Abstract
Long noncoding RNAs (lncRNAs) have important regulatory roles and can function at the level of chromatin. To determine where lncRNAs bind to chromatin, we developed capture hybridization analysis of RNA targets (CHART), a hybridization-based technique that specifically enriches endogenous RNAs along with their targets from reversibly cross-linked chromatin extracts. CHART was used to enrich the DNA and protein targets of endogenous lncRNAs from flies and humans. This analysis was extended to genome-wide mapping of roX2, a well-studied ncRNA involved in dosage compensation in Drosophila. CHART revealed that roX2 binds at specific genomic sites that coincide with the binding sites of proteins from the male-specific lethal complex that affects dosage compensation. These results reveal the genomic targets of roX2 and demonstrate how CHART can be used to study RNAs in a manner analogous to chromatin immunoprecipitation for proteins.
Keywords: chromatin-associated RNAs, chromatin-modifying complexes, RNase H mapping
Generating cellular diversity from genetic information requires the regulatory interplay between cis-acting elements encoded at specific loci in chromatin and trans-acting factors that bind them (1). Although the importance of trans-acting proteins (e.g., transcription factors) has long been appreciated, there is growing interest in the role of long noncoding RNAs (lncRNAs) (2) as factors that can regulate specific chromatin loci. This interest is enhanced by the recent discovery that the majority of eukaryotic genomes are transcribed (3) and that many of the resulting transcripts are developmentally regulated (4) but do not encode proteins. Although the functional scope of these RNAs remains unknown (5–7), several lncRNAs play important regulatory roles at the level of chromatin (8). Determining where these ncRNAs bind on the genome is central to determining their function.
Examples of lncRNAs that influence chromatin include the roX ncRNAs in flies and Xist in mammals, both having well-established roles in dosage compensation (8, 9); Kcnq1ot1 and Air ncRNAs, which are expressed from genomically imprinted loci and affect chromatin silencing (10–13); Evf2, HSR1, and other ncRNAs that positively regulate transcription (14–16); lncRNAs that target the dihydrofolate reductase promoter and the rDNA promoters through triplex formation (17, 18); and the human HOTAIR and HOTTIP lncRNAs, which regulate polycomb-repressed and trithorax-activated chromatin, respectively (19, 20). Dysregulation of several of these lncRNAs has been associated with disease (21, 22). Our understanding of the biochemical roles of these RNAs comes largely from their interactions with specific proteins—insights gained from classical biochemical techniques developed for studying translation and RNA-processing complexes and also more recent technological advances using RNA immunoprecipitation (23) and cross-linking and immunoprecipitation (24–26). These experiments suggest that several lncRNAs specifically interact with chromatin-m odifying machinery and may act as scaffolds for multiple complexes (27) or as targeting modules to direct these complexes to specific chromatin loci (reviewed in refs. 28 and 29). There are various modes by which an RNA can interact with a chromatin locus, including direct interactions with the DNA (through canonical Watson-Crick base pairing or nonconical structures such as triple helices) or indirect interactions mediated through a nascent RNA or protein (28).
Determining the direct functions of lncRNAs requires knowledge of where they act. This requirement motivates the development of technology to generate genomic binding profiles of lncRNAs in chromatin that is analogous to chromatin immunoprecipitation (ChIP) for proteins. Ideally, this technology would (i) provide enrichments and resolution similar to ChIP, (ii) use cross-linking conditions that are reversible and allow for analysis of RNA, DNA, and protein from the same enriched sample, and (iii) provide adequate controls to distinguish RNA targets from the background signal. Although there are several techniques that localize RNAs on chromatin, none fulfill all these criteria. For example, both fluorescence in situ hybridization (FISH) (30) and a related biochemical approach (31), which relies on indirect biotinylation of biomolecules near the target RNA, are important techniques that localize RNAs to genomic loci, but neither has demonstrated high resolution across the genome. The ability of nucleic-acid probes to retrieve lncRNAs from cross-linked extracts has been shown (32), but it is unclear if the signal was RNA-mediated or rather due to direct interactions of the long capture oligos with complementary regions found in nearby DNA. Either way, the efficiency and specificity of these technologies have not allowed the precision required for high-resolution genome-wide profiling.
