Abstract
The genetic and demographic impact of European contact with Native Americans has remained unclear despite recent interest. Whereas archeological and historical records indicate that European contact resulted in widespread mortality from various sources, genetic studies have found little evidence of a recent contraction in Native American population size. In this study we use a large dataset including both ancient and contemporary mitochondrial DNA to construct a high-resolution portrait of the Holocene and late Pleistocene population size of indigenous Americans. Our reconstruction suggests that Native Americans suffered a significant, although transient, contraction in population size some 500 y before the present, during which female effective size was reduced by ∼50%. These results support analyses of historical records indicating that European colonization induced widespread mortality among indigenous Americans.
Keywords: Bayesian analysis, coalescent analysis, population genetics
Despite considerable interest from both archaeologists and evolutionary geneticists, the demographic history of indigenous Americans remains contentious. One source of debate concerns the impact of European contact, where archaeological and genetic studies have painted contrasting stories. Both archaeological and historical records indicate that European contact and colonialism initiated a significant reduction in the indigenous population size through warfare, enslavement, societal disruption, and especially widespread epidemic disease (1–3), although the magnitude of population decline remains in dispute (4, 5). However, few genetic analyses have found evidence for a substantial population contraction during this period (but see refs. 6–8), and several recent, relatively large-scale demographic reconstructions have found no signal of a significant decline (9–12). Thus, genetic studies appear at odds with historical evidence, and the demographic impact of European colonization on indigenous Americans has remained unclear.
Although historical evidence regarding population size during the 16th century is fragmentary, surviving personal and governmental accounts document substantial, if not catastrophic, declines during the period. For instance, working in Mexico, the Franciscan friar Fray Toribio de Benavente wrote that “At this time [Mexico] was extremely full of people, and when the smallpox began to attack the Indians it became so great a pestilence among them… that in most provinces more than half the population died.” (13) Governmental taxation and tribute records have allowed for a more quantitative accounting in some areas. For instance, taxation records for 24 districts of Peru suggest that the taxable population size decreased by about 30% (from 466,748 to 324,895) between 1573 and 1602, although the region had previously endured two smallpox epidemics during 1546 and 1558 (3). Nonetheless, although historical records provide insights into demographic processes in specific regions, the overall extent of population decline remains in dispute. Some authors have suggested that the total Native American population size decreased by more than 90%, whereas others have argued that significant population losses were more localized and the magnitude of the reduction relatively modest (e.g., ref. 4).
Genetic studies have the potential to inform the debate regarding the full extent of population losses. So far, demographic reconstructions have found little evidence for a recent reduction in population size, thus supporting the view that the impact of European colonization on Native Americans was relatively modest. For instance, Fagundes et al. (12) using Bayesian demographic reconstruction methods, found that population sizes have remained essentially constant for the last 15 ky, if not longer. Similarly, Mulligan et al. (10) found evidence for three distinct demographic epochs, but little evidence of a significant contraction coincident with European contact. Ho and Endicott (11), using an alternative calibration methodology, also found that population size has not significantly changed during the last several thousand years.
While genetic studies have suggested that the impact of European contact was minimal, recent increases in the amount of genetic data available, including high-quality, accurately dated ancient sequences, have significantly increased the potential resolution of demographic reconstructions. The use of ancient DNA in demographic inference is particularly beneficial for two reasons. First, the addition of ancient sequences increases the statistical power to infer sizes at deeper points in the genealogy, where resolution is likely to be low. Second, when ancient sequences are accurately dated, evolutionary rate can be estimated simultaneously with other model parameters, providing a method of molecular clock calibration that appropriately incorporates uncertainty in the evolutionary rate. In the absence of ancient DNA, calibration of the molecular clock requires use of an exogenously computed rate, a potentially error-prone procedure (11).
Here, we use a dataset containing both ancient and contemporary mitochondrial sequences to reconstruct the demographic history of indigenous Americans. We use a Bayesian Markov chain Monte Carlo (MCMC) technique to coestimate the ancestry of the sequences, the model of sequence evolution, and a time-dependent effective population size. Because our sequences are taken from multiple points in time our dataset is internally calibrated, and we estimate the full posterior distribution of evolutionary rate in a manner identical to other model parameters. We find that indigenous Americans experienced a significant contraction in population size some 500 years before the present (ypb), during which female effective size was reduced by ∼50%, thus suggesting that the impact of European colonization was both widespread and severe.
