Abstract
New drugs for preserving and restoring pancreatic β-cell function are critically needed for the worldwide epidemic of type 2 diabetes and the cure for type 1 diabetes. We previously identified a family of neurogenic 3,5-disubstituted isoxazoles (Isx) that increased expression of neurogenic differentiation 1 (NeuroD1, also known as BETA2); this transcription factor functions in neuronal and pancreatic β-cell differentiation and is essential for insulin gene transcription. Here, we probed effects of Isx on human cadaveric islets and MIN6 pancreatic β cells. Isx increased the expression and secretion of insulin in islets that made little insulin after prolonged ex vivo culture and increased expression of neurogenic differentiation 1 and other regulators of islet differentiation and insulin gene transcription. Within the first few hours of exposure, Isx caused biphasic activation of ERK1/2 and increased bulk histone acetylation. Although there was little effect on histone deacetylase activity, Isx increased histone acetyl transferase activity in nuclear extracts. Reconstitution assays indicated that Isx increased the activity of the histone acetyl transferase p300 through an ERK1/2-dependent mechanism. In summary, we have identified a small molecule with antidiabetic activity, providing a tool for exploring islet function and a possible lead for therapeutic intervention in diabetes.
Keywords: differentiation, glucose-stimulated insulin secretion, human islets, p300
Nearly one-third of the adult population of the United States is at risk for type 2 diabetes because of abnormal glucose tolerance or abnormally high fasting glucose (1). Type 2 diabetes is now found not only in adults but also in chronically overweight children. Pancreatic β cells play a unique role in glucose homeostasis by secreting insulin when the concentrations of glucose and other nutrients in the circulation rise (2). Insulin facilitates proper nutrient utilization and storage by most tissues. β cells are vulnerable to persistent nutrient excess from secretory stress. During the development of type 2 diabetes, pancreatic β cells become progressively unable to produce and secrete sufficient insulin to prevent hyperglycemia (1, 3, 4). Identifying strategies to maintain euglycemia is essential to limit diabetes and its destructive consequences (5, 6).
Nutrients regulate insulin production at several steps in the biosynthetic pathway in addition to its secretion, including cleavage of the preprohormone, translation, and transcription (7–11). The immediate events to replenish secreted insulin involve translation of preexisting mRNA and hormone processing. On a longer time scale, new insulin gene transcription maintains the pool of mRNA for translation on demand.
Insulin gene transcription is regulated by the cooperation of a group of glucose-sensitive transcription factors expressed in a tissue-restricted manner (12, 13). Among the most important of these transcription factors are neurogenic differentiation 1 (NeuroD1, also known as BETA2), pancreatic and duodenal homeobox 1 (PDX-1), and v-maf musculoaponeurotic fibrosarcoma oncogene homolog A (MafA), which activate the insulin gene promoter synergistically and are essential for glucose-stimulated insulin gene transcription. Mutations in PDX-1 and NeuroD1 have been linked to maturity-onset diabetes of the young (MODY) and are classified as MODY4 and MODY6 genes, respectively (14, 15).
NeuroD1 is required for the development of neurons and neuroendocrine cells in other organs including lung, intestine, and pancreas (16, 17). In the adult, NeuroD1 functions primarily in pancreatic β cells and in continued neurogenesis in hippocampal CA1 neurons. Neurogenins (Ngn) are direct transcriptional regulators of NeuroD1 during neuronal and pancreatic development (18, 19). Although Ngn1 and 2 act exclusively in neuronal lineages, Ngn3 induces NeuroD1 expression in pancreatic β cells. Expression of one or more of these factors helps promote differentiation of pancreatic endocrine cells from various stem cell populations (6, 20).
We previously identified a family of 3,5-disubstituted isoxazoles (Isx) by screening a chemical library in mouse pluripotent stem cells for activators of the gene encoding the homeodomain transcription factor, NK2 transcription factor-related, locus 5 (Nkx2.5) (21). We discovered in collateral studies that Isx also had strong neurogenic activity in several types of neural progenitor cells (22). This activity was mediated in part through induction of NeuroD1 expression.
Because of the importance of NeuroD1 in the development of the pancreas and in insulin production in pancreatic β cells, we examined effects of Isx on the properties of β cells. We find that this molecule increases insulin production and restores insulin production by human islets following long-term ex vivo culture. We provide the initial characterization of the changes elicited by Isx to improve the essential behaviors of β cells.
Results
Isx Increases Glucose-Induced Insulin Secretion and Enhances Expres-sion of Factors Important for Insulin Gene Transcription in Human Islets.
We examined the effect of Isx on the function of β cells within human islets maintained in culture for up to 1 y. Based on earlier studies and concentration effects shown later (21, 22), most studies used 20 or 40 μM Isx. In addition to β cells, islets contain other cell types, including a, δ, and γ or pp cells which secrete glucagon, somatostatin, and pancreatic polypeptide, respectively (23). Shared and cell-specific transcription factors mediate their distinct nutrient-regulated hormone secretions, and their interplay is important for islet function. After months in culture, islets display reduced expression of β-cell–restricted transcription factors and become less able to secrete insulin in response to glucose (24, 25).
