Abstract
SRC-3 is an important coactivator of nuclear receptors including the retinoic acid (RA) receptor α. Most of SRC-3 functions are facilitated by changes in the posttranslational code of the protein that involves mainly phosphorylation and ubiquitination. We recently reported that SRC-3 is degraded by the proteasome in response to RA. Here, by using an RNAi E3-ubiquitin ligase entry screen, we identified CUL-3 and RBX1 as components of the E3 ubiquitin ligase involved in the RA-induced ubiquitination and subsequent degradation of SRC-3. We also show that the RA-induced ubiquitination of SRC-3 depends on its prior phosphorylation at serine 860 that promotes binding of the CUL-3–based E3 ligase in the nucleus. Finally, phosphorylation, ubiquitination, and degradation of SRC-3 cooperate to control the dynamics of transcription. In all, this process participates to the antiproliferative effect of RA.
Retinoic acid (RA) influences cell differentiation, proliferation, and apoptosis through modifications in the expression of target genes. The transcription of RA target genes is a highly coordinated process that requires a well-defined cross-talk among RA nuclear receptors (RARs), basal transcription machinery, and several transcriptional coregulators including the p160 family of coactivators (SRC-1, SRC-2, and SRC-3) (1). For each transcriptional component, there is a fine-tuned code of posttranslational modifications that control their activity, partners’ association/dissociation, localization, and turnover (2, 3). This regulation is especially true for the coactivator SRC-3, which is a key regulator of nuclear receptors, metabolic homeostasis, and cell proliferation. Indeed, much of its function is facilitated through changes in the posttranslational code of the protein including phosphorylation and several types of posttranslational modifications (2, 4, 5).
In response to RA, SRC-3 binds to RARs and then recruits a battery of coregulatory proteins such as chromatin remodelers and modifiers that act in a coordinated and combinatorial manner to decompact chromatin and direct the transcriptional machinery to the promoter. Recently, we demonstrated that, in response to RA, SRC-3 is degraded by the proteasome (6, 7). However, the underlying mechanism of SRC-3 degradation and its link with the transcription of RA target genes was still unclear. Here, in a high-throughput screen based on the use of a siRNA thematic library and chemical transfection to create transient gene knockdown in MCF7 cells, we identified cullin 3 (CUL-3) and the Ring protein RBX1 as components of the E3 ligase complex involved in SRC-3 ubiquitination and degradation. We also show that SRC-3 degradation is involved in the transcription of RAR target genes and in the antiproliferative action of RA, through a phosphorylation-dependent ubiquitination “code.”
Results
CUL-3–Based E3 Ligase Controls RA-Induced Degradation of SRC-3.
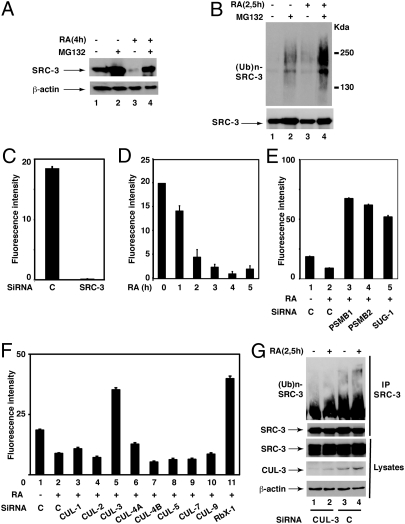
Given that in human MCF7 breast cancer cells, SRC-3 is degraded in response to RA by the 26S proteasome (Fig. 1A) (6), we addressed whether this process is regulated by ubiquitination. In immunoprecipitation experiments, SRC-3 was constitutively ubiquitinated in agreement with other reports (4) and ubiquitinated SRC-3 accumulated in the presence of the proteasome inhibitor MG132 (Fig. 1B). Ubiquitination was also enhanced in response to RA, either in the absence or presence of MG132 (Fig. 1B).
Fig. 1.
Screening of the E3 ligase involved in the RA-induced degradation and ubiquitination of SRC-3. (A and B) Extracts from MCF7 cells treated or not with RA (0.1 μM) and MG132 (4 μM) were analyzed by immunoblotting for SRC-3 degradation and for SRC-3 ubiquitination after immunoprecipitation. (C) Silencing of SRC-3 abrogates the immunofluorescence signal obtained with SRC-3 antibodies. (D) RA induces the degradation of SRC-3 as assessed by the disappearance of the fluorescence signal. (E) The RA-induced degradation of SRC-3 is reversed with siRNAs targeting proteasome subunits. (F) In the high-throughput screen, siRNAs against CUL-3 and RBX1 reverse the degradation of SRC-3. Values are the mean ± SD of at least three different experiments. (G) Analysis of SRC-3 ubiquitination as in B, after CUL-3 silencing with specific siRNAs (50 nM).
