Abstract
Histone demethylase JHDM1D (also known as KDM7A) modifies the level of methylation in histone and participates in epigenetic gene regulation; however, the role of JHDM1D in tumor progression is unknown. Here, we show that JHDM1D plays a tumor-suppressive role by regulating angiogenesis. Expression of JHDM1D was increased in mouse and human cancer cells under long-term nutrient starvation in vitro. Expression of JHDM1D mRNA was increased within avascular tumor tissue at the preangiogenic switch, along with increased expression of angiogenesis-regulating genes such as Vegf-A. Stable expression of JHDM1D cDNA or siRNA silencing of JHDM1D in cancer cells did not affect cell proliferation, anchorage-independent cell growth, or cell cycle progression in vitro. Notably, JHDM1D-expressing mouse melanoma (B16) and human cervical carcinoma (HeLa) cells exhibited significantly slower tumor growth in vivo compared with the original cells. This reduction in tumor growth was associated with decreased formation of CD31+ blood vessels and reduced infiltration of CD11b+ macrophage linage cells into tumor tissues. Expression of multiple angiogenic factors such as VEGF-B and angiopoietins was decreased in tumor xenografts of JHDM1D-expressing B16 and HeLa cells. Our results provide evidence that increased JHDM1D expression suppressed tumor growth by down-regulating angiogenesis under nutrient starvation.
Keywords: antiangiogenesis, epigenetics, nutrient deprivation, tumor microenvironment
Angiogenesis plays a critical role in tumor progression, invasion, and metastasis (1, 2). Angiogenic factors such as vascular endothelial growth factor (VEGF)-A have been reported to stimulate angiogenesis under hypoxia (3, 4). However, the regulation of angiogenesis under nutrient starvation remains to be clarified. In addition, epigenetic regulation of angiogenesis can be essential but has not been elucidated to date.
Histone demethylase JHDM1D (also known as KDM7A) is a member of the plant homeodomain (PHD) finger protein (PHF) family of PHD- and JmjC domain-containing histone demethylases, and it participates in epigenetic regulation (5). PHF2 recognizes histone trimethyl H3K4 (H3K4me3) through its PHD, and this interaction is essential for histone H3K9me1 demethylation (6). PHF8 demethylates monomethyl H4K20 (H4K20me1) with additional demethylase activities of H3K9me1 and H3K9me2 (7). PHF2 is highly expressed in embryonic neural tube and ganglia (8). PHF8 regulates neural development, and mutations in PHF8 cause X-linked mental retardation (7, 9). JHDM1D demethylates histone H3K9me2 and H3K27me2 (10). Similar to PHF2/PHF8, JHDM1D may regulate neural differentiation and development in mammals (11, 12). However, the mechanism of induction of JHDM1D and its role in tumor progression are unknown.
Antiangiogenic therapy effectively suppresses the growth of solid tumors, and it has been widely used clinically (13–15). However, tumor responses to antiangiogenic therapy are not fully understood. We previously developed a simple model system to maintain cancer cells under hypoxia and nutrient starvation and demonstrated that long-term hypoxia and nutrient starvation may stimulate tumor aggressiveness (16) and affect host cells, causing leukemia in mice (17). Thus, we hypothesized that a tumor microenvironment associated with nutrient starvation can be important for the regulation of tumor progression. In this study, our aim was to elucidate the epigenetic regulation of cancer cells under nutrient starvation to improve antiangiogenic treatment. We examined the role of JHDM1D in tumor progression by using cancer cells stably expressing JHDM1D and found that JHDM1D suppresses tumor growth by regulating, at least in part, tumor angiogenesis under nutrient starvation.
Results
Histone Demethylase JHDM1D Was Highly Expressed Under Nutrition Starvation.