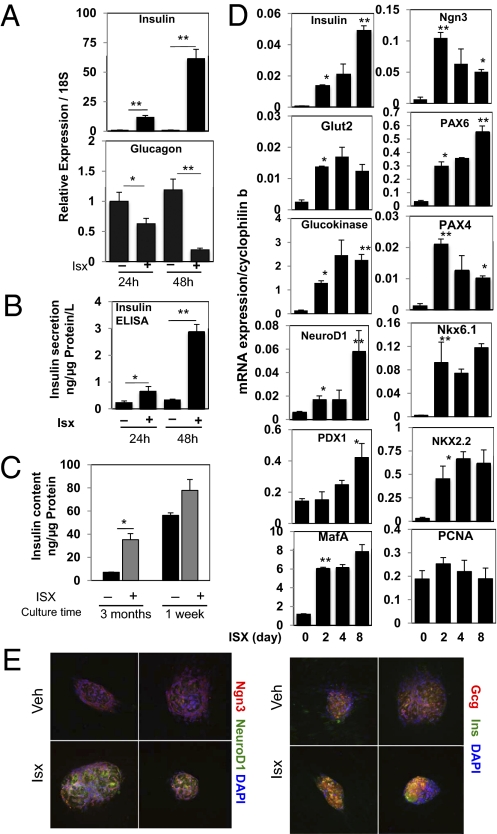
Treatment of 6-mo-old human islets with Isx for 1–2 d induced a large increase in preproinsulin mRNA and an 80% decrease in glucagon mRNA (Fig. 1A). Insulin secreted from these islets over a 24-h period also was increased markedly (Fig. 1B). To determine the relative significance of these changes, we compared the effects of Isx on human islets cultured for 3 mo and on newly obtained human islets. Insulin content in the 3-mo-old islets was increased ∼10-fold by Isx, to a value nearly 75% of the insulin content of islets isolated only 1 wk earlier (Fig. 1C). In addition, a 2-d exposure to Isx increased the insulin content of the fresh islets by nearly 25%. Islets in culture for 2 mo and treated with Isx for 2 d revealed an obvious increase in Ngn3, NeuroD1, and insulin immunostaining (Fig. 1E).
Fig. 1.
Isx induces expression of insulin and transcription factors in primary cultured human islets. (A) Primary human islets cultured in RPMI 160 for 6 mo were treated with vehicle or 40 μM Isx for 1 or 2 d. Expression of human insulin and glucagon mRNA was assessed. (B) Insulin secretion measured by ELISA over 24 h from islets treated with Isx or vehicle for 1 or 2 d. (C) Total insulin content of islets cultured for 3 mo and fresh islets, each treated for 48 h with Isx or DMSO. (D) Time course of pancreatic gene induction in islets cultured for 1 y and treated with DMSO for 8 d or with 40 μM Isx for 2, 4, or 8 d. (E) Immunohistochemical staining of insulin, NeuroD1, Ngn3, and MafA of islets cultured for 2 mo and treated with Isx or vehicle for 48 h. Nuclei were stained with DAPI. Vehicle- and Isx-treated samples were compared using Student's t test (*P < 0.05; **P < 0.01).
A time course of Isx action on mRNAs in islets cultured for 1 y showed that preproinsulin mRNA increased by 10-fold at 24 h and by 50-fold after several days of Isx exposure (Fig. 1D). Both glucokinase and glucose transporter 2 (Glut2) mRNAs increased, consistent with the greater glucose responsiveness. Along with NeuroD1, we examined the expression of transcription factors associated with insulin gene transcription and differentiation of β cells. The majority displayed one of two expression patterns temporally. Several transcription factors increased throughout the time course or increased until reaching a plateau; these included insulin, NeuroD1, MafA, PDX-1, paired box gene 6, Nkx6.1, Nkx2.2, forkhead box A2 (Foxa2), hepatic nuclear factor (Hnf) 6, Hnf1α, Hnf1β, Hnf4α, and islet 1 (Isl1) (Fig. 1D and Fig. S1B) (26–30). Others, which increased rapidly and then decreased at longer times, included Ngn3 and paired box gene 4 (Pax4). Increases in PDX-1, the Hnfs, and Isl1 were relatively small, but increases in the other factors generally were more than 10-fold. Neither proliferating cell nuclear antigen, an indicator of proliferation, nor octamer-binding transcription factor 4, a factor associated with stem cells, showed consistent or substantial changes in mRNA expression.
Isx Effects on MIN6 β Cells.
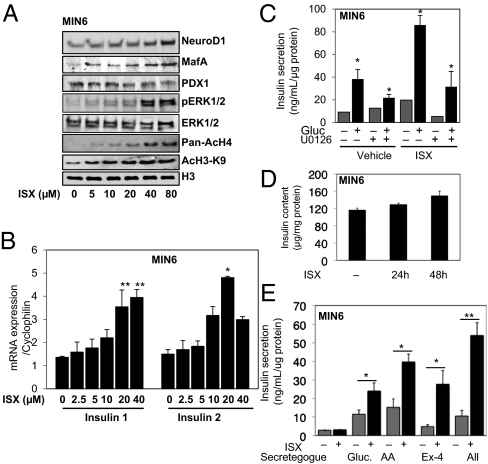
We next asked if Isx induced similar changes in isolated ΜΙΝ6 β cells. After 24 h, immunoreactive NeuroD1 increased at all concentrations of Isx tested (Fig. 2A). MafA expression also increased, but there was little change in PDX-1.
Fig. 2.
Isx activates insulin expression in MIN6 cells. (A) MIN6 cells were treated with 5–80 μM Isx for 24 h or with DMSO (0.08%). Immunoblots of NeuroD1, MafA, PDX-1, pERK1/2 (pT183/pY185), acetylated histones AcH4-K5K8K12K16 and acetylated histone 3 lysine 9 (AcH3K9), and total histone H3 in 40 μg of cell lysate. (B) Quantitative RT-PCR of insulin 1 and insulin 2 gene expression in MIN6 cells treated with Isx for 24 h as in A. (C) GSIS in 15 min from MIN6 cells pretreated for 48 h with Isx or vehicle with or without the MEK1/2 inhibitor U0126 for 24 h. (D) Insulin content in MIN6 cells treated with Isx for 24 or 48 h. (E) Insulin secretion induced by glucose, amino acids (AA), and exendin-4 (Ex-4) singly or in combination (All) in MIN6 cells pretreated with Isx for 48 h. Comparisons were made using Student's t test as in Fig. 1 (*P < 0.05; **P < 0.01).