We report the development of CHART (capture hybridization analysis of RNA targets), a hybridization-based purification strategy that can be used to map the genomic binding sites for endogenous RNAs. We used CHART to purify lncRNAs and their associated protein and DNA targets and to determine the genome-wide localization of roX2 RNA in chromatin. We began by identifying regions of the target RNA available for hybridization to short, complementary oligonucleotides. We then designed affinity-tagged versions of these oligonucleotides to retrieve the target RNA along with its associated factors from reversibly cross-linked chromatin extracts under optimized CHART conditions. By isolating and purifying the CHART-enriched DNA fragments, analogous to ChIP, CHART allows the identification of the genomic binding sites of endogenous RNAs (Fig. 1). These data definitively demonstrate that a lncRNA, roX2, localizes to the same sites across the genome as the chromatin-modifying protein complex with which it is proposed to act. Together, these data demonstrate the utility of CHART as a tool in the study of RNAs.
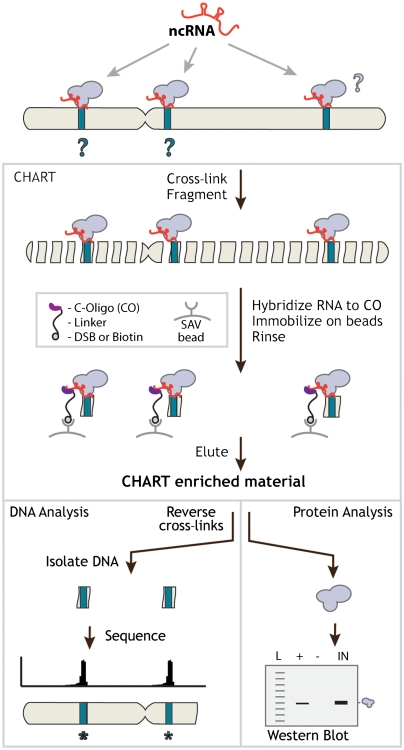
Fig. 1.
CHART is a hybridization-based strategy that uses complementary oligonucleotides to purify the RNA together with its targets from reversibly cross-linked extracts. The cartoon here shows the scenario where the RNA is bound in direct contact with the DNA together with proteins, but other configurations are also possible (see the text). CHART-enriched material can be analyzed in various ways; the two examples depicted here are (Left) sequencing the DNA to determine genomic loci where the RNA is bound and (Right) analyzing the protein content by Western blot analysis.
Results
Design and Development of CHART.
We sought to affinity purify an RNA together with its targets by using oligonucleotides that are complementary to the RNA sequence and developed this technology for roX2, an approximately 600-nt ncRNA that regulates dosage compensation in Drosophila (9). Guided by the success of a chromatin-purification strategy that uses short, affinity-tagged oligonucleotides (C-oligos) to enrich genomic loci through hybridization to DNA in cross-linked extracts (33), we pursued a similar strategy using C-oligos to capture endogenous roX2 RNA along with its associated targets in reversibly cross-linked extracts (Fig. 1).
We first sought to ensure that these C-oligos would target stretches of roX2 RNA available for hybridization and not occluded by protein binding or secondary structure. We adapted an RNase-H mapping assay (34–36) to probe sites on roX2 available to hybridization in the context of cross-linked chromatin extracts. RNase-H specifically hydrolyzes the RNA strand of a DNA-RNA hybrid (37). As RNase-H is not active when exposed to the detergents present in many chromatin extraction procedures, we determined assay conditions ideal for both solubilization of the chromatin and RNase-H mapping (Fig. S1A). Exposing chromatin extracts to 20-mer DNA oligonucleotides one at a time and measuring hybridization to roX2 by sensitivity to RNase-H revealed regions of roX2 that were significantly and reproducibly more available for C-oligo hybridization than others (Fig. S1 B and C). These differences could be due to differences in accessibility of roX2 or to factors independent of roX2, such as other competing sequences in the extract. Because both roX2-dependent and roX2-independent mechanisms that lead to low RNase-H sensitivity could also interfere with efficient hybridization to C-oligos in the context of CHART enrichment, we focused on accessible sites with high RNase-H sensitivity for C-oligo design.
We next sought conditions to specifically enrich roX2 RNA together with its associated targets and tested a range of hybridization conditions and C-oligo chemistries (including O2′-methylated ribonucleotides and locked nucleic acids) on the basis of related applications (33, 35, 38, 39). In these experiments we used desthiobiotin-conjugated C-oligos (Fig. S1D), which allow for gentle biotin elution (33, 40). Determining CHART hybridization conditions required balancing the solubility of the chromatin extract, the stability of duplex formed upon C-oligo binding to RNA, and the stringency required to directly capture only the desired RNA. Using the design illustrated in Fig. S1D, we found that a cocktail of three approximately 25-mer DNA-based C-oligos provided a low background signal and high specific yields of roX2 in a buffer with high ionic strength and high concentrations of denaturants (Fig. 2A). Approximately half of roX2 RNA input could be retrieved from the cross-linked chromatin extract. This enrichment was specific; CHART using a scrambled control C-oligo did not enrich roX2, and control RNAs were not enriched by roX2 CHART. We conclude that DNA-based C-oligos hybridizing to RNase-H–sensitive locations on a target RNA can specifically enrich the RNAs from a cross-linked chromatin extract.