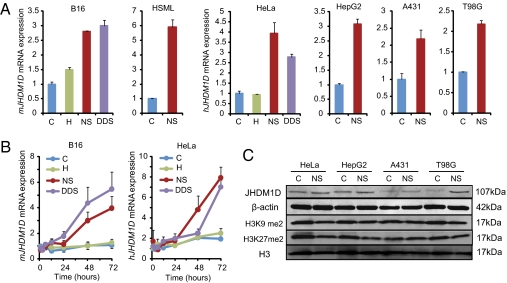
To determine the effect of nutrient starvation on the specific up-regulation of histone demethylase genes, microarray screening was conducted under hypoxia and nutrition starvation. We found that JHDM1D was commonly up-regulated under nutrient starvation, and hypoxia and nutrient starvation double-deprivation stress (DDS) in human cancer cells. To more clearly investigate whether JHDM1D is induced in response to nutrient starvation, we examined the mRNA expression of JHDM1D under normoxia, hypoxia, nutrient starvation, and DDS in mouse (B16 and HSML) and human (HeLa, HepG2, A431, and T98G) cancer cells. JHDM1D mRNA expression was highly up-regulated in response to nutrient starvation and DDS in various cancer cells (Fig. 1A), in addition to normal cells (HLMVEC and THP-1) (Fig. S1A), in vitro. JHDM1D mRNA expression was prominently increased under long-term nutrient starvation (48–72 h) in mouse and human cancer cells (Fig. 1B). Because JHDM1D mRNA expression was increased under nutrient starvation, we examined the protein expression levels of JHDM1D in response to nutrition starvation. JHDM1D expression was increased in response to nutrient starvation in cancer cells (approximately twofold in HeLa, HepG2, and A431 cells and threefold in T98G cells) (Fig. 1C), but the total methylation levels of histone H3K9me2 and H3K27me2 (Fig. 1C) were not significantly decreased.
Fig. 1.
JHDM1D was highly expressed in cancer cells under long-term nutrient starvation. (A) Relative mRNA expression of JHDM1D was increased in response to nutrient starvation in mouse and human cancer cell lines examined by real-time PCR analysis. C, control; H, hypoxia; NS, nutrient starvation; DDS, double-deprivation stress. (B) Long-term exposure of cancer cells to nutrient starvation stimulated mRNA expression of JHDM1D. (C) JHDM1D expression was increased under nutrient starvation in various human cancer cells examined by Western blotting. Cell lysates were obtained in control (C) and nutrient starvation (NS) conditions and were subjected to Western blotting for hJHDM1D, β-actin, histone H3K9 me2, H3K27me2, and H3.
JHDM1D Was Expressed in Avascular Tumor Tissue at the Preangiogenic Switch.
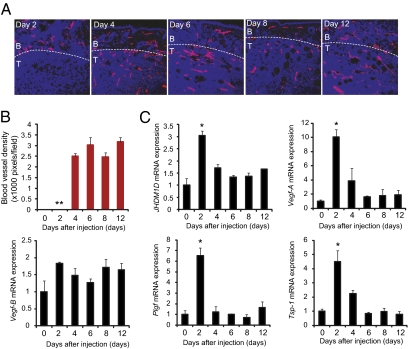
To investigate whether JHDM1D is up-regulated in avascular tumor tissue, mouse uterine cancer (HSML) cells were s.c. inoculated into C57BL/6 mice, and tumor samples for days 2, 4, 6, 8, and 12 (n = 3 for each time point) were prepared. Tumor angiogenesis was evident between days 2 and 4, with the formation of CD31+ blood vessels (Fig. 2 A and B). We found that mRNA expression of JHDM1D was significantly up-regulated in avascular tumor tissue at the preangiogenic switch on day 2 (Fig. 2C). Hypoxia-inducible proangiogenic factors such as Vegf-A (4) were highly expressed in the avascular tumor tissue on day 2 (Fig. 2C). The expression of several proangiogenic genes was not significantly increased in avascular tissue (Fig. 2C and Fig. S1B). Notably, expression of the antiangiogenic factor Tsp-1 was also significantly up-regulated in avascular tissue on day 2. Together, these data indicate that the balance between proangiogenic and antiangiogenic factors is important for the angiogenic switch. The avascular tumor tissue is not only hypoxic but also nutrient-starved, and JHDM1D may play a role at this stage (Fig. 2C).