Results
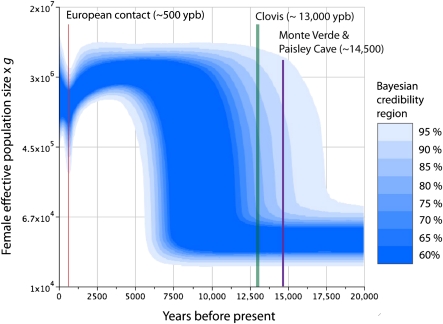
Using the genealogy sampler BEAST to implement an extended Bayesian skyline plot (EBSP) analysis, our reconstruction delineates several distinct demographic epochs, including a significant and transient contraction in population size some 500 ypb (Fig. 1). Although some uncertainty accompanies our reconstruction, the population size appears markedly reduced for several hundred years, although it has since returned to levels similar to those before the event. When all demographic functions sampled by the Markov chain are pooled, the mean inferred population sizes suggest a decrease in female effective size of roughly 50% from the predecline maximum some 5000 ypb to the population nadir roughly 500 ypb.
Fig. 1.
Extended Bayesian skyline plot of female effective population size scaled by generation time (g), with archaeological sites and historical events as shown. Colored regions indicate Bayesian credibility intervals.
Although ancient DNA provides a powerful means of evolutionary clock calibration, the sampling of this parameter introduces additional uncertainty into reconstruction efforts, particularly when dated sequences span a short period relative to the full depth of the genealogy. This uncertainty can partially obscure demographic features, hence discarding calibration data may increase resolution in some cases, although information regarding the calendrical timing of events will be lost. Given that the majority of our ancient sequences have dates that are less than 10% of the total tree depth, the amount of uncertainty introduced by the rate estimation procedure for our dataset may be considerable. To assess the impact of this source of uncertainty and examine demographic properties in an uncalibrated context, we discarded all ancient sequences and performed another EBSP analysis (Fig. 2). Whereas the general form of skyline plot appears similar to the calibrated analyses, the population size reduction is somewhat more pronounced. Additionally, the time (in substitutional units, subs.) at which the initial phase of population growth began is identified much more clearly.
Fig. 2.
Extended Bayesian skyline plot constructed without calibrating information from ancient sequences. Darker colors indicate regions of reduced certainty.
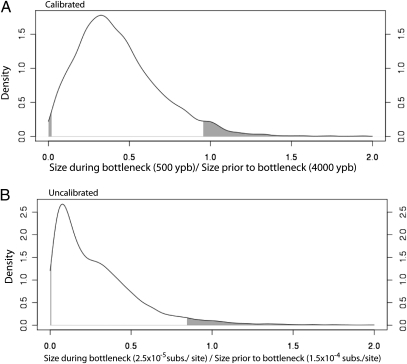
To examine the severity of the bottleneck in a more quantitative fashion, we examined the ratio of the population size before the bottleneck (at 4000 ypb in the calibrated analysis, or 10−4 subs. per site in the uncalibrated analysis) to the size at the nadir (500 ypb calibrated, 2 × 10−5 subs. per site uncalibrated) for each demographic function sampled by the Markov chain. The densities of these ratios then reflect the relative probability that the bottleneck reduced the population size by a particular amount, with 1.0 representing the null hypothesis that no size reduction occurred. For both the calibrated and uncalibrated analyses, the distributions are strongly biased toward values less than one and 95% highest posterior density (HPD) intervals exclude 1.0 (0.015–0.954 calibrated, 0.003–0.843 uncalibrated) (Fig. 3). As an additional check for statistical significance, we computed the fraction of states sampled by the chain in which the ratio was 1.0 or greater. For both analyses, this value was less than 0.05 (0.041 and 0.013 for the calibrated and uncalibrated analyses, respectively), indicating a significance at the α = 0.05 level.
Fig. 3.
Density of ratios of population size during the bottleneck (500 ypb for calibrated analysis, 2.5 × 10−5 subs. per site for uncalibrated analysis) to size before bottleneck (4000 ypb for calibrated analysis, 1.5 × 10−4 subs. per site for uncalibrated analysis). Shaded regions indicate 95% highest posterior density (HPD) intervals.