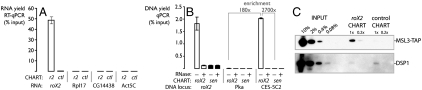
Fig. 2.
CHART allows specific enrichment of roX2 along with its associated targets. (A) Enrichment of RNAs by roX2 CHART (using C-oligos listed in Table S2) as measured by RT-qPCR. (B) Enrichment of DNA loci by roX2 CHART. CES-5C2 is a regulatory site enriched by roX2 CHART. The enrichment values are labeled for comparison of CES-5C2 by roX2 CHART with sense-oligo CHART and also with roX2 CHART at a control site, Pka. RNase-positive lanes represent CHART enrichment from extracts pretreated with RNase to eliminate RNA-mediated signal. Error bars represent ±SEM for three qPCR experiments. Primers are listed in Table S3. (C) Specific enrichment of a tagged MSL subunit, MSL3-TAP, by roX2 CHART. DSP1 antisera (64) is used as a negative control because of its sensitivity.
CHART Enrichment of roX2 Targets.
Having established CHART enrichment of roX2 RNA itself, we tested whether proteins and DNA loci associated with roX2 were also enriched. We first examined candidate genomic sites of roX2 binding. We found that DNA was enriched for both the endogenous roX2 locus and a known regulatory site of dosage compensation, chromatin entry site 5C2 (CES-5C2) (41) but not control sites (Fig. 2B). To test whether the CHART-enrichment of DNA was RNA-dependent, and not an artifact caused by hybridization of the C-oligos with cognate genomic DNA, we treated the extract with RNase prior to C-oligo hybridization. The majority of the enrichment at the endogenous locus (approximately 93%), and essentially all of the enrichment at the trans-acting locus (> 99%), was RNA-mediated (Fig. 2B); only a minority of the DNA enriched at the endogenous roX2 locus (approximately 7%) could be accounted for by direct binding of the C-oligos to DNA. The RNA-mediated enrichment of the regulatory site (CES-5C2) was substantial (> 100-fold over a control locus and > 1000-fold over the sense-oligo control), and the yields (approximately 1–2%) were similar to those retrieved by ChIP.
As further support of the specificity of CHART, a control experiment using sense oligos (therefore not complementary to roX2 RNA) did not enrich the DNA locus where roX2 binds in trans and displayed low levels of enrichment of the endogenous roX2 locus (consistent with the levels of direct DNA binding observed in the RNase control). Also, individual C-oligos were each successful at specifically enriching the appropriate loci (Fig. S2). Using individual C-oligos led to substantially lower yields, however, demonstrating that the cocktail of three C-oligos acts synergistically (Fig. S2).
In addition to DNA, we analyzed the proteins enriched by roX2 CHART and found that a subunit of the male-specific lethal complex (MSL3) was enriched relative to a scrambled control by roX2 CHART (Fig. 2C). The yield of MSL3 protein (approximately 1%) is similar to the enrichment observed for the DNA targets and significantly greater than the yield of a negative control protein, DSP1 (yield < 0.1%), which was not enriched in the roX2 CHART compared with a scrambled control CHART. We conclude that enrichment of roX2 by CHART simultaneously enriches protein and DNA representing roX2 targets, and this enrichment is specific.
Extending CHART to a Mammalian RNA.
Because roX2 CHART successfully enriched roX2-associated targets, we tested whether these same conditions are general for enrichment of other RNAs, including longer mammalian lncRNAs. We applied CHART to endogenous NEAT1 (3.8 kb), a lncRNA found in human cells, and compared the enrichment to another human lncRNA, MALAT1 (> 6.5 kb) in two different cell lines (42–48). Although these lncRNAs are both retained in the nucleus, undergo similar processing, and are encoded next to each other in the genome, they have distinct localizations in the nucleus, NEAT1 localizing to paraspeckles and MALAT1 to nuclear speckles (49, 50). By RNase-H mapping these RNAs from HeLa cells to reveal regions available for hybridization (Fig. S3A) and applying the CHART protocol developed for roX2, we found that both RNAs could be enriched from cross-linked chromatin extracts derived from two human cell lines (Fig. 3A and Fig. S3B). These RNA yields were lower than observed for roX2, which may be due to differences in vetting of C-oligos (only subregions of these RNAs were mapped by RNase-H), shearing of longer RNAs, or in the complexity or age of the chromatin extract. Regardless of the reason for the modest (approximately threefold) differences in RNA yield, both extracts led to similar CHART enrichment of MALAT1- and NEAT1-associated DNA (see below).