Fig. 2.
JHDM1D was highly expressed in avascular tumor tissue. (A) The angiogenic switch occurs between days 2 and 4 in a tumor tissue of HSML cells, as determined by immunohistochemical staining of CD31+ blood vessels in tumor tissue. B, border area of tumor; T, tumor tissue. (B) CD31+ blood vessels were not formed within tumor tissue on day 2. Quantitative analysis of CD31+ blood vessels. (C) mRNA expression of JMJD1D and proangiogenic factors were increased in avascular tumor tissue on day 2, as measured by quantitative real-time PCR analysis (*P < 0.05).
Induction of JHDM1D Significantly Suppressed in Vivo Tumor Growth, Although JHDM1D Had a Minor Effect on in Vitro Cell Growth.
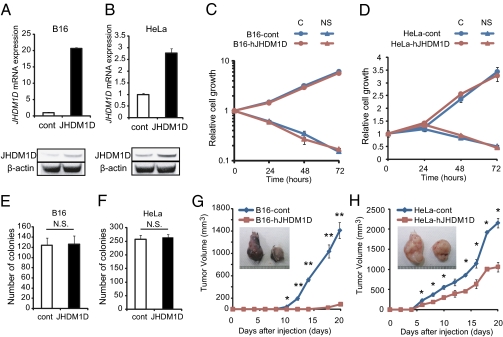
To investigate the role of hJHDM1D in tumor progression, we generated B16 and HeLa cells stably expressing hJHDM1D (B16-hJHDM1D and HeLa-hJHDM1D, respectively) by retroviral transfection (Fig. 3 A and B). The introduction of hJHDM1D into B16 and HeLa cells did not affect cell proliferation under both growth-rich and nutrient-starved conditions in vitro (Fig. 3 C and D). We further examined the fate of JHDM1D-expressing cells under starvation over 72 h. Cell survival or death was not affected for 96 or 120 h, suggesting that there was no significant difference in autophagy between the cells expressing JHDM1D and control cells for 120 h (Fig. S2A). Stable expression of hJHDM1D in B16 and HeLa cells did not affect anchorage-independent cell growth in vitro (Fig. 3 E and F). Consistent with these data, silencing of hJHDM1D by two different siRNAs against hJHDM1D had no effect on the proliferation of HeLa cells under both growth-rich and nutrient-starved conditions (Fig. S2B). In addition, cell cycle histograms of JHDM1D-expressing cells were examined, because a family member, PHF8, has been reported to regulate cell cycle progression (18). JHDM1D-expressing B16 and HeLa cells exhibited minor alterations in cell cycle progression under both growth-rich and nutrient-starved conditions (Fig. S3). Although JHDM1D had only a small effect on in vitro cell growth, JHDM1D may play a role in in vivo tumor growth (Fig. 2). To investigate the role of JHDM1D in in vivo tumor growth, 1 × 107 B16-hJHDM1D and HeLa-hJHDM1D cells were s.c. inoculated into C57BL/6J (n = 8) and C.B17/Icr-scidJcl scid/scid mice (n = 3), respectively. We confirmed that JHDM1D overexpression was maintained in vivo (Fig. S4A). Tumor growth was significantly suppressed in mice inoculated with both B16-hJHDM1D and HeLa-hJHDM1D cells compared with the control cells (Fig. 3 G and H). Conversely, JHDM1D depletion by siRNA significantly stimulated tumor growth in vivo (Fig. S5). In addition, adenoviral infection of mVEGF164 decreased the inhibitory effect of JHDM1D on the tumor growth of HeLa cells in vivo (Fig. S6).
Fig. 3.