Isx-enhanced ERK1/2 phosphorylation was detectable after 24 h. A time course of treatment with 20 μM Isx indicated a biphasic effect on ERK1/2, with a small initial activation followed by a slower sustained activation from ∼2 h of Isx exposure (Fig. S2). Acetylation of histone H3 and H4 also was increased by Isx in β cells, as was found in other cell types treated with this compound (21). These changes were accompanied by as much as a fourfold increase in preproinsulin mRNA (Fig. 2B).
Glucose-stimulated insulin secretion (GSIS) was increased nearly fourfold by Isx pretreatment (Fig. 2C), with only a small increase in insulin content (Fig. 2D). The increase in GSIS was partially blocked by inhibiting ERK1/2 activation using a MAP kinase/extracellular signal-regulated kinase kinase (MEK) inhibitor. Follow-up experiments showed that Isx increased stimulation of insulin secretion by glucose, amino acids, and exendin-4 (a long-acting glucagon-like peptide 1 agonist), individually and in combination, by two- to fivefold (Fig. 2E). The calcineurin inhibitor FK506, which blocks ERK1/2 activation (31), the PI3-kinase inhibitor wortmannin, and nifedipine, an inhibitor of L-type voltage-dependent calcium channels, together with the chelator 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA) all reduced the increase in GSIS caused by Isx (Fig. S1E).
Epigenetic Mechanisms Contributing to Isx Action.
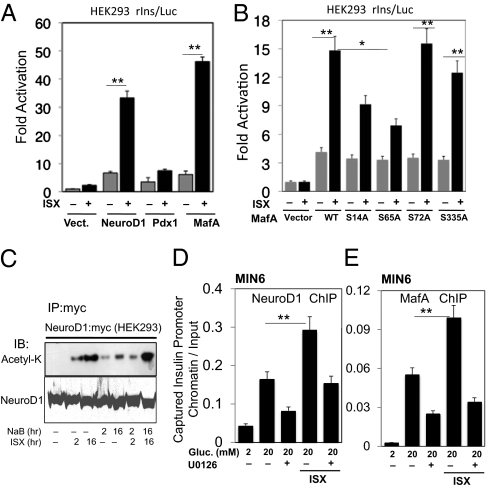
We used reporter assays in MIN6 and HEK-293 cells to determine if the activities of factors that were induced also were impacted by Isx. Insulin gene reporter activity was increased by fourfold or more by NeuroD1 or MafA in the presence of Isx (Fig. 3A and Fig. S3). Activity caused by PDX-1 was increased by twofold or less by Isx. Comparable or greater chromatin binding of NeuroD1 and MafA to the insulin gene promoter was caused by Isx and was inhibited by blocking ERK1/2 (Fig. 3 D and E), as was shown previously for glucose-stimulated binding of these factors (31). Isx also caused acetylation of NeuroD1 more strongly than the histone deacetylase (HDAC) inhibitor sodium butyrate (Fig. 3C). Although NeuroD1 is an ERK1/2 substrate (32), we have been unable to show that MafA is an ERK1/2 substrate. MafA regulatory phosphorylation on serine-proline sites by other enzymes has been reported (33, 34). Consistent with a role for serine modification, mutation of serine 65 and, to a lesser extent, serine 14 limited the ability of Isx to enhance MafA transcriptional activity (Fig. 3B).
Fig. 3.
Isx activates transcription factors that regulate insulin gene expression. (A) Insulin promoter reporter activity in HEK-293 cells transfected with NeuroD1, MafA, PDX-1, or (B) transfected with WT or mutant MafA, treated with Isx or vehicle for 24 h. (C) Myc-NeuroD1 was expressed in HEK-293 cells. Myc immunoprecipitates were blotted with anti-acetyl lysine (Acetyl-K) and anti-NeuroD1. (D and E) ChIP of NeuroD1 (D) and MafA (E) binding to the insulin promoter in MIN6 cells after 10 min in glucose. Comparisons were made using Student's t test as in Fig. 1 (*P < 0.05 and **P < 0.01).
Isx Regulates Histone Acetylation by Enhancing Histone Acetyl-transferase Activity.
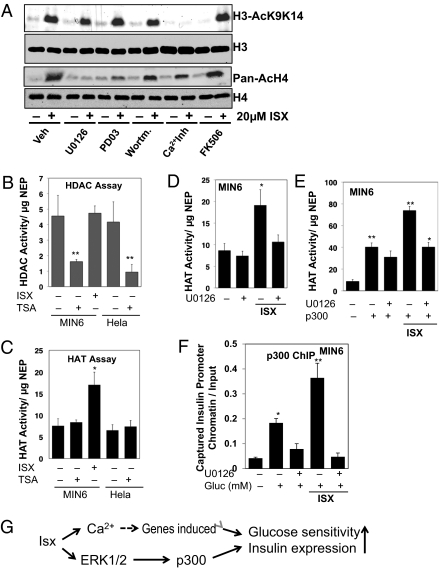
Previously we showed that Isx decreased phosphorylation of HDAC5 and enhanced its nuclear export, possibly accounting for changes in transcription of Nkx2.5 (22). Here we find that, in addition to an effect on the acetylation of NeuroD1, Isx also had a marked effect on the acetylation of histones (Fig. 4A). Nifedipine, a calcium-channel blocker, in combination with the chelating agent BAPTA, decreased acetylation of H3 on lysines 9 and 14 induced by Isx but had little effect on acetylation of H4. The MEK/ERK pathway inhibitors U0126 and PD325901 inhibited Isx-induced acetylation of H4, as did the calcineurin inhibitor FK506, but had little effect on H3 acetylation. In contrast, the PI3-kinase inhibitor wortmannin had no effect on either of these readouts.
Fig. 4.