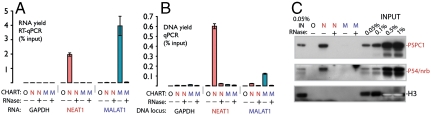
Fig. 3.
NEAT1 CHART, but not MALAT1 CHART, specifically enriches NEAT1 RNA along with its protein and DNA targets. (A) Enrichment of the indicated RNAs from HeLa chromatin extracts by either N, NEAT1 CHART; M, MALAT1 CHART; or O, a mock (no C-oligo) control as measured by RT-qPCR. (B) Similar to A, but enrichment of associated DNA loci as determined by qPCR. Error bars represent ±SEM for three independent CHART experiments. (C) Specific enrichment of two paraspeckle proteins, p54/nrb and PSPC1, by NEAT1 CHART from MCF7 extract. Histone H3 was chosen as a negative control because it is a highly sensitive antiserum and NEAT1 is not expected to be predominantly chromatin bound.
NEAT1 assembles cotranscriptionally with paraspeckle proteins, and fluorescence-imaging experiments suggest that NEAT1 is retained at its endogenous locus (51). Both NEAT1 and MALAT1 CHART demonstrated specific enrichment of their own endogenous genomic loci but not the other’s (Fig. 3B and Fig. S3C). Pretreatment of the extract with RNase abrogates the CHART signal (Fig. 3B and Fig. S3C), demonstrating that CHART enrichment is RNA-mediated. In addition to retrieving the endogenous NEAT1 locus, we expected NEAT1 CHART to enrich paraspeckle proteins. Indeed, we found robust and specific RNA-dependent enrichment of both PSPC1 and p54/nrb (Fig. 3C), two proteins found in paraspeckles that interact with NEAT1 (43, 46, 47). Thus, the analysis of the DNA and proteins enriched by NEAT1 CHART demonstrates that the conditions developed for roX2 CHART also work for a longer endogenous lncRNA from human cells, supporting the generality of CHART.
The observed enrichment of RNAs together with their targets indicate that CHART might be combined with high-throughput sequencing to determine the genome-wide binding profile of an RNA. We tested this conjecture by sequencing the DNA enriched by roX2 CHART to study its genome-wide localization.
Extension of roX2 CHART to Genome-Wide Analysis.
We sequenced the roX2 CHART-enriched DNA to generate a genome-wide binding profile for roX2. roX2 is known to localize to the X chromosome (chrX) (52–54), where it acts together with the MSL complex (including protein subunits MSL1, MSL2, MSL3, MLE, and MOF) (9). The MSL complex affects dosage compensation, at least in part, by regulating acetylation of histone H4 lysine 16 (H4K16) in the bodies of active genes (55–60) and influencing transcriptional elongation (61). Therefore we expected strong enrichment of the roX2 CHART-seq signal on chrX and sought to learn more about roX2 by examining its distribution in comparison with ChIP results for proteins and modifications associated with dosage compensation.
Upon aligning the sequenced reads to the fly genome, the predominant signals from roX2 CHART-seq were a series of intense peaks on chrX (Fig. 4A), consistent with FISH data (52, 53). Some roX2 CHART signals, however, coincide with the peaks in the control sense-oligo profiles (for examples, see Fig. 4A and the autosomal signals in Fig. S4A). We interpret these peaks as sites where the C-oligos directly enrich DNA. When normalized by the sense-oligo control and ordered by significance, the top 173 roX2 CHART peaks were all found on chrX (Fig. S4B). This strong enrichment of roX2 CHART signals on chrX is consistent with the role of roX2 in dosage compensation.
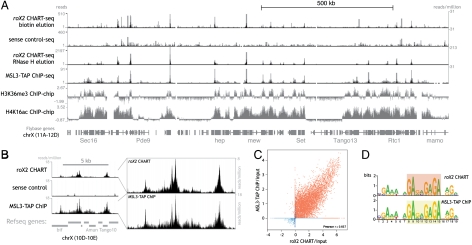
Fig. 4.