Induction of JHDM1D suppressed in vivo tumor growth but played a minor role in cell growth in vitro. (A–D) Stable expression of hJHDM1D in B16 (A) and HeLa (B) cells did not affect cell proliferation in vitro (C and D) under both growth-rich and nutrient-starved conditions. (E and F) hJHDM1D expression in B16 (E) and HeLa (F) cells did not affect the anchorage-independent growth of tumor cells. (G and H) JHDM1D-expressing B16 and HeLa cells suppressed tumor growth in vivo. Tumor volumes of hJHDM1D-expressing B16 (n = 8) (G) and hJHDM1D-expressing HeLa (n = 3) (H) cells compared with the control cells.
JHDM1D Suppressed Tumor Angiogenesis and Infiltration of CD11b+ Cells by Regulating Angiogenic Factors.
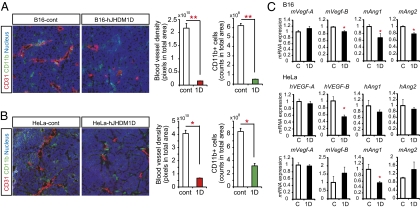
To investigate the tumor-suppressive role of JHDM1D in vivo, immunohistochemical examination was performed on the tumor tissues of B16-hJHDM1D and HeLa-hJHDM1D cells in comparison with empty-vector control cells. The formation of CD31+ blood vessels and infiltration of CD11b+ and F4/80+ macrophage lineage cells were significantly decreased in a tumor xenograft of hJHDM1D-expressing cells (Fig. 4 A and B and Fig. S7A). Blood vessel functionality and coverage of pericytes (αSMA1+) on CD31+ blood vessels were not significantly different between the control and JHDM1D-expressing tumor vasculatures (Fig. S7B), suggesting that JHDM1D participates in antiangiogenic effects. As a result, tumors expressing JHDM1D were more apoptotic than control cells (Fig. S8).
Fig. 4.
JHDM1D decreased tumor angiogenesis and the infiltration of CD11b+ cells in vivo. Immunohistochemical staining revealed that JHDM1D decreased the formation of CD31+ blood vessels and the number of CD11b+ cells in tumors. (A and B) Quantitative analysis of CD31+ blood vessels and CD11b+ cells in B16 (A) and HeLa (B) cells. (C) JHDM1D-expressing cell-xenografted tumor tissues had decreased expression of proangiogenic factors. Expression of proangiogenic factors (VEGF-A, VEGF-B, Ang1, and Ang2) in tumor tissues was measured by quantitative real-time PCR analysis (*P < 0.05).
To investigate whether the antiangiogenic effects of the JHDM1D-expressing tumor were attributable to the regulation of angiogenic factors, the mRNA expression of major angiogenic factors in tumor xenograft samples of hJHDM1D-expressing B16 and HeLa cells were examined. The expression of multiple proangiogenic factors such as VEGF-B, Ang1, and Ang2, as well as other factors, were significantly decreased in tumor samples of hJHDM1D-expressing B16 cells compared with control cells (Fig. 4C and Fig. S9 A and B). The expression levels of tumor (human origin)- or host (mouse origin)-derived angiogenic factors were separately examined using human- and mouse-specific primers against major angiogenic factors in tumor xenograft samples of hJHDM1D-expressing HeLa cells. The expression levels of tumor-derived hVEGF-B, hVEGF-C, hVEGF-D, and hFGF-2, as well as host-derived mAng1, were significantly decreased in tumor xenograft samples of hJHDM1D-expressing HeLa cells compared with that in control cells (Fig. 4C and Fig. S9A), suggesting that JHDM1D plays a role in the regulation of multiple angiogenic factors in the nutrient-starved tumor microenvironment, resulting in the suppression of tumor growth.
Discussion
We have shown that the histone demethylase JHDM1D is highly expressed in various cancer cells in response to nutrient starvation, and that subsequent suppression of solid tumor growth was associated with the down-regulation of several proangiogenic factors such as VEGF-B and angiopoietins.