The signature of the bottleneck is also apparent in the genealogical structure of the dataset. A majority-rule consensus genealogy constructed from the trees sampled by the Markov chain demonstrates a transiently increased rate of coalescence near the tips of the genealogy (Fig. 4), particularly in haplogroups C and D. Because the rate at which lineages coalesce is inversely proportional to population size, the preponderance of coalescent events near the tips in these haplogroups suggests that the bottleneck affected haplogroups C and D disproportionately. However, the broad distribution of haplogroups in modern indigenous American populations complicates interpretation of this observation with regards to an area most affected by the population decline. Insight into the magnitude of the decline at local scales will likely involve direct comparison between pre- and postcontact populations, a task we leave to future investigations. Despite these early coalescences, the overall genealogical structure is similar to that found in other studies, with five well-defined clades corresponding to the major haplogroups A–D and X. Haplogroups have similar ages, with the mean value for the MRCA of each haplogroup occurring roughly 15 ky. Additionally, the ancient sequences appear relatively well mixed with contemporary samples, and with few exceptions ancient lineages do not form isolated or divergent clades.
Fig. 4.
Majority-rule consensus tree constructed from genealogies sampled by the Markov chain. Gray bars indicate one SD in node height. Red lineages are ancient samples from North America; blue lineages are ancient samples from South America.
The population contraction ended a long period of demographic stability that began ∼7–10 ka, following a period of rapid expansion that increased effective size nearly 100-fold. The time at which this period of expansion began remains somewhat uncertain, with the most likely dates ranging from 8 to 12 ka. Thus, whereas archeological data indicate a human presence as early as 14.5 ka at the Monte Verde site in southern South America (14, 15) and perhaps as early at 15.5 ka at the Debra L. Friedkin site in Texas (16), our analysis suggests that population densities remained low during this period, and may not have increased in earnest until after the disappearance of the Clovis complex. While statistical power to infer size is low deep in the genealogy, we find the initial number of New World founders to be on the order of several thousand (assuming a generation time of 25 y), a figure in agreement with some recent analyses (9–12), although larger than several others (17, 18).
Discussion
Using a dataset combining both ancient and contemporary mtDNA, we have demonstrated that the effective size of the Native American population underwent a detectable, although transient, contraction some 500 ypb. Although some uncertainty exists, the inferred population size 500 ypb is ∼50% of the predecline maximum some 3000 y before present. Although our analysis contrasts with several recent studies of Native American demographic history, our results are consistent with historical records suggesting that epidemics, warfare, enslavement, and famines resulted in significant population declines among Native Americans during the 16th century. Additionally, the scale of the contraction suggests that the depopulation was not localized to particular regions or communities, and instead, was likely to have been widespread or to have had an especially severe impact on the most populous regions.
Although our reconstruction supports a relatively strong population decline, we cannot rule out the possibility that changes in population structure have influenced our analysis. For instance, if population size were inflated due to structure effects, and the arrival of Europeans induced a period of strong mixing that removed the structure-related bias, then a spurious signal of population size change may have been produced. Although we do not formally test this possibility, several lines of evidence suggest that structure did not play an important role. First, as noted above, examination of the consensus genealogy shows little evidence for increased coalescence rates among samples from the same physical location, and previous studies have documented substantial gene flow between populations (e.g., ref. 8). Second, if the size of Native American subpopulations was variable and not strictly regulated, population structure would likely have decreased effective size (19), contrary to the scenario above. Finally, as mentioned in the introduction, historical records corroborate the mortality-related crash hypothesis.
Although our analysis does not directly address the arrival time of humans into the New World, it does indicate that population size remained low for a period somewhat longer than other studies have found (9–12, 20, 21). These earlier studies have argued that rapid population growth may have begun shortly after the last glacial maximum (LGM, ∼20 kya), leading to high population densities significantly predating Clovis, perhaps as early at 16 kya (12). However, little archaeological evidence supports a significant human presence before 15 kya. In contrast, our reconstruction indicates that population densities most likely did not significantly increase until well after the arrival to the New World, suggesting that factors other than access to new territory promoted population growth. Our analysis is therefore consistent with models suggesting an early coastal and riverine spread of the initial settlers, followed by a prolonged period of niche establishment, cultural differentiation, and a later increase in demographic magnitude and complexity (22–24).