roX2 CHART-seq reveals robust enrichment of roX2 on chrX and precise localization to sites of MSL binding. (A) Top four rows, mapped sequencing reads from roX2 and sense-oligo CHART data (performed from S2 cells expressing MSL3-TAP) (55) compared to MSL3-TAP ChIP data from MSL3-TAP Clone 8 (41). Both mapped read numbers and normalized read numbers are listed. Note the RNase-H–eluted roX2 CHART has higher peaks signals at roX2 binding sites and required a different scale than the other three sequencing tracks. Below, ChIP-chip data for the indicated histone modifications are shown (S2 cells, ModENCODE) (65). (B) Finer-scale examples and comparisons of roX2 CHART data, with normalized read depth, except Far Right where normalized for peak height. (C) Correlation between the roX2 CHART signal and MSL3-TAP ChIP signal (41) by plotting the conservative enrichment magnitudes (relative to corresponding inputs) on a log 2 scale of roX2 CHART peaks (from combined RNase-H-elution replicates) and MSL3-TAP ChIP peaks. Peaks from chrX are shown in red and autosomal peaks in blue, but the Pearson r was determined including both sets of peaks. (D) A motif identified from the top roX2 CHART peaks, depicted here as a motif logo in comparison with a nearly identical motif previously determined from MSL3-TAP ChIP-chip data (41).
The enrichment of peaks on chrX was encouraging, but the autosomal signals revealed that CHART eluant contains contaminants from nonspecific hybridization. Many of the artifactual peaks could be filtered by using extra controls and post hoc computational approaches. In this case, setting the appropriate thresholds was viable given our strong expectation of chrX enrichment, but ideally CHART could be performed and interpreted for lncRNAs without such expectations. We sought to minimize retrieval of these contaminants by increasing the biochemical specificity of CHART, thereby increasing the interpretability of the raw mapped CHART reads.
To improve the CHART protocol and minimize purification of products from direct binding of C-oligos to DNA, we removed the heated hybridization step to avoid denaturing the DNA and eluted the roX2 CHART material enzymatically with RNase-H. In this alternative to biotin elution, the DNA bound via roX2 RNA should elute from the resin, but DNA directly bound to the C-oligo should not elute. Because we were no longer using a biotin elution, we used biotinylated rather than desthiobiotinylated C-oligos (Fig. S1E). These modifications maintained the specific enrichment of the endogenous roX2 locus and CES-5C2 (Fig. S4C), leading us to sequence two independent RNase-H–eluted roX2 CHART replicates.
It was immediately evident from the raw, mapped sequencing reads that the RNase-H–eluted CHART samples greatly reduced background from nonspecific hybridization (Fig. 4A). This conclusion is supported by statistical analyses that reveal a decrease in raw autosomal read intensities in comparison to the previous biotin-eluted CHART sample (Fig. S4D). The two RNase-H–eluted CHART samples showed excellent agreement (Fig. S4E), and ordering the peaks by input-normalized significance (i.e., without other CHART controls) demonstrated that the top 214 peaks from these data are on chrX (Fig. S4F).
These data demonstrate that roX2 CHART can be combined with sequencing to map the binding sites of a lncRNA, as exemplified by the robust, RNA-mediated enrichment of a series of sites highly enriched on chrX found by roX2 CHART. To further validate the CHART-seq technology and explore the localization of roX2, we used these data to test at molecular resolution whether roX2 localization coincides with specific features of dosage-compensated chromatin, especially sites bound by the MSL complex.
Analysis of roX2 CHART-seq–Enriched Sites.
The MSL complex is thought to find its binding sites through at least two different mechanisms. Genetic and molecular experiments have revealed a set of 150–300 high-occupancy sites containing a GA-rich sequence motif (41, 62). These sites may act as chromatin entry sites for initial, sequence-specific recognition, followed by spreading to sites on the chrX located in active genes (9). This second class of sites is thought to be recognized through general marks of active transcription, such as H3K36me3, because active autosomal genes can acquire MSL binding when inserted on X (63). Genome-wide CHART of roX2 allowed us to test whether roX2 RNA has the same preference for chromatin entry sites as the MSL complex. When compared to ChIP-chip or ChIP-seq for chromatin modifications associated with dosage compensation (H4K16ac and H3K36me3) or with the ChIP signal observed for a tagged version of MSL3, the roX2 CHART signal was notable for its coincidence with MSL high-occupancy sites (Fig. 4 A and B). The lower significance autosomal signals did not line up with previously proposed MSL binding sites (62) and were not enriched for MSL binding, which suggests they are unlikely to be real roX2 binding sites. Statistical analyses demonstrated that the top roX2 CHART peaks are all enriched for MSL binding (Fig. S5A), and known MSL binding sites have a higher roX2 CHART signal than non-MSL-binding sites (Fig. S5B). Not only does the roX2 CHART signal overlap with the MSL-ChIP signal, but also the datasets correlate (Fig. 4C) and the intense peaks align precisely (Fig. S5C). Inspection of the data also reveals that the CHART signal typically mirrors the contours of the MSL3-ChIP signal (Fig. 4B). These data are consistent with roX2 acting as an integral subunit of the MSL complex while the complex is bound to chromatin.