Then we aimed at investigating which E3-ubiquitin ligase is involved in the RA-induced ubiquitination and degradation of SRC-3. We performed a high-throughput screen based on the use of a siRNA thematic library to create transient gene knockdown in MCF7 cells. The screen was based on the immunofluorescence analysis of SRC-3 with specific antibodies. Through combining the imaging of cells in microtiter plates with powerful image analysis algorithms, the screen determines whether silencing of a specific E3 ligase reverses the RA-induced degradation of SRC-3.
First, the technique was validated by checking that the signal disappears upon knockdown of SRC-3 with specific siRNAs (Fig. 1C). Then kinetic experiments performed after RA addition indicated that SRC-3 degradation occurs within 3–5 h (Fig. 1D). This degradation process was reversed by siRNAs targeting subunits of the 20S core proteasome (PSMB1 and PSMB2) or the SUG-1 subunit of the 19S subcomplex (Fig. 1E) corroborating that it involves the 26S proteasome.
For the screen, we used a library of 111 siRNAs with four different siRNAs per target (Dataset S1). Upon statistical analysis of SRC-3 nuclear intensities displayed in the transfected cells, we determined two lists of candidate genes, differing by the level of selection stringency (α = 1.5 and α = 2 for maximum stringency). Seven potential hits validated by at least two siRNAs (α = 2) were found, among which CUL-3 and RBX1 were highly significant (P values) and validated by 3 and 4 siRNAs, respectively (Dataset S1 and Fig. 1F). However, the screen did not identify with high confidence any of the other E3 ligases (CUL-1-skp1-Fbw7α and E6AP) previously reported to regulate SRC-3 degradation (4, 8) (Fig. 1F). Finally, CUL-3 silencing also reversed the increase in SRC-3 ubiquitination observed at 2.5 h after RA addition (before degradation) (Fig. 1G). In conclusion, our screen indicates that the RA-induced ubiquitination and degradation of SRC-3 involves a cullin-RING Ligase (CRL) assembled with CUL-3 (CRL3).
SRC-3 Is Phosphorylated at S860 Before Degradation.
CRLs consist of three core components: a cullin scaffold protein, a RING domain protein (RBX1) that recruits the E2 conjugating enzyme, and a substrate-adaptor protein that binds target proteins (9). Given that CRL3s recognize substrates containing serine-rich domains that can be phosphorylated (10–12), we investigated whether the RA-induced ubiquitination and degradation of SRC-3 is controlled by phosphorylation.
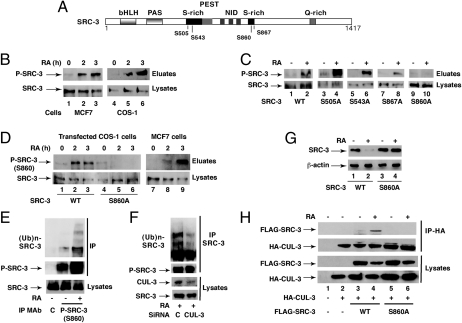
SRC-3 depicts serine-rich motifs (Fig. 2A), and the amount of endogenous phosphorylated SRC-3 markedly increased 2 h after RA addition to MCF7 cells or transfected COS-1 cells before SRC-3 degradation (Fig. 2B). According to our previous studies (6, 13), p38MAPK is rapidly activated in response to RA through nongenomic effects (14) and phosphorylates SRC-3 (6) and several other targets. SRC-3 depicts 4 p38MAPK consensus phosphorylation sites: S505, S543, S860, and S867 (Fig. 2A) (5). Therefore, we investigated whether one of these residues was phosphorylated in response to RA. FLAG-tagged SRC-3 mutants with S505, S543, S860, or S867 substituted with alanines were constructed and overexpressed in COS-1 cells. The S505A, S543A, and S867A mutants were phosphorylated in response to RA as efficiently as WT SRC-3 (Fig. 2C). However, the S860A mutant was not phosphorylated (Fig. 2C, lanes 9 and 10), indicating that S860 is a target for RA signaling.
Fig. 2.