Isx is a HAT activator. (A) Immunoblots of acetylated histones in MIN6 cells pretreated with the indicated inhibitors for 16 h and then treated with vehicle or Isx. (B and C) HDAC (B) and HAT (C) activities in 50 μg of nuclear extract protein from MIN6 cells treated with Isx or DMSO for 24 h. Trichostatin A (TSA; 2 μM) was added at as a positive control. HeLa nuclear extract also was included as a positive control. (D and E) HAT activity from nuclear extracts of MIN6 cells without (D) or with (E) expression of p300 for 48 h, pretreated with Isx or 10 μM U0126 for 24 h. (F) ChIP analysis of the association of p300 with the insulin gene promoter in MIN6 cells upon glucose (Gluc) stimulation after pretreatment with Isx and/or U0126 for 24 h. Statistical analysis was performed using Student's t test as in Fig. 1. *P < 0.05; **P < 0.01. (G) Simplified view of the actions of Isx.
To determine the basis for increased histone acetylation in β cells, HDAC and histone acetyltransferase (HAT) activities were measured in nuclear extracts from MIN6 or HeLa cells treated with vehicle or Isx for 24 h (Fig. 4 B and C). HDAC activity in MIN6 cells was not affected by the compound, although it was inhibited by the HDAC inhibitor trichostatin A (35). Units of activity were similar in MIN6 and HeLa nuclear extracts. On the other hand, HAT activity in nuclear extracts from Isx-treated MIN6 cells was increased approximately twofold by Isx. HAT activity in the Isx-treated cells was partially dependent on ERK1/2, as suggested by MEK inhibitor sensitivity (Fig. 4D). Effects of Isx on HATs expressed in HEK-293 cells indicated that p300 and cAMP response element-binding (CREB)-binding protein (CBP), but not p300/CBP-associated factor or general control non-derepressible (GCN) 5, activities were increased by a combination of ERK2 expression and Isx but were blocked by expression of kinase-dead ERK2 (Fig. S4). To validate the apparent regulation of p300 by Isx, p300 was expressed in MIN6 cells. Isx increased HAT activity in the nuclear extracts over and above that from expressed p300; U0126 inhibited the Isx-induced increase in HAT activity (Fig. 4E). Chromatin recruitment of p300 to the insulin promoter upon stimulation by glucose was increased by Isx pretreatment in an ERK1/2-dependent manner (Fig. 4F). These findings are consistent with the conclusion that Isx stimulates p300 activity and that Isx-induced changes in histone and transcription factor acetylation are caused at least in part by regulation of p300.
Discussion
Isx increased the expression of transcription factors that enhance β-cell differentiation and control nutrient-responsive insulin gene transcription, resulting in an increase in preproinsulin mRNA and intracellular insulin content. Isx potentiated insulin secretion induced by nutrient and hormonal secretagogues before the increase in intracellular insulin content, indicating that it also adjusts the efficiency of the secretory machinery. The end result is a substantial increase in characteristics essential for mature β-cell function, insulin biosynthesis, and release in response to secretory challenge.
To create these changes in culture-aged islets, Isx induced a phalanx of factors that direct β-cell differentiation. These transcription factors have functions, often overlapping, in β-cell development, and several have overlapping functions in mature β cells as well (28). Ngn3 induces NeuroD1 expression in pancreatic precursors and suppresses cell division through induction of a cyclin-dependent kinase inhibitor (36). Ngn3 arises early in islet differentiation before the distinction between α and β cells; in contrast to some of the other factors, Ngn3 becomes undetectable in the adult pancreas (18, 37). Both NeuroD1 and Ngn3 can drive islet differentiation and induce Nkx2.2 (30, 38, 39); Ngn3 also induces Pax4. Nkx2.2 and Pax4 are required for development of the β-cell lineage (14, 40–42). Along with Foxa2 and PDX-1, Nkx2.2 directly regulates MafA expression (43–46).
Nkx6.1, also downstream of Nkx2.2, is required for the development of β-cell precursors during the secondary wave of β-cell development (47, 48). Nkx6.1 also suppresses glucagon expression, perhaps accounting for repression of glucagon by Isx. Foxa2 also is important in glucose metabolism and insulin secretion, regulating genes including the ATP-sensitive K+ channel subunits, inward rectifier potassium channel Kir6.2 and the sulfonyl urea receptor Sur-1 (49–52). MODY can be caused by mutations in Hnf4α, Hnf1α, and Hnf1β, which also regulate a number of molecules important for glucose sensing and other β-cell functions (53, 54). Among MafA targets are glucokinase, Glut2, the glucagon-like peptide 1 receptor, and prohormone convertase, which processes proinsulin (55). Consistent with the control of these genes by MafA, its overexpression caused a left shift in glucose sensitivity of insulin secretion (55). Induction of these proteins by MafA is likely to contribute to enhanced insulin production and the improved secretory responsiveness of β cells and islets exposed to Isx.
Isx stimulated acetylation of nuclear proteins through its action on acetyltransferases, including p300. p300 has a major impact on β-cell function, including transcription of the insulin gene (56, 57). p300 can acetylate not only histones but also other proteins that support β cells, such as NeuroD1 (58). Acetylation of NeuroD1 may affect both DNA binding and activation functions. Mutations in the Kruppel-like factor 11 (KLF11; MODY 7) revealed an association with early-onset type 2 diabetes (59). KLF11 is activated by p300, as are a large fraction of other MODY genes (60). Thus, activation of p300 is likely to be one of the significant mechanisms of Isx action.
Isx induced biphasic activation of ERK1/2 that is temporally distinct from that induced by nutrients. Several glucose-sensitive insulin gene transcription factors are regulated by ERK1/2. Although MafA apparently is not an ERK1/2 substrate, chromatin binding of MafA along with NeuroD1 and PDX-1 is ERK1/2 dependent (31, 61). ERK1/2 also phosphorylate p300 and control its activity and chromatin association, perhaps explaining the decrease in Isx-enhanced p300 activity caused by blocking ERK1/2 (62, 63). Loss of p300 from the insulin gene promoter is accompanied by loss of these three essential factors, suggesting that an ERK1/2-regulated event is acetylation, which is thought to control access of the three factors to the proximal promoter.