If roX2 is binding to the same spectrum of chromatin entry sites as MSL3, one prediction is that the locations of roX2 CHART peaks can be used to find a DNA motif associated with roX2 binding, and this motif should be similar to the motif previously derived for sites of MSL3 binding. Indeed, motif analysis of the CHART data for roX2 yields a nearly identical motif to that derived from the ChIP analysis of MSL3 (Fig. 4D). This sequence can attract local MSL activity when inserted onto an autosome (41). In sum, these data demonstrate that CHART allows the determination of the genome-wide binding sites of a ncRNA.
Discussion
Although recent advances have demonstrated the importance of lncRNAs as regulatory factors and revealed that many of these lncRNAs can act in concert with chromatin-modifying machinery, our understanding of where these lncRNAs directly act on chromatin has progressed more slowly. We developed CHART and used it to examine the genomic binding sites of a lncRNA. Because this approach is analogous to ChIP, we present a comparison of these techniques. Depending on the antibody, useful ChIP enrichments range from a fewfold to up to 3 orders of magnitude for the best antisera. roX2 CHART achieves enrichments on the high end of this range, at times exceeding 3 orders of magnitude (Fig. 2B). In theory, the resolution of CHART-seq could have proven significantly worse than ChIP-seq because CHART requires a higher degree of cross-linking. In practice, any loss of resolution observed for CHART-seq is minor as can be seen by comparing MSL3-TAP (where TAP is a tandem affinity purification epitope tag) ChIP-seq signals to roX2 CHART-seq signals (Fig. 4 A and B). Therefore CHART appears similar to ChIP in enrichment and resolution.
The limitations of CHART also overlap with those of ChIP. Neither provides information regarding the stoichiometry of binding at each genomic locus—only enrichment values. Also like ChIP, there is no guarantee that different target loci will be enriched with equal efficiency, because at some loci the C-oligos may have less access (e.g., if they are occluded from binding, similar to epitope masking with ChIP). Given the utility and importance of ChIP despite these caveats, it is reasonable to expect similar utility from CHART. Importantly, both ChIP and CHART provide information about the localization of the factor to chromatin loci but do not reveal the molecular basis of the interaction; CHART-enriched targets could either be directly bound to the RNA or bound through other factors such as bridging proteins or RNAs. We found no evidence that roX2 binding sites are enriched for sequences with Watson-Crick complementarily to roX2 (Table S1), which suggests that the interactions between roX2 and these loci are indirect, very short, or based on non-Watson-Crick interactions.
Also similar to ChIP, CHART-enriched material can be used to examine either candidate genomic loci or genome-wide binding profiles. We applied both to roX2 and found roX2 localized to dosage-compensated regions on chrX, as expected. Comparison of the high-resolution map from roX2 CHART with published data for the MSL complex achieved by using ChIP revealed that roX2 binds at the same sites in chromatin as the MSL complex. Because many lncRNAs are thought to act together with chromatin-modifying machinery, this comparison allowed us to validate the previously untested inference that a lncRNA can act at the same sites on chromatin across the genome as its associated chromatin-modifying complex.
CHART was used successfully for a longer mammalian ncRNA from two different cell lines (Fig. 3 and Fig. S3). Few lncRNAs are known to bind to specific genomic sites, but RNAs can be retained near their endogenous loci, serving as a positive control for CHART enrichment without previous knowledge of trans-acting sites. We found that CHART analysis of endogenous loci can be complicated by the direct DNA binding of the C-oligos, but using RNase-pretreated extract allows this artifactual signal to be distinguished from the desired RNA-mediated CHART signal. Analysis of the RNAs examined here shows that CHART may be successfully applied to RNAs of different lengths and origin.
Despite the successful mapping of genomic binding sites using roX2 CHART-enriched samples, it is not yet clear how roX2 compares to other chromatin-bound lncRNAs in binding mode and stoichiometry, and therefore the generality of CHART will be determined as it is applied to more RNAs. Although the strength of roX2 CHART signals allows them to be easily distinguished from nonspecific background, the use of oligonucleotides as affinity reagents will always raise the potential of direct or indirect off-target hybridization. From analysis of the autosomal biotin-eluted roX2 CHART peaks, we found that particular caution is required when interpreting sharp peaks (< 600 bp) and peaks that contain motifs with homology to the target RNA; this pattern is indicative of likely artifacts and therefore requires further experimentation. In the case of roX2 CHART, the CHART-identified binding sites were not found to have homology to the RNA, which demonstrates that these potential artifacts were avoided.