Studies that produce or examine ancient DNA must take into account both the possibility of contamination by modern sources as well as postmortem damage to the molecule (25). Several lines of reasoning suggest that such errors did not significantly impact our results. First, repeated analysis of subsets of our data yielded consistent results. When sequences were grouped on the basis of physical origin (Peru vs. North America), and the analysis rerun on each subset together with the contemporary sequences, results remained largely unchanged. If either dataset contained contaminated or inaccurately dated sequences, the two subanalyses would likely have produced divergent results. Additionally, all ancient sequences fell into previously established haplogroups and, with few exceptions, did not form isolated or divergent clades (Fig. 4), as might be expected if sequences had been significantly damaged. Similarly, the bottleneck is still apparent even when all ancient data are removed. Second, we conducted an additional test using BEAST with a model specifically designed to detect age-dependent sequencing errors, which yielded negative results. Finally, a preliminary analysis with another set of ancient sequences from an independent source (26) also yielded results similar to those described here, further suggesting that our findings were not influenced by idiosyncratic features of the ancient DNA used.
Methods
Our dataset contained 137 contemporary full mitochondrial genomes representing all five major North American mtDNA haplogroups and 63 ancient sequences from the first hypervariable segment (HVS1) of the mitochondrial genome, obtained from two published sources, and supplemented with two previously unpublished sequences (Table 1). The first set of ancient DNA comprises 32 sequences isolated from burial sites in southern Ontario and western Illinois, with estimated ages ranging from 800 to 2900 y before present (ybp). Sample isolation, preparation, and dating are described elsewhere (8). The second set of ancient DNA consists of 29 HVS1 sequences obtained from the Palpa region in southern Peru, with dates spanning 800–2700 y before present. Sequences were isolated and dated as described in refs. 27 and 28. Dates were assigned using a combination of accelerator mass spectrometry (AMS) 14C radiocarbon dating and contextual information. Because the power to infer evolutionary rate increases with the difference in age among the samples, we also included two previously unpublished sequences from the Late Archaic period, ∼5000 ypb, from the site Pernil Alto, close to the modern city of Palpa, Peru. Sequences were obtained using methods similar to refs. 27 and 28. Additional information about collection site, dates, and molecular protocol enhancements are provided in SI Text.
Table 1.
Ancient mtDNA used in this study, with source locality and approximate age
| Site | Region | Count | Approximate date | Source |
| Great Western Park | SW Ontario | 4 | A.D. 1200 | 25 |
| Hind | SW Ontario | 12 | 900 B.C. | 25 |
| Morse | Fulton County, Illinois | 6 | 700 B.C. | 25 |
| Orendorf | Fulton County, Illinois | 10 | A.D. 1200 | 25 |
| Zimmer | SW Ontario | 1 | 900 B.C. | 25 |
| Pernil Alto | Palpa, Peru | 4 | 700–3000 B.C. | This study, 26 |
| Botigiriayocc | Palpa, Peru | 3 | A.D. 1200 | 8 |
| Chillo | Palpa, Peru | 1 | A.D. 1300 | 26 |
| Pacapaccari | Palpa, Peru | 1 | A.D. 1200 | 8 |
| Huayuncalla | Palpa, Peru | 2 | A.D. 1000 | 8 |
| Jauranga | Palpa, Peru | 5 | 200 B.C.–A.D.400 | 26 |
| Layuni | Palpa, Peru | 1 | A.D. 1200 | 26 |
| Mollake Chico | Palpa, Peru | 1 | 700 B.C. | 26 |
| Los Molinos | Palpa, Peru | 5 | A.D. 150–850 | 26 |
| Hanaq Pacha | Palpa, Peru | 1 | A.D. 400 | 26 |
| Cueva Yacatogia | Palpa, Peru | 4 | A.D. 800–850 | 8 |
Some sites (Pernil Alto, Juaranga, and Los Molinos) included samples from multiple periods.
To reconstruct the demographic history of our genetic samples we used the Bayesian MCMC machinery in BEAST (29). BEAST estimates the posterior distribution of a wide range of model parameters, including genealogical structure, substitution model, and population size functions given a set of genetic sequences. After aligning all sequences using the online version of MaFFT (30), sequences were divided into two partitions corresponding to the coding region (sites 600–16,000 of the Cambridge reference sequence) and the HVS1, sites 16,000–16,589. Partitions were analyzed using independent clock and substitution models but a shared genealogy. Separate TN93 (31) substitutional models were used for both partitions, with γ-distributed rate variation across four categories and base frequencies estimated from the data. To ameliorate the potential effects of time-dependent molecular rates, an uncorrelated relaxed clock model was used for both partitions, which allows evolutionary rate to vary across branches in the genealogy. Demographic history was reconstructed using the extended Bayesian skyline model of Heled and Drummond (32), which models population size as a series of piecewise linear functions, with the number of change points estimated from the data.