Several strategies have been described for generating β cells from stem cells, pancreatic duct cells, and other differentiated cell types (6). The majority of these strategies involved the heterologous expression of groups of transcription factors or staged groups of hormonal factors that induce β-cell differentiation (64–68). A small molecule was identified that induced pancreatic progenitors from embryonic stem cells (69). Compounds that enhanced β-cell proliferation also have been reported (70). The plant alkaloid, conophylline, can induce β-cell differentiation from rat pancreatic acinar cells and fetal pancreatic tissue over 3–6 wk (71). It has a slower time course of action and a larger effect on PDX-1, suggesting mechanisms of action distinct from Isx.
In conclusion, Isx is among relatively few single molecules known that can improve β-cell function dramatically. Preliminary experiments in mice suggest that the compounds have little toxicity. Because they require micromolar concentrations to exert their effects, the current compounds may not be useful in patients but nevertheless are valuable aids to develop noninvasive ap-proaches to improve β-cell function.
Experimental Procedures
Materials.
Antibody sources are as follows: ERK1/2 and pERK1/2 as described (61); NeuroD1 (N-19), cMaf (M-153), PDX-1 (N-18), and p300 (C-20) were from Santa Cruz Biotechnology; pan-AcH4 K5K8K12K16 (06-866), AcH3K9 (07-352), AcH3K14 (07-353), and AcH3K9K14 (07-353) were from Upstate/Millipore; acetyl lysine (catalog no. 9441) was from Cell Signaling Technology; Myc was from the National Cell Culture Center. Inhibitors were from the following sources: U0126 and FK506 were from LC Laboratories; PD0325901 was from Stemgent; nifedipine, BAPTA was from Calbiochem; wortmannin and trichostatin A were from Sigma; 3.5-disubstituted Isx was described previously (Fig. S1A) (21, 22).
Cell Culture and Treatments.
MIN6 cells were cultured in DMEM (Gibco), containing 25 mM glucose, 10% FBS, 10 mM Hepes (pH 7.4), 10.2 mM l-glutamine, 50 mM sodium pyruvate, 2.5 mM β-mercaptoethanol, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in 10% CO2. Human islets were obtained from the Islet Cell Resource Basic Science Islet Distribution and from the University of Alabama, Birmingham Islet Resource Facility and were cultured in RPMI 1640 with 11 mM glucose for up to 1 y The medium was changed twice weekly, and the islets were subcultured when they reached 90% confluence. For Isx treat-ments, cells were incubated in medium containing either Isx or the equivalent volume of DMSO vehicle (0.04%). To measure GSIS, cells were placed for 2 h in Krebs–Ringer bicarbonate/Hepes containing 0.1% BSA and 2 mM glucose and were stimulated for 15 min with 20 mM glucose, a 1× amino acid mixture (concentrations as in DMEM), or 50 nM exendin-4. U0126 (10 μM), nifedipine (3 μM), BAPTA (10 μM), wortmannin (0.5 μM), or FK506 (0.1μM) was added for 1–24 h as indicated.
DNA Constructs.
Rat insulin 1 promoter-luciferase reporter construct (pGL3-rIns -410, +1) was described previously (31). Vectors encoding MafA:myc, PDX-1:myc and NeuroD1:myc were from Michael German (University of California, San Francisco, CA); p300 was from Joseph Garcia (University of Texas Southwestern Medical Center, Dallas, TX). MafA:myc mutants were generated by Quik-Change mutagenesis (Agilent Technologies).
ChIP and Quantitative PCR.
ChIP was performed as described (61) with the following modifications. Chromatin was cross-linked with 1% formaldehyde and sonicated with a Bioruptor 200 (Diagenode) on ice. ChIP products were analyzed by real-time quantitative PCR as previously described for real-time PCR (61) using the primers 5′-CAGACCTAGCACCAGGG-3′ and 5′-GGACTTTG-CTGTTTGTCCC-3′ to amplify the −157/−50 fragment of the mouse insulin promoter. Results were expressed relative to input and are presented as the mean ± SEM of at least two independent experiments in triplicate.
HAT and HDAC Assays.
Activities were measured in 50 μg of nuclear protein with HAT and HDAC colorimetric assay kits from Biovision Biotechnology.
Gene Expression.
Total RNA was extracted from human islets with TRI reagent (Ambion). cDNA was prepared from total RNA by a mixture of random hexamer- and oligo-dT–primed reverse transcription (iScript; Bio-Rad). Expression of insulin and glucagon mRNAs relative to 18S RNA was evaluated by TaqMan assays (Applied Biosystems). Relative expression of pancreatic factors was determined using Power SYBR Green PCR Master Mix (Applied Biosystems). Primer sequences are given in Tables S1 and S2. SYBR Green and TaqMan probe-based PCR was performed using the ABI 7500 DNA Sequence Detection System with standard fluorescent chemistries and thermal cycling: 50 °C for 2 min, 95 °C for 10 min for one cycle, an additional 40 cycles at 95 °C for 15 s, and then 58 °C for 1 min.
Luciferase Reporter Gene Assays.
HEK-293 cells (0.15 × 106 cells) in triplicate were transfected with pGL3-rIns reporter (0.1 μg per well) (Stratagene) and were cotransfected with 0.5 μg per well of either empty vector (pcDNA3.1) or vectors encoding NeuroD1:myc, MafA:myc or PDX-1:myc using Lipofectamine 2000 (Invitrogen). After 24 h, cells were stimulated with 20 μM Isx or DMSO for another 24 h. Activity was measured using the dual luciferase reporter system (Promega) with Renilla luciferase as internal control.
Statistics.