Phosphorylation at S860 is the signal for SRC-3 ubiquitination and degradation in response to RA. (A) Schematic representation of SRC-3 with the main phosphorylation sites. (B) Kinetics of SRC-3 phosphorylation in RA-treated MCF7 and transfected COS-1 cells, after phosphoprotein affinity purification and immunoblotting. (C) Analysis as in B of the phosphorylation of the SRC-3 mutants in transfected COS-1 cells. (D) Kinetics of SRC-3 phosphorylation at S860 after phosphoprotein affinity purification and immunoblotting with antibodies recognizing specifically SRC-3 phosphorylated at this residue. (E) S860 phosphorylation controls SRC-3 ubiquitination, as assessed by immunoprecipitation of MCF7 cells extracts with the phospho-antibodies. (F) Knockdown of CUL-3 diminishes the ubiquitination of phosphorylated SRC-3 immunoprecipitated as in E. (G) Immunoblots showing that in transfected COS-1 cells, SRC-3WT but not SRC-3 (S860A) is degraded. (H) In transfected COS-1 cells, HA-CUL-3 coimmunoprecipitates with FLAG-SRC-3 WT but not with the S860A mutant.
Next, antibodies recognizing specifically SRC-3 phosphorylated at S860 were generated and used in immunoblotting experiments after phosphoprotein affinity purification: A signal at the right position was detected in MCF7 cells and in COS-1 cells overexpressing SRC-3 WT (Fig. 2D). No signal was obtained with SRC-3 (S860A) (Fig. 2D, lanes 4–6), validating the specificity of the phosphospecific antibodies. Collectively these results indicate that SRC-3 becomes phosphorylated at S860 in response to RA.
Phosphorylation at S860 Is the Signal for SRC-3 Ubiquitination/Degradation and for CUL-3 Binding.
Next, we analyzed whether S860 phosphorylation controls the ability of SRC-3 to be ubiquitinated and degraded in response to RA. Immunoprecipitation experiments were performed with MCF7 cells and our antibodies recognizing specifically SRC-3 phosphorylated at S860. The amount of phosphorylated SRC-3 increased in response to RA as well as the amount of ubiquitinated SRC-3 (Fig. 2E), and this effect was inhibited upon knockdown of CUL-3 (Fig. 2F). Finally, in contrast to SRC-3 WT, the S860A mutant was not degraded after RA addition (Fig. 2G). Thus, ubiquitination by CRL3 and degradation concern SRC-3 phosphorylated at S860.
Then we compared the ability of FLAG-SRC-3 to coimmunoprecipitate with HA-CUL-3 in transfected COS cells. SRC-3WT but not SRC-3 (S860A) was pulled down with CUL-3 in response to RA (Fig. 2H). Altogether these results indicate that the RA-induced ubiquitination and degradation of SRC-3 require a priming phosphorylation at S860 that controls the binding of CUL-3 complexes.
CUL-3 Migrates to the Nucleus in Response to RA.
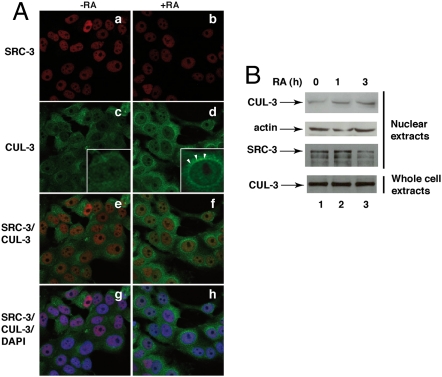
Then we analyzed the intracellular distribution of CUL-3 and SRC-3 in MCF7 cells by immunofluorescence and confocal analysis (Fig. 3A). In the absence of RA, SRC-3 was present essentially in nuclei (Fig. 3A, a) and CUL-3 in the cytoplasm (Fig. 3A, c). After RA treatment, CUL-3 accumulated at the perinuclear surface and in nuclei where it colocalized with SRC-3 (Fig. 3A, d, f, and h). That CUL-3 migrates to nuclei was corroborated by immunoblotting of nuclear extracts (Fig. 3B). Most interestingly, the amount of SRC-3 phosphorylated at S860 also increased in nuclei (Fig. 4A).
Fig. 3.
CUL-3 migrates to nuclei in response to RA. (A) MCF7 cells treated (Right) or not (Left) with RA for 2.5 h were triple stained with DAPI (blue), SRC-3 (red; a and b), and CUL-3 (green; c and d) antibodies and examined by confocal microscopy. The merge images overlapping the red and green (e and f) or the red, green, and blue fluorescence (g and h) are shown. (B) Immunoblots showing that CUL-3 levels increase in the nuclei of RA-treated cells. The arrows in d show the accumulation of CUL-3 at the perinuclear surface.
