA). Dom34 was eluted at 4 °C overnight using elution buffer (20 mM HEPES-KOH pH 7.4, 0.5 M KCl, 10% glycerol, 1 mM EDTA, 50 mM DTT). Eluted protein was further purified over an S100 sizing column (GE Healthcare) in 20 mM Tris-Cl pH 7.5, 500 mM NaCl, 5% glycerol, and 5 mM beta-mercaptoethanol.
Untagged Hbs1 Purification.
Hbs1 was purified from pCS21 as described previously (26) with the following modifications. Following purification over an S100 size exclusion column, 1 mM DTT, 0.5 mM EDTA and 20 ug/mL TEV protease were added to pooled Hbs1-His6. Cleavage was allowed to proceed for 3 h at room temperature, after which TEV protease and uncleaved Hbs1-His6 were removed by passage over Ni-NTA resin (Qiagen). Cleaved Hbs1 was purified further on a RESOURCE Q column (GE Healthcare) using Ion Wash Buffer (20 mM Tris-Cl pH 7.5, 50 mM NaCl, 5 mM beta-mercaptoethanol, 5% glycerol) and a 0–100% gradient of Ion Elution Buffer (20 mM Tris-Cl pH 7.5, 500 mM NaCl, 5 mM beta-mercaptoethanol, 5% glycerol).
Mutagenesis of Hbs1/eRF3.
Hbs1 and eRF3 mutants were derived using the primers indicated in Table S1. Proteins were induced at 16 °C overnight and purified over successive HisTrap FF, S100 size exclusion and RESOURCE Q ion exchange columns (GE Healthcare) using the buffers and protocols detailed in the sections above.
ATPase Assay.
ATP hydrolysis assays were performed in 1X Buffer E (20 mM Tris-Cl pH 7.5, 2.5 mM Mg(OAc)2, 100 mM KOAc pH 7.6, 2 mM DTT, 0.25 mM spermidine) containing 1.5 μM Dom34, 2 μM Hbs1, 0.67 μM 80S ribosomes, 0.25 μM Rli1, 1 mM GTP, 1 mM ATP and 5 nM 32P-αATP. All time points were quenched in 30% formic acid and analyzed on a prerun PEI-cellulose TLC plate (EMD Chemicals, Inc.) in 0.5 M KH2PO4 pH 3.5.
In Vitro Reconstituted Yeast Translation Ssystem.
Ribosome complexes encoding Met-Phe-tRNAphe in the P site and a stop or nonstop codon in the A site were formed as described by Eyler and Green (37). All factors necessary for the in vitro reconstituted yeast translation system were purified as previously described (37, 38). Unless otherwise indicated, complexes were formed on mRNAs containing 18 nucleotides downstream of the P site. Variable-length mRNAs (Table S2: +0, +9, +18, +23, +30, +36, +47, +60) were in vitro transcribed from complementary ssDNA templates (Integrated DNA Technologies) using T7 RNA polymerase and a short clamp oligo. All mRNAs transcripts were purified from a denaturing 1X TBE (89 mM Tris, 89 mM Boric acid, 2 mM EDTA) urea gel to ensure homogeneity.
Subunit Separation and Release Assays.
Both native gel and electrophoretic TLC measurements of subunit dissociation and release were performed as previously described (26, 37). For all native gel experiments, TIF6 was used to trap dissociated subunits. 35S-Met-labeled peptidyl-tRNAs were used to visualize subunit dissociation in all cases except for Fig. 5A, where 32P-pCp-labeled mRNA was used. Native gels and TLC plates were developed using a Typhoon 9410 phosphoimager system (GE Healthcare) and quantitated using ImageQuantTL (GE Healthcare). Time courses were fit to single exponential kinetics.
Ribosome Pelleting.