The ability of cancer cells to adapt to nutrient deprivation is important for tumor progression (19, 20); this ability is partly derived from alterations in tumor metabolism, including an enrichment of glycolysis (Warburg effect) (21, 22). Adaptation of prokaryotic cells to nutrient starvation has been reported as a stringent control in which cells terminate cell proliferation to synthesize required nutrient components (23, 24). Similar control in mammalian cells could be very important, but this control has not yet been identified. We found that JHDM1D, a member of the PHF2/PHF8 histone demethylase family, is highly expressed under nutrient-deprivation conditions in various cancer cell lines and normal cells in vitro (Fig. 1) and in avascular tumor tissue in vivo (Fig. 2). Thus, JHDM1D appears to be important for the epigenetic control of mammalian cells under nutrient starvation.
JHDM1D has been shown to be involved in neural development (11, 12), but its role in tumor progression has not been elucidated. Histone demethylase family proteins such as UTX (also known as KDM6A), JMJD3 (also known as KDM6B), and JMJD5 (also known as KDM8) are involved in tumor progression (25, 26). KDM6A and KDM6B are induced by the human papillomavirus (HPV), and stimulate the expression of cyclin-dependent kinase inhibitor p16INK4A (25). KDM8 demethylases H3K36me2 and modulates cell cycle progression by regulating cyclin A (26). Recently, PHF8 closely related to JHDM1D (5) was shown to regulate cell cycle progression (18). We examined the role of JHDM1D in cell growth, anchorage-independent tumor growth, and cell cycle progression. JHDM1D had a minor effect on cell growth and cell cycle progression, but it significantly suppressed tumor growth in vivo (Fig. 3 and Fig. S3). These data suggest an alternative mechanism of JHDM1D in cancer cells, other than cell proliferation and cell cycle regulation.
Angiogenesis is regulated by multiple proangiogenic and antiangiogenic factors (1, 2). Under hypoxia, stabilization of hypoxia-inducible factor (HIF)-1α promotes angiogenesis by transcriptional up-regulation of VEGF-A and Glut1 (27). Hypoxia alters tumor metabolism (28) and may maintain the stemness of cancer cells (29). On the other hand, few studies have reported the regulation of angiogenesis under nutrient starvation. We previously demonstrated that multiple angiogenic genes such as Ang2 and Tie2 were regulated under hypoxia and nutrient starvation, suggesting an importance of the epigenetic regulation of angiogenesis in the extreme tumor microenvironment (16). Here, we identified JHDM1D as an epigenetic regulator of tumor angiogenesis under nutrient starvation; angiogenesis (i.e., formation of CD31+ blood vessels) and the infiltration of CD11b+ and F4/80+ cells into tumor xenografts of JHDM1D-expressing cells were significantly decreased (Fig. 4 A and B and Fig. S7A). In addition, the expression of several proangiogenic factors, including VEGF-B and angiopoietins, was decreased by the induction of JHDM1D within tumor tissues (Fig. 4C). The expression of JHDM1D decreased the formation of CD31+ blood vessels and the infiltration of CD11b+ and F4/80+ macrophages. JHDM1D expression also induced a delay in the initiation of tumor growth possibly by suppressing the process of angiogenic switch in tumor tissue (Fig. S4B). Thus, JHDM1D plays a negative role in angiogenesis under nutrient starvation, and may lead to tumor mass dormancy until proliferation is reactivated. In the reduced tumor growth of JHDM1D-expressing cells, there are some possibilities such as angiocrine effect (30), reduced dependence on blood vessels (31), and other effects (32).
We found that in response to nutrient starvation, cancer cells commonly up-regulated JHDM1D and suppressed angiogenesis. Continuous up-regulation of JHDM1D in tumor cells using an expression vector could result in effective suppression of tumor progression. On the basis of our results, tumor-oriented promoters or nutrient starvation-dependent promoters/enhancers could be used for small-molecule screening of agents that effectively induce JHDM1D. Following antiangiogenic therapy, up-regulation of JHDM1D could be used as an anticancer strategy agent to improve antiangiogenic effects.
Materials and Methods
Cell Culture and Nutrient Starvation.