Fig. 4.
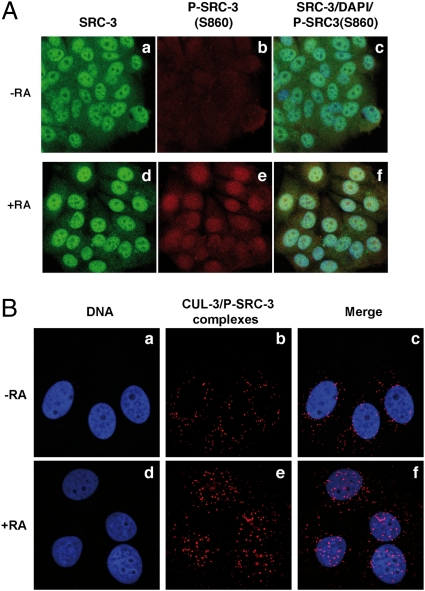
SRC-3 phosphorylated at S860 interacts with CUL-3 in nuclei. (A) MCF7 cells treated (d–f) or not (a–c) with RA for 2 h were triple-stained with DAPI (blue), and antibodies recognizing SRC-3 (green) or its phosphorylated form (red) and examined by confocal microscopy. The merge images overlapping the red, green, and blue fluorescence are shown (c and f). (B) Proximity ligation assay showing the CUL-3/P-SRC-3(S860) complexes (red; b and e) in MCF7 cells. The merge between blue and red is shown (c and f).
Next, to explore further the interaction between CUL-3 and SRC-3 phosphorylated at S860, we used a proximity ligation assay (PLA) (14, 15), which allows the in situ detection of interacting endogenous proteins. Rabbit anti–CUL-3 and mouse anti–phospho-SRC-3 antibodies were used followed by species-specific secondary antibodies, called PLA probes, each attached with a unique short DNA strand. When in close proximity, the DNA strands can be joined by a circle-forming DNA oligonucleotide. After amplification and revelation with labeled complementary oligonucleotide probes, the complexes are easily visible as bright red spots under a fluorescence microscope.
A few CUL-3/P-SRC-3 complexes were seen in the cytosol of control cells (Fig. 4B, b and c), in line with some constitutive phosphorylation of SRC-3. After RA treatment, the number of complexes increased in nuclei (Fig. 4B, e and f), suggesting that SRC-3 phosphorylation that occurs in nuclei would target CUL-3 to nuclei (16).
In contrast, the other cullins (CUL-1 and CUL-2) were present essentially in nuclei and RA did not affect their localization (Fig. S1), corroborating the specific role of CUL-3 in the RA response. RBX1 was nuclear either in the absence or presence of RA, in line with its participation to all CRL complexes (Fig. S2).
CUL-3 and SRC-3 Phosphorylation Are Required for the Transcription of RA-Target Genes.
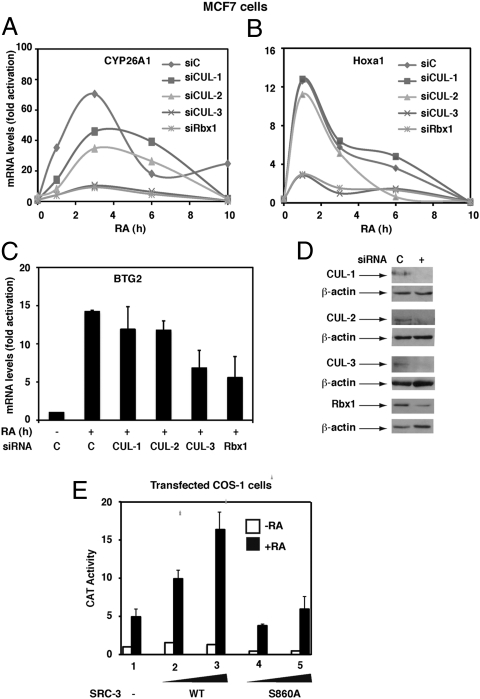
SRC-3 contributes to the transcription of RA receptor α (RARα)-target genes in several cell types (6, 7). Because the ubiquitin/proteasome pathway is implicated in the transcriptional functions of nuclear receptors and their coactivators (1, 4, 6), we investigated whether CUL-3 complexes are involved in the transcription of endogenous RARα target genes exemplified by the Cyp26A