Initial MCMC runs mixed poorly, prompting us to adopt a metropolis-coupled MCMC scheme as implemented in BEAST-MC3. Eight simultaneous chains were run, with swapping of chains every 100 steps and a δ-heating parameter of 0.003. Chains were run for 5 × 107 steps, with sampling of parameters every 5,000 steps. The initial 5 × 106 steps were discarded as burn-in. MCMC convergence was assessed by examination of parameter traces, effective sample sizes, and similarity of multiple repeated runs. The complete BEAST input file is available on request from the corresponding author. Visualization of the extended Bayesian skyline plot was constructed with in-house software, available on request from the corresponding author.
To help improve mixing characteristics we specified several informative priors. First, the total height of the genealogy was restricted to between 20,000 and 150,000 y. Although this restriction likely did not significantly impact the shape of the estimated posterior (whose mean was roughly 40,000), it facilitated mixing by preventing the chains from occasionally sampling extremely deep trees. Second, because all ancient sequences were from HVS1, relatively little calibration information was available for the coding region. We therefore specified as a prior on the mean evolutionary rate of the coding region a Γ-distribution with shape 2 and scale 8 × 10−9 (mean 1.6 × 10−8), values picked on the basis of previous examination of the coding region mutation rate (e.g., refs. 33 and 34). Finally, we increased the expected number of population change points from 0.69 (the default value) to 2.0, in accordance with our prior expectation that population size has changed multiple times during the period under consideration.
Supplementary Material
Acknowledgments
B.D.O. thanks Joe Felsenstein, Mary Kuhner, and the members of the Felsenstein/Kuhner laboratory for helpful discussions of the results and manuscript, and National Science Foundation Postdoctoral Fellowship DBI-0906018 for funding. L.F.-S. thanks the German Research Foundation (DFG) for funding (Grant FE 1161/1-1). We also thank M. Reindel (German Archaeological Institute, DAI) and H. Gorbahn (University of Kiel) for supplying samples and archaeological information.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.R.R. is a guest editor invited by the Editorial Board.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. JF904865 and JF904866).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112563108/-/DCSupplemental.
References
- 1.Dobyns HF. Disease transfer at contact. Annu Rev Anthropol. 1993;22:273–291. [Google Scholar]
- 2.Crosby AW. Ecological Imperialism: The Biological Expansion of Europe. New York: Cambridge Univ Press; 2004. [Google Scholar]
- 3.Livi-Bacci M. The depopulation of Hispanic America after the conquest. Popul Dev Rev. 2006;32:199–232. [Google Scholar]
- 4.Larsen CS. In the wake of Columbus: Native population biology in the postcontact Americas. Am J Phys Anthropol. 1994;37:109–154. [Google Scholar]
- 5.Thornton R. Aboriginal North American population and rates of decline, c.a. AD 1500-1900. Curr Anthropol. 1997;38:310–315. [Google Scholar]
- 6.Bolnick DA, Smith DG. Unexpected patterns of mitochondrial DNA variation among Native Americans from the southeastern United States. Am J Phys Anthropol. 2003;122:336–354. doi: 10.1002/ajpa.10284. [DOI] [PubMed] [Google Scholar]
- 7.Wang N, et al. Genetic signatures of pre-expansion bottleneck in the Choctaw population of Oklahoma. Am J Phys Anthropol. 2004;124:373–379. doi: 10.1002/ajpa.10363. [DOI] [PubMed] [Google Scholar]
- 8.Shook BA, Smith DG. Using ancient mtDNA to reconstruct the population history of northeastern North America. Am J Phys Anthropol. 2008;137:14–29. doi: 10.1002/ajpa.20835. [DOI] [PubMed] [Google Scholar]
- 9.Atkinson QD, Gray RD, Drummond AJ. mtDNA variation predicts population size in humans and reveals a major Southern Asian chapter in human prehistory. Mol Biol Evol. 2008;25:468–474. doi: 10.1093/molbev/msm277. [DOI] [PubMed] [Google Scholar]