Human cervical carcinoma (HeLa), hepatocellular carcinoma (HepG2), epidermoid carcinoma (A431), glioblastoma (T98G), rhabdomyosarcoma (A673), and murine melanoma (B16) cell lines were purchased from the American Type Culture Collection. The murine uterine cancer cell line HSML was kindly provided by Dr. Kudoh (Hirosaki University, Hirosaki, Japan). All cells were maintained in DMEM (Nacalai Tesque) supplemented with 10% FBS at 37 °C in an atmosphere of 5% CO2, excluding HSML cells, which were maintained in RPMI medium 1640 (Nacalai Tesque). Nutrient deprivation was conducted as previously described and as indicated in SI Materials and Methods (16).
Cell Proliferation Assay.
Cell proliferation was measured by the sulforhodamine B assay as previously described (16).
Gene Expression Analysis Using Real-Time PCR.
Total RNA was extracted from cells using the Isogen reagent (Nippon Gene) and converted to cDNA with Prime Script reverse transcriptase (Takara) according to the instructions of the manufacturer and used for quantitative real-time PCR amplification using SYBR Green (Takara) (Table S1).
Western Blotting.
Cell lysates were applied to a 10% or 12.5% polyacrylamide gel and transferred to a poly(vinylidene difluoride) membrane (Invitrogen). The membrane was incubated with rabbit polyclonal anti-JHDM1D antibody, histone dimethyl H3K27 and histone H3 (Abcam), or mouse monoclonal anti-histone dimethyl H3K9 (Abcam) and β-actin antibody (1:1,000; Millipore), followed by horseradish peroxidase-conjugated secondary antibodies (GE Healthcare). Signals were detected using enhanced chemiluminescence detection reagents (GE Healthcare) and were acquired with a luminescent image analyzer (LAS-3000; Fujifilm).
Colony Formation Assay.
Cells were suspended in DMEM containing 0.6% methylcellulose (Wako) supplemented with 10% FBS and were seeded onto six-well plates at a density of 1–2.5 × 104 cells per well in triplicate. After 7–10 d, anchorage-independent colonies were counted using a microscope.
Retroviral Transfection of JHDM1D.
A PMX-puro retroviral vector containing full-length human JHDM1D was transfected by the pantropic platinum-GP retroviral packaging cell line (PlatGP) (33) using Fugene 6 transfection reagent (Roche). Target cells were infected overnight with the vs.v-virus/polybrene-containing supernatant. After infection, the colonies were selected in a medium containing 1.5 μg/mL puromycin.
Animal Studies and Tumor Xenograft.
Murine B16, HSML, or human HeLa cells (1 × 107) were s.c. injected into C57BL/6J or C.B17/Icr-scidJcl scid/scid mice. Tumor volume was measured and analyzed using the Student t test. All animal care procedures were in accordance with institutional guidelines approved by Tokyo Medical and Dental University.
Immunohistochemical Staining and Analysis.
Freshly frozen tumor samples were cut at a thickness of 14 μm by a cryostat (Leica) and were stained with hamster anti-CD31 (BD Biosciences) and rat anti-CD11b (BD Biosciences). Sections were incubated with the appropriate secondary antibody and the nuclear-staining dye To-Pro-3 (Invitrogen), and then analyzed with a confocal microscope (Radiance 2000; Bio-Rad).
Supplementary Material
Acknowledgments
We thank Drs. Y. Kanki and J. Suehiro and the members of the Department of Molecular Oncology, Tokyo Dental and Medical University and Research Center for Advanced Science and Technology, University of Tokyo, for helpful discussions, advice, and support. This work was supported by a Grant-in-Aid Special Project Research on Cancer-Bioscience (17014020) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108462109/-/DCSupplemental.