Experiments with two groups were analyzed for statistical significance using an unpaired two-tailed Student's t test. Error bars show SD unless otherwise stated.
Supplementary Material
Acknowledgments
We thank Michael Lawrence and other members of the M.H.C. laboratory for comments about the data and manuscript, Kathy McGlynn Tucker for technical assistance, and Dionne Ware for administrative assistance. For many of the islets used in these studies special thanks go to the University of Alabama, Birmingham Islet Resource Facility supported by the University of Alabama, Birmingham Comprehensive Diabetes Center, partially supported by National Institutes of Health Grant P60 DK079626 awarded to the UCDC. This work was supported by National Institutes of Health Grants R01 DK55310 and R37 DK34128 and Welch Foundation Grant I1243 (to M.H.C.), by a grant from the American Heart Association Jon Holden DeHaan Cardiac Myogenesis Research Center, and by National Heart, Lung, and Blood Institute Progenitor Cell Biology Consortium Grant U01 HL100401 (to J.W.S.). J.K.O. was supported by National Institute of General Medical Sciences Pharmacological Sciences Training Grant 5-T32 GM007062. E.M.D. was supported by a National Institute of Diabetes and Digestive and Kidney Diseases minority supplement during early parts of this work and later by a postdoctoral fellowship from the Juvenile Diabetes Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118526109/-/DCSupplemental.
References
- 1.Genuth S, et al. Expert Committee on the Diagnosis and Classification of Diabetes Mellitus Follow-up report on the diagnosis of diabetes mellitus. Diabetes Care. 2003;26:3160–3167. doi: 10.2337/diacare.26.11.3160. [DOI] [PubMed] [Google Scholar]
- 2.Newgard CB, McGarry JD. Metabolic coupling factors in pancreatic β-cell signal transduction. Annu Rev Biochem. 1995;64:689–719. doi: 10.1146/annurev.bi.64.070195.003353. [DOI] [PubMed] [Google Scholar]
- 3.Muoio DM, Newgard CB. Mechanisms of disease: Molecular and metabolic mechanisms of insulin resistance and β-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:193–205. doi: 10.1038/nrm2327. [DOI] [PubMed] [Google Scholar]
- 4.Rutter GA, Parton LE. The β-cell in type 2 diabetes and in obesity. Front Horm Res. 2008;36:118–134. doi: 10.1159/000115360. [DOI] [PubMed] [Google Scholar]
- 5.Halban PA, German MS, Kahn SE, Weir GC. Current status of islet cell replacement and regeneration therapy. J Clin Endocrinol Metab. 2010;95:1034–1043. doi: 10.1210/jc.2009-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borowiak M, Melton DA. How to make β cells? Curr Opin Cell Biol. 2009;21:727–732. doi: 10.1016/j.ceb.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Redmon JB, Towle HC, Robertson RP. Regulation of human insulin gene transcription by glucose, epinephrine, and somatostatin. Diabetes. 1994;43:546–551. doi: 10.2337/diab.43.4.546. [DOI] [PubMed] [Google Scholar]
- 8.Xu G, et al. Insulin mediates glucose-stimulated phosphorylation of PHAS-I by pancreatic β cells. An insulin-receptor mechanism for autoregulation of protein synthesis by translation. J Biol Chem. 1998;273:4485–4491. doi: 10.1074/jbc.273.8.4485. [DOI] [PubMed] [Google Scholar]
- 9.Wicksteed B, et al. A cis-element in the 5′ untranslated region of the preproinsulin mRNA (ppIGE) is required for glucose regulation of proinsulin translation. Cell Metab. 2007;5:221–227. doi: 10.1016/j.cmet.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Steiner DF, Park SY, Støy J, Philipson LH, Bell GI. A brief perspective on insulin production. Diabetes Obes Metab. 2009;11(Suppl 4):189–196. doi: 10.1111/j.1463-1326.2009.01106.x. [DOI] [PubMed] [Google Scholar]
- 11.Goodge KA, Hutton JC. Translational regulation of proinsulin biosynthesis and proinsulin conversion in the pancreatic β-cell. Semin Cell Dev Biol. 2000;11:235–242. doi: 10.1006/scdb.2000.0172. [DOI] [PubMed] [Google Scholar]
- 12.Ohneda K, Ee H, German M. Regulation of insulin gene transcription. Semin Cell Dev Biol. 2000;11:227–233. doi: 10.1006/scdb.2000.0171. [DOI] [PubMed] [Google Scholar]
- 13.Aramata S, Han SI, Yasuda K, Kataoka K. Synergistic activation of the insulin gene promoter by the β-cell enriched transcription factors MafA, β2, and Pdx1. Biochim Biophys Acta. 2005;1730:41–46. doi: 10.1016/j.bbaexp.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Habener JF, Stoffers DA. A newly discovered role of transcription factors involved in pancreas development and the pathogenesis of diabetes mellitus. Proc Assoc Am Physicians. 1998;110:12–21. [PubMed] [Google Scholar]
- 15.Vaxillaire M, Froguel P. Monogenic diabetes in the young, pharmacogenetics and relevance to multifactorial forms of type 2 diabetes. Endocr Rev. 2008;29:254–264. doi: 10.1210/er.2007-0024. [DOI] [PubMed] [Google Scholar]
- 16.Kageyama R, Ishibashi M, Takebayashi K, Tomita K. bHLH transcription factors and mammalian neuronal differentiation. Int J Biochem Cell Biol. 1997;29:1389–1399. doi: 10.1016/s1357-2725(97)89968-2. [DOI] [PubMed] [Google Scholar]
- 17.Chae JH, Stein GH, Lee JE. NeuroD: The predicted and the surprising. Mol Cells. 2004;18:271–288. [PubMed] [Google Scholar]