In addition to locating the genomic targets of an RNA, CHART can also be used to examine other RNA associated factors; we have demonstrated this point by analyzing CHART-enriched material by Western blot for protein targets (Figs. 2C and 3C). Because CHART involves reversible cross-linking, the enriched material can be used for the reciprocal of an RNA-IP; instead of pulling down protein and looking for RNA, CHART allows enrichment of the RNA and examination of which proteins copurify by Western blot. Therefore, although we focused here on the use of CHART to examine DNA targets, CHART-enriched material can also be analyzed for other factors, and we anticipate the extension of CHART to proteomic analyses to uncover RNA-associated proteins.
In summary, we have developed CHART, a technique that allows determination of RNA targets. CHART was successfully applied to lncRNAs of different lengths from two different organisms. We were able to extend CHART to robust genome-wide analysis and from this analysis address the previously untested inference that a lncRNA can act across the genome at the same sites as an associated chromatin-modifying complex. Given the intense interest in the functionality of lncRNAs, including their roles regulating chromatin structure and gene expression, CHART provides a valuable tool to identify the genomic loci directly regulated by an RNA, as exemplified here with roX2.
Materials and Methods
To accomplish CHART enrichment, extract (250 μL, 8 × 107 cell equivalent) was adjusted to hybridization conditions (20 mM Hepes pH 7.5, 817 mM NaCl, 1.9 M urea, 0.4% SDS, 5.7 mM EDTA, 0.3 mM EGTA, 0.03% sodium deoxycholate, 5× Denhardt’s solution) and precleared with ultralink-streptavidin resin (Pierce). C-oligos (800 nM each R2.1–3) were added and hybridized (55 °C for 20 min; 37 °C for 10 min; 45 °C for 60 min; 37 °C for 30 min). The bound material was captured by using streptavidin beads [(MyOne C1; Invitrogen, overnight, room temperature (RT)], rinsed five times with WB250 (250 mM NaCl, 10 mM Hepes pH 7.5, 2 mM EDTA, 1 mM EGTA, 0.2% SDS, 0.1% N-lauroylsarcosine), and eluted with 12.5 mM biotin in WB250 for 1 h at RT. For RNase-pretreated extract, RNase (Roche, DNase-free, 1 μL) was added to the initial extract and allowed to incubate for 10 min at RT prior to adjusting to hybridization conditions. RNase-H–eluted CHART was performed similarly, except we omitted the prebinding to ultralink-streptavidin resin and used higher concentrations C-oligos (1.3 μM each). For the RNase-H elution, the final rinse was with RNase-H rinse buffer (50 mM Hepes pH 7.5, 75 mM NaCl, 3 mM MgCl2, 0.125% N-lauroylsarcosine, 0.025% sodium deoxycholate, 20 u/mL SUPERasIN, 5 mM DTT). The CHART-enriched material was then resuspended in RNase-H rinse buffer (100 μL) and RNase H (10 U) was added. The elution was allowed to proceed for 10 min with gentle shaking at RT. The beads were captured and the reaction stopped with EDTA before proceeding to analyze the CHART-enriched proteins or nucleic acids. For detailed methods, see SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Jose Antao for suggesting cell culture conditions; Daniel Locker for providing the DSP1 antisera; Shangtao Liu, Jerome Dejardin, and members of the Kingston lab for helpful discussions; Sarah K. Bowman for critical reading of this manuscript; and the Szostak Lab for assistance and use of their DNA synthesizer. This research was funded by grants from the National Institutes of Health National Institute of General Medical Sciences (5R01GM043901 to R.E.K. and GM045744 to M.I.K) and a Helen Hay Whitney Foundation postdoctoral fellowship (M.D.S.).
Note.
During revision of this manuscript, another approach was published describing a strategy to map the binding sites of RNAs (66).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data from genome-wide CHART analyses have been deposited at the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE28180).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113536108/-/DCSupplemental.