- 10.Mulligan CJ, Kitchen A, Miyamoto MM. Updated three-stage model for the peopling of the Americas. PLoS ONE. 2008;3:e3199. doi: 10.1371/journal.pone.0003199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho SY, Endicott P. The crucial role of calibration in molecular date estimates for the peopling of the Americas. Am J Hum Genet. 2008;83:142–146. doi: 10.1016/j.ajhg.2008.06.014. author reply 146–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagundes NJ, et al. Mitochondrial population genomics supports a single pre-Clovis origin with a coastal route for the peopling of the Americas. Am J Hum Genet. 2008;82:583–592. doi: 10.1016/j.ajhg.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster EA, editor. Motolinia's History of the Indians of New Spain. Westport, CT: Greenwood Press; 1973. [Google Scholar]
- 14.Dillehay TD, et al. Monte Verde: Seaweed, food, medicine, and the peopling of South America. Science. 2008;320:784–786. doi: 10.1126/science.1156533. [DOI] [PubMed] [Google Scholar]
- 15.Dillehay TD. Monte Verde: A Late Pleistocene Settlement in Chile. Vol 1. Washington, DC: Smithonian Institution Press; 1989. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert MT, et al. DNA from pre-Clovis human coprolites in Oregon, North America. Science. 2008;320:786–789. doi: 10.1126/science.1154116. [DOI] [PubMed] [Google Scholar]
- 17.Hey J. On the number of New World founders: A population genetic portrait of the peopling of the Americas. PLoS Biol. 2005;3:e193. doi: 10.1371/journal.pbio.0030193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ray N, et al. A statistical evaluation of models for the initial settlement of the American continent emphasizes the importance of gene flow with Asia. Mol Biol Evol. 2010;27:337–345. doi: 10.1093/molbev/msp238. [DOI] [PubMed] [Google Scholar]
- 19.Whitlock MC, Barton NH. The effective size of a subdivided population. Genetics. 1997;146:427–441. doi: 10.1093/genetics/146.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva WA, Jr, et al. Mitochondrial genome diversity of Native Americans supports a single early entry of founder populations into America. Am J Hum Genet. 2002;71:187–192. doi: 10.1086/341358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fagundes NJ, Kanitz R, Bonatto SL. A reevaluation of the Native American mtDNA genome diversity and its bearing on the models of early colonization of Beringia. PLoS ONE. 2008;3:e3157. doi: 10.1371/journal.pone.0003157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon EJ. Human colonization of the Americas: Timing, technology and process. Quat Sci Rev. 2001;20:277–299. [Google Scholar]
- 23.Rothhammer F, Dillehay TD. The late Pleistocene colonization of South America: An interdisciplinary perspective. Ann Hum Genet. 2009;73:540–549. doi: 10.1111/j.1469-1809.2009.00537.x. [DOI] [PubMed] [Google Scholar]
- 24.Dillehay TD. Probing deeper into first American studies. Proc Natl Acad Sci USA. 2009;106:971–978. doi: 10.1073/pnas.0808424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bandelt HJ. Mosaics of ancient mitochondrial DNA: Positive indicators of nonauthenticity. Eur J Hum Genet. 2005;13:1106–1112. doi: 10.1038/sj.ejhg.5201476. [DOI] [PubMed] [Google Scholar]
- 26.Bolnick DA, Smith DG. Migration and social structure among the Hopewell: Evidence from ancient DNA. Am Antiq. 2007;72:627–644. [Google Scholar]
- 27.Fehren-Schmitz L, Reindel M, Cagigao ET, Hummel S, Herrmann B. Pre-Columbian population dynamics in coastal southern Peru: A diachronic investigation of mtDNA patterns in the Palpa region by ancient DNA analysis. Am J Phys Anthropol. 2010;141:208–221. doi: 10.1002/ajpa.21135. [DOI] [PubMed] [Google Scholar]
- 28.Fehren-Schmitz L, et al. Diachronic investigations of mitochondrial and Y-chromosomal genetic markers in pre-Columbian Andean highlanders from South Peru. Ann Hum Genet. 2011;75:266–283. doi: 10.1111/j.1469-1809.2010.00620.x. [DOI] [PubMed] [Google Scholar]
- 29.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 32.Heled J, Drummond AJ. Bayesian inference of population size history from multiple loci. BMC Evol Biol. 2008;8:289. doi: 10.1186/1471-2148-8-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kivisild T, et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishmar D, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.