- 18.Huang HP, et al. Regulation of the pancreatic islet-specific gene BETA2 (neuroD) by neurogenin 3. Mol Cell Biol. 2000;20:3292–3307. doi: 10.1128/mcb.20.9.3292-3307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sommer L, Ma Q, Anderson DJ. neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- 20.Gasa R, et al. Proendocrine genes coordinate the pancreatic islet differentiation program in vitro. Proc Natl Acad Sci USA. 2004;101:13245–13250. doi: 10.1073/pnas.0405301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadek H, et al. Cardiogenic small molecules that enhance myocardial repair by stem cells. Proc Natl Acad Sci USA. 2008;105:6063–6068. doi: 10.1073/pnas.0711507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider JW, et al. Small-molecule activation of neuronal cell fate. Nat Chem Biol. 2008;4:408–410. doi: 10.1038/nchembio.95. [DOI] [PubMed] [Google Scholar]
- 23.Steiner DJ, Kim A, Miller K, Hara M. Pancreatic islet plasticity: Interspecies comparison of islet architecture and composition. Islets. 2010;2:135–145. doi: 10.4161/isl.2.3.11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buitrago A, Gylfe E, Hellman B, Idahl LA, Johansson M. Function of microdissected pancreatic islets cultured in a chemically defined medium. I. Insulin content and release. Diabetologia. 1975;11:535–540. doi: 10.1007/BF01222103. [DOI] [PubMed] [Google Scholar]
- 25.Hollande E, Giron BJ, Reyt F, Dutrillaux MC. Insulin secretion by human pancreas cultured for one year. J Physiol (Paris) 1976;72:815–832. [PubMed] [Google Scholar]
- 26.Wilson ME, Scheel D, German MS. Gene expression cascades in pancreatic development. Mech Dev. 2003;120:65–80. doi: 10.1016/s0925-4773(02)00333-7. [DOI] [PubMed] [Google Scholar]
- 27.Lyttle BM, et al. Transcription factor expression in the developing human fetal endocrine pancreas. Diabetologia. 2008;51:1169–1180. doi: 10.1007/s00125-008-1006-z. [DOI] [PubMed] [Google Scholar]
- 28.Oliver-Krasinski JM, Stoffers DA. On the origin of the β cell. Genes Dev. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scearce LM, et al. Functional genomics of the endocrine pancreas: The pancreas clone set and PancChip, new resources for diabetes research. Diabetes. 2002;51:1997–2004. doi: 10.2337/diabetes.51.7.1997. [DOI] [PubMed] [Google Scholar]
- 30.White P, May CL, Lamounier RN, Brestelli JE, Kaestner KH. Defining pancreatic endocrine precursors and their descendants. Diabetes. 2008;57:654–668. doi: 10.2337/db07-1362. [DOI] [PubMed] [Google Scholar]
- 31.Lawrence MC, McGlynn K, Park BH, Cobb MH. ERK1/2-dependent activation of transcription factors required for acute and chronic effects of glucose on the insulin gene promoter. J Biol Chem. 2005;280:26751–26759. doi: 10.1074/jbc.M503158200. [DOI] [PubMed] [Google Scholar]
- 32.Khoo S, et al. Regulation of insulin gene transcription by extracellular-signal regulated protein kinases (ERK) 1 and 2 in pancreatic β cells. J Biol Chem. 2003;278:32969–32977. doi: 10.1074/jbc.M301198200. [DOI] [PubMed] [Google Scholar]
- 33.Benkhelifa S, et al. Phosphorylation of MafA is essential for its transcriptional and biological properties. Mol Cell Biol. 2001;21:4441–4452. doi: 10.1128/MCB.21.14.4441-4452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han SI, Aramata S, Yasuda K, Kataoka K. MafA stability in pancreatic β cells is regulated by glucose and is dependent on its constitutive phosphorylation at multiple sites by glycogen synthase kinase 3. Mol Cell Biol. 2007;27:6593–6605. doi: 10.1128/MCB.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bieliauskas AV, Pflum MK. Isoform-selective histone deacetylase inhibitors. Chem Soc Rev. 2008;37:1402–1413. doi: 10.1039/b703830p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyatsuka T, Kosaka Y, Kim H, German MS. Neurogenin3 inhibits proliferation in endocrine progenitors by inducing Cdkn1a. Proc Natl Acad Sci USA. 2011;108:185–190. doi: 10.1073/pnas.1004842108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwitzgebel VM, et al. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- 38.Gasa R, et al. Induction of pancreatic islet cell differentiation by the neurogenin-neuroD cascade. Differentiation. 2008;76:381–391. doi: 10.1111/j.1432-0436.2007.00228.x. [DOI] [PubMed] [Google Scholar]
- 39.Gu C, et al. Pancreatic β cells require NeuroD to achieve and maintain functional maturity. Cell Metab. 2010;11:298–310. doi: 10.1016/j.cmet.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith SB, Watada H, Scheel DW, Mrejen C, German MS. Autoregulation and maturity onset diabetes of the young transcription factors control the human PAX4 promoter. J Biol Chem. 2000;275:36910–36919. doi: 10.1074/jbc.M005202200. [DOI] [PubMed] [Google Scholar]
- 41.Smith SB, et al. Neurogenin3 and hepatic nuclear factor 1 cooperate in activating pancreatic expression of Pax4. J Biol Chem. 2003;278:38254–38259. doi: 10.1074/jbc.M302229200. [DOI] [PubMed] [Google Scholar]
- 42.Brun T, Gauthier BR. A focus on the role of Pax4 in mature pancreatic islet β-cell expansion and survival in health and disease. J Mol Endocrinol. 2008;40:37–45. doi: 10.1677/JME-07-0134. [DOI] [PubMed] [Google Scholar]