Prior to pelleting, reactions were incubated in Buffer E+Mg [20 mM Tris-Cl pH 7.5, 10 mM Mg(OAc)2, 100 mM KOAc pH 7.6, 2 mM DTT, 0.25 mM spermidine] containing 0.5 mM nucleotide (as indicated for each figure) and 0.3–0.6 μM ribosomes; 0.35–0.7 μM Dom34-His6, 0.5–1.5 μM Hbs1-His6, and/or 1.2 μM Rli1-His6 were preincubated with ribosomes for 5 min. After preincubation, Hbs1-His6 was chased with 9 μM unlabeled Hbs1, or Dom34-His6 was chased with 5 μM unlabeled Dom34. Chase times ranged from 5–20 min. Reactions were pelleted through 600 μL ribosome pelleting buffer (20 mM Tris-Cl pH 7.5, 5 mM Mg(OAc)2, 100 mM KOAc pH 7.6, 2 mM DTT, 0.25 mM spermidine) for 60 min at 75 krpm in an MLA-130 rotor. Resulting pellets were resuspended in 1X Buffer E and analyzed via Western blot analysis.
Supplementary Material
Acknowledgments.
We thank J. Lorsch and his lab members for valuable discussion, and D. Eyler and N. Guydosh for discussion and thoughtful comments on the manuscript. This work was funded by Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 20283.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113956108/-/DCSupplemental.
References
- 1.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 2.Rodnina MV, Wintermeyer W. Recent mechanistic insights into eukaryotic ribosomes. Curr Opin Cell Biol. 2009;21:435–443. doi: 10.1016/j.ceb.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 3.Zavialov AV, Hauryliuk VV, Ehrenberg M. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol Cell. 2005;18:675–686. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 4.Peske F, Rodnina MV, Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Freistroffer DV, Pavlov MY, MacDougall J, Buckingham RH, Ehrenberg M. Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J. 1997;16:4126–4133. doi: 10.1093/emboj/16.13.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pisarev AV, et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barthelme D, et al. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc Natl Acad Sci USA. 2011;108:3228–3233. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barthelme D, et al. Structural organization of essential iron-sulfur clusters in the evolutionarily highly conserved ATP-binding cassette protein ABCE1. J Biol Chem. 2007;282:14598–14607. doi: 10.1074/jbc.M700825200. [DOI] [PubMed] [Google Scholar]
- 9.Karcher A, Buttner K, Martens B, Jansen RP, Hopfner KP. X-ray structure of RLI, an essential twin cassette ABC ATPase involved in ribosome biogenesis and HIV capsid assembly. Structure. 2005;13:649–659. doi: 10.1016/j.str.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Karcher A, Schele A, Hopfner KP. X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi. J Biol Chem. 2008;283:7962–7971. doi: 10.1074/jbc.M707347200. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Dean M, Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet. 2005;6:123–142. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- 13.Kerr ID. Sequence analysis of twin ATP binding cassette proteins involved in translational control, antibiotic resistance, and ribonuclease L inhibition. Biochem Biophys Res Commun. 2004;315:166–173. doi: 10.1016/j.bbrc.2004.01.044. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Z, Fang LL, Johnsen R, Baillie DL. ATP-binding cassette protein E is involved in gene transcription and translation in Caenorhabditis elegans. Biochem Biophys Res Commun. 2004;323:104–111. doi: 10.1016/j.bbrc.2004.08.068. [DOI] [PubMed] [Google Scholar]
- 15.Andersen DS, Leevers SJ. The essential Drosophila ATP-binding cassette domain protein, pixie, binds the 40S ribosome in an ATP-dependent manner and is required for translation initiation. J Biol Chem. 2007;282:14752–14760. doi: 10.1074/jbc.M701361200. [DOI] [PubMed] [Google Scholar]
- 16.Dong J, et al. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem. 2004;279:42157–42168. doi: 10.1074/jbc.M404502200. [DOI] [PubMed] [Google Scholar]
- 17.Kispal G, et al. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 2005;24:589–598. doi: 10.1038/sj.emboj.7600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yarunin A, et al. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 2005;24:580–588. doi: 10.1038/sj.emboj.7600540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen ZQ, et al. The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. J Biol Chem. 2006;281:7452–7457. doi: 10.1074/jbc.M510603200. [DOI] [PubMed] [Google Scholar]