References
- 1.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 2.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Clark MB, et al. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JT. Lessons from X-chromosome inactivation: Long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelbart ME, Kuroda MI. Drosophila dosage compensation: A complex voyage to the X chromosome. Development. 2009;136:1399–1410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murakami K, Oshimura M, Kugoh H. Suggestive evidence for chromosomal localization of non-coding RNA from imprinted LIT1. J Hum Genet. 2007;52:926–933. doi: 10.1007/s10038-007-0196-4. [DOI] [PubMed] [Google Scholar]
- 11.Mohammad F, et al. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol Cell Biol. 2008;28:3713–3728. doi: 10.1128/MCB.02263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 13.Nagano T, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 14.Feng J, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 16.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 18.Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: An answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 22.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: Regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert C, Svejstrup JQ. RNA immunoprecipitation for determining RNA-protein associations in vivo. Curr Protoc Mol Biol. 2006 doi: 10.1002/0471142727.mb2704s75. Chapter 27: Unit 27 24. [DOI] [PubMed] [Google Scholar]
- 24.Darnell RB. Interdiscip Rev RNA. Vol 1. New York: Wiley; 2010. HITS-CLIP: Panoramic views of protein-RNA regulation in living cells; pp. 266–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ule J, Jensen K, Mele A, Darnell RB. CLIP: A method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 27.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr Opin Genet Dev. 2010;20:142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Levsky JM, Singer RH. Fluorescence in situ hybridization: Past, present and future. J Cell Sci. 2003;116:2833–2838. doi: 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- 31.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 32.Mariner PD, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rinke J, Appel B, Blocker H, Frank R, Luhrmann R. The 5′-terminal sequence of U1 RNA complementary to the consensus 5′ splice site of hnRNA is single-stranded in intact U1 snRNP particles. Nucleic Acids Res. 1984;12:4111–4126. doi: 10.1093/nar/12.10.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wassarman DA, Steitz JA. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol Cell Biol. 1991;11:3432–3445. doi: 10.1128/mcb.11.7.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- 37.Stein H, Hausen P. Enzyme from calf thymus degrading the RNA moiety of DNA-RNA Hybrids: Effect on DNA-dependent RNA polymerase. Science. 1969;166:393–395. doi: 10.1126/science.166.3903.393. [DOI] [PubMed] [Google Scholar]
- 38.Ryder U, Sproat BS, Lamond AI. Sequence-specific affinity selection of mammalian splicing complexes. Nucleic Acids Res. 1990;18:7373–7379. doi: 10.1093/nar/18.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: Requirement of a primer 3′ overhang. Proc Natl Acad Sci USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirsch JD, et al. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: Uses for protein labeling, detection, and isolation. Anal Biochem. 2002;308:343–357. doi: 10.1016/s0003-2697(02)00201-4. [DOI] [PubMed] [Google Scholar]
- 41.Alekseyenko AA, et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clemson CM, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hutchinson JN, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saha S, Datta K, Rangarajan P. Characterization of mouse neuronal Ca2+/calmodulin kinase II inhibitor alpha. Brain Res. 2007;1148:38–42. doi: 10.1016/j.brainres.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 46.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sunwoo H, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2010;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franke A, Baker BS. The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol Cell. 1999;4:117–122. doi: 10.1016/s1097-2765(00)80193-8. [DOI] [PubMed] [Google Scholar]
- 53.Kelley RL, et al. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- 54.Meller VH, et al. Ordered assembly of roX RNAs into MSL complexes on the dosage-compensated X chromosome in Drosophila. Curr Biol. 2000;10:136–143. doi: 10.1016/s0960-9822(00)00311-0. [DOI] [PubMed] [Google Scholar]
- 55.Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gelbart ME, Larschan E, Peng S, Park PJ, Kuroda MI. Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat Struct Mol Biol. 2009;16:825–832. doi: 10.1038/nsmb.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilfillan GD, et al. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev. 2006;20:858–870. doi: 10.1101/gad.1399406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kind J, et al. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell. 2008;133:813–828. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 59.Smith ER, Allis CD, Lucchesi JC. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J Biol Chem. 2001;276:31483–31486. doi: 10.1074/jbc.C100351200. [DOI] [PubMed] [Google Scholar]
- 60.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larschan E, et al. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 2011;471:115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 2008;4:e1000302. doi: 10.1371/journal.pgen.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gorchakov AA, Alekseyenko AA, Kharchenko P, Park PJ, Kuroda MI. Long-range spreading of dosage compensation in Drosophila captures transcribed autosomal genes inserted on X. Genes Dev. 2009;23:2266–2271. doi: 10.1101/gad.1840409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mosrin-Huaman C, Canaple L, Locker D, Decoville M. DSP1 gene of Drosophila melanogaster encodes an HMG-domain protein that plays multiple roles in development. Dev Genet. 1998;23:324–334. doi: 10.1002/(SICI)1520-6408(1998)23:4<324::AID-DVG7>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 65.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chu C, et al. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol Cell. 2011;44:667–648. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.