- 43.Anderson KR, White P, Kaestner KH, Sussel L. Identification of known and novel pancreas genes expressed downstream of Nkx2.2 during development. BMC Dev Biol. 2009;9:65. doi: 10.1186/1471-213X-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doyle MJ, Sussel L. Nkx2.2 regulates β-cell function in the mature islet. Diabetes. 2007;56:1999–2007. doi: 10.2337/db06-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raum JC, et al. FoxA2, Nkx2.2, and PDX-1 regulate islet β-cell-specific mafA expression through conserved sequences located between base pairs -8118 and -7750 upstream from the transcription start site. Mol Cell Biol. 2006;26:5735–5743. doi: 10.1128/MCB.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lynn FC, et al. Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc Natl Acad Sci USA. 2007;104:10500–10505. doi: 10.1073/pnas.0704054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sander M, et al. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of β-cell formation in the pancreas. Development. 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- 48.Schisler JC, et al. The Nkx6.1 homeodomain transcription factor suppresses glucagon expression and regulates glucose-stimulated insulin secretion in islet β cells. Proc Natl Acad Sci USA. 2005;102:7297–7302. doi: 10.1073/pnas.0502168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Gauthier BR, Hagenfeldt-Johansson KA, Iezzi M, Wollheim CB. Foxa2 (HNF3β) controls multiple genes implicated in metabolism-secretion coupling of glucose-induced insulin release. J Biol Chem. 2002;277:17564–17570. doi: 10.1074/jbc.M111037200. [DOI] [PubMed] [Google Scholar]
- 50.Lee CS, et al. Foxa2 controls Pdx1 gene expression in pancreatic β-cells in vivo. Diabetes. 2002;51:2546–2551. doi: 10.2337/diabetes.51.8.2546. [DOI] [PubMed] [Google Scholar]
- 51.Lantz KA, et al. Foxa2 regulates multiple pathways of insulin secretion. J Clin Invest. 2004;114:512–520. doi: 10.1172/JCI21149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao N, et al. Foxa2 controls vesicle docking and insulin secretion in mature β cells. Cell Metab. 2007;6:267–279. doi: 10.1016/j.cmet.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 53.Eeckhoute J, Formstecher P, Laine B. Maturity-onset diabetes of the young Type 1 (MODY1)-associated mutations R154X and E276Q in hepatocyte nuclear factor 4alpha (HNF4alpha) gene impair recruitment of p300, a key transcriptional co-activator. Mol Endocrinol. 2001;15:1200–1210. doi: 10.1210/mend.15.7.0670. [DOI] [PubMed] [Google Scholar]
- 54.Gupta RK, et al. The MODY1 gene HNF-4alpha regulates selected genes involved in insulin secretion. J Clin Invest. 2005;115:1006–1015. doi: 10.1172/JCI200522365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H, Brun T, Kataoka K, Sharma AJ, Wollheim CB. MAFA controls genes implicated in insulin biosynthesis and secretion. Diabetologia. 2007;50:348–358. doi: 10.1007/s00125-006-0490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma A, et al. The NeuroD1/BETA2 sequences essential for insulin gene transcription colocalize with those necessary for neurogenesis and p300/CREB binding protein binding. Mol Cell Biol. 1999;19:704–713. doi: 10.1128/mcb.19.1.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiu Y, Sharma A, Stein R. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol Cell Biol. 1998;18:2957–2964. doi: 10.1128/mcb.18.5.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qiu Y, Guo M, Huang S, Stein R. Acetylation of the BETA2 transcription factor by p300-associated factor is important in insulin gene expression. J Biol Chem. 2004;279:9796–9802. doi: 10.1074/jbc.M307577200. [DOI] [PubMed] [Google Scholar]
- 59.Neve B, et al. Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic β cell function. Proc Natl Acad Sci USA. 2005;102:4807–4812. doi: 10.1073/pnas.0409177102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandez-Zapico ME, et al. MODY7 gene, KLF11, is a novel p300-dependent regulator of Pdx-1 (MODY4) transcription in pancreatic islet β cells. J Biol Chem. 2009;284:36482–36490. doi: 10.1074/jbc.M109.028852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lawrence MC, et al. Chromatin-bound mitogen-activated protein kinases transmit dynamic signals in transcription complexes in β-cells. Proc Natl Acad Sci USA. 2008;105:13315–13320. doi: 10.1073/pnas.0806465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen YJ, Wang YN, Chang WC. ERK2-mediated C-terminal serine phosphorylation of p300 is vital to the regulation of epidermal growth factor-induced keratin 16 gene expression. J Biol Chem. 2007;282:27215–27228. doi: 10.1074/jbc.M700264200. [DOI] [PubMed] [Google Scholar]
- 63.Foulds CE, Nelson ML, Blaszczak AG, Graves BJ. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol Cell Biol. 2004;24:10954–10964. doi: 10.1128/MCB.24.24.10954-10964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kobinger GP, et al. Pharmacologically regulated regeneration of functional human pancreatic islets. Mol Ther. 2005;11:105–111. doi: 10.1016/j.ymthe.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 65.Zhang D, et al. Highly efficient differentiation of human ES cells and iPS cells into mature pancreatic insulin-producing cells. Cell Res. 2009;19:429–438. doi: 10.1038/cr.2009.28. [DOI] [PubMed] [Google Scholar]
- 66.D'Amour KA, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 67.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kroon E, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 69.Chen S, et al. A small molecule that directs differentiation of human ESCs into the pancreatic lineage. Nat Chem Biol. 2009;5:258–265. doi: 10.1038/nchembio.154. [DOI] [PubMed] [Google Scholar]
- 70.Wang W, et al. Identification of small-molecule inducers of pancreatic β-cell expansion. Proc Natl Acad Sci USA. 2009;106:1427–1432. doi: 10.1073/pnas.0811848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kawakami M, et al. Promotion of β-cell differentiation by the alkaloid conophylline in porcine pancreatic endocrine cells. Biomed Pharmacother. 2010;64:226–231. doi: 10.1016/j.biopha.2009.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.