References
- 1.Ferrara N, Kerbel RS. Angiogenesis as a therapeutic target. Nature. 2005;438:967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 3.Shibuya M, Claesson-Welsh L. Signal transduction by VEGF receptors in regulation of angiogenesis and lymphangiogenesis. Exp Cell Res. 2006;312:549–560. doi: 10.1016/j.yexcr.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Harris AL. Hypoxia—a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 5.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 6.Wen H, et al. Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J Biol Chem. 2010;285:9322–9326. doi: 10.1074/jbc.C109.097667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi HH, et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasenpusch-Theil K, et al. PHF2, a novel PHD finger gene located on human chromosome 9q22. Mamm Genome. 1999;10:294–298. doi: 10.1007/s003359900989. [DOI] [PubMed] [Google Scholar]
- 9.Laumonnier F, et al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J Med Genet. 2005;42:780–786. doi: 10.1136/jmg.2004.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton JR, et al. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol. 2010;17:38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C, et al. Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res. 2010;20:154–165. doi: 10.1038/cr.2010.5. [DOI] [PubMed] [Google Scholar]
- 12.Tsukada Y, Ishitani T, Nakayama KI. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 2010;24:432–437. doi: 10.1101/gad.1864410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurwitz H, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 14.Sandler A, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 15.Escudier B, et al. TARGET Study Group Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356:125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 16.Osawa T, Muramatsu M, Watanabe M, Shibuya M. Hypoxia and low-nutrition double stress induces aggressiveness in a murine model of melanoma. Cancer Sci. 2009;100:844–851. doi: 10.1111/j.1349-7006.2009.01105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osawa T, Tsuchida R, Muramatsu M, Yuasa Y, Shibuya M. Human glioblastoma cells exposed to long-term hypoxia and nutrient starvation stimulated induction of secondary T-cell leukemia in mice. Blood Cancer J. 2011;1:e6. doi: 10.1038/bcj.2011.5. , 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 20.Takasu M, Tada Y, Wang JO, Tagawa M, Takenaga K. Resistance to apoptosis induced by microenvironmental stresses is correlated with metastatic potential in Lewis lung carcinoma. Clin Exp Metastasis. 1999;17:409–416. doi: 10.1023/a:1006632819086. [DOI] [PubMed] [Google Scholar]
- 21.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 23.Sokawa Y, Sokawa J, Kaziro Y. Role of rel gene in translation during amino acid starvation in Escherichia coli. Nature. 1974;249:59–62. doi: 10.1038/249059a0. [DOI] [PubMed] [Google Scholar]
- 24.Shibuya M, Kaziro Y. Studies on stringent control in a cell-free system. Regulation by guanosine-5′-diphosphate-3′-diphosphate of the synthesis of elongation factor Tu. J Biochem. 1979;86:403–411. doi: 10.1093/oxfordjournals.jbchem.a132539. [DOI] [PubMed] [Google Scholar]
- 25.McLaughlin-Drubin ME, Crum CP, Münger K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci USA. 2011;108:2130–2135. doi: 10.1073/pnas.1009933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsia DA, et al. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc Natl Acad Sci USA. 2010;107:9671–9676. doi: 10.1073/pnas.1000401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda E, Achen MG, Breier G, Risau W. Hypoxia-induced transcriptional activation and increased mRNA stability of vascular endothelial growth factor in C6 glioma cells. J Biol Chem. 1995;270:19761–19766. doi: 10.1074/jbc.270.34.19761. [DOI] [PubMed] [Google Scholar]
- 28.Pouysségur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–443. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- 29.Das B, et al. A hypoxia-driven vascular endothelial growth factor/Flt1 autocrine loop interacts with hypoxia-inducible factor-1alpha through mitogen-activated protein kinase/extracellular signal-regulated kinase 1/2 pathway in neuroblastoma. Cancer Res. 2005;65:7267–7275. doi: 10.1158/0008-5472.CAN-04-4575. [DOI] [PubMed] [Google Scholar]
- 30.Butler JM, Kobayashi H, Rafii S. Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat Rev Cancer. 2010;10:138–146. doi: 10.1038/nrc2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu JL, Rak JW, Coomber BL, Hicklin DJ, Kerbel RS. Effect of p53 status on tumor response to antiangiogenic therapy. Science. 2002;295:1526–1528. doi: 10.1126/science.1068327. [DOI] [PubMed] [Google Scholar]
- 32.Ricci-Vitiani L, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 33.Morita S, Kojima T, Kitamura T. Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.