- 20.Khoshnevis S, et al. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 2010;11:214–219. doi: 10.1038/embor.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dooher JE, Schneider BL, Reed JC, Lingappa JR. Host ABCE1 is at plasma membrane HIV assembly sites and its dissociation from Gag is linked to subsequent events of virus production. Traffic. 2007;8:195–211. doi: 10.1111/j.1600-0854.2006.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmerman C, et al. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature. 2002;415:88–92. doi: 10.1038/415088a. [DOI] [PubMed] [Google Scholar]
- 23.Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T. Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2–5 A pathway. J Biol Chem. 1995;270:13308–13317. doi: 10.1074/jbc.270.22.13308. [DOI] [PubMed] [Google Scholar]
- 24.Bisbal C, et al. The 2–5 A/RNase L pathway and inhibition by RNase L inhibitor (RLI) Methods Mol Biol. 2001;160:183–198. doi: 10.1385/1-59259-233-3:183. [DOI] [PubMed] [Google Scholar]
- 25.Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shoemaker CJ, Eyler DE, Green R. Dom34∶Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L, et al. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat Struct Mol Biol. 2010;17:1233–1240. doi: 10.1038/nsmb.1922. [DOI] [PubMed] [Google Scholar]
- 29.Graille M, Chaillet M, van Tilbeurgh H. Structure of yeast Dom34: A protein related to translation termination factor Erf1 and involved in No-Go decay. J Biol Chem. 2008;283:7145–7154. doi: 10.1074/jbc.M708224200. [DOI] [PubMed] [Google Scholar]
- 30.Lee HH, et al. Structural and functional insights into Dom34, a key component of no-go mRNA decay. Mol Cell. 2007;27:938–950. doi: 10.1016/j.molcel.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 31.Song H, et al. The crystal structure of human eukaryotic release factor eRF1—Mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 32.Frolova LY, et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi K, et al. Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1alpha complex. Proc Natl Acad Sci USA. 2010;107:17575–17579. doi: 10.1073/pnas.1009598107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito K, et al. Omnipotent role of archaeal elongation factor 1 alpha (EF1alpha in translational elongation and termination, and quality control of protein synthesis. Proc Natl Acad Sci USA. 2010;107:19242–19247. doi: 10.1073/pnas.1009599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharya A, McIntosh KB, Willis IM, Warner JR. Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Mol Cell Biol. 2010;30:5562–5571. doi: 10.1128/MCB.00618-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatchikian EC, et al. Isolation, characterization, and biological activity of the Methanococcus thermolithotrophicus ferredoxin. J Bacteriol. 1989;171:2384–2390. doi: 10.1128/jb.171.5.2384-2390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eyler DE, Green R. Distinct response of yeast ribosomes to a miscoding event during translation. RNA. 2011;17:925–932. doi: 10.1261/rna.2623711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–145. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- 39.Carr-Schmid A, Pfund C, Craig EA, Kinzy TG. Novel G-protein complex whose requirement is linked to the translational status of the cell. Mol Cell Biol. 2002;22:2564–2574. doi: 10.1128/MCB.22.8.2564-2574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng Z, et al. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009;23:1106–1118. doi: 10.1101/gad.1770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodnina MV, Gromadski KB, Kothe U, Wieden HJ. Recognition and selection of tRNA in translation. FEBS Lett. 2005;579:938–942. doi: 10.1016/j.febslet.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 42.Atkinson GC, Baldauf SL, Hauryliuk V. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol Biol. 2008;8:290. doi: 10.1186/1471-2148-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becker T, et al. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 44.Keeling KM, Salas-Marco J, Osherovich LZ, Bedwell DM. Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:5237–5248. doi: 10.1128/MCB.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gross T, et al. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–649. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- 46.Bolger TA, Folkmann AW, Tran EJ, Wente SR. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008;134:624–633. doi: 10.1016/j.cell.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cosson B, et al. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol Cell Biol. 2002;22:3301–3315. doi: 10.1128/MCB.22.10.3301-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hosoda N, et al. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J Biol Chem. 2003;278:38287–38291. doi: 10.1074/jbc.C300300200. [DOI] [PubMed] [Google Scholar]
- 49.Ter-Avanesyan MD, et al. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 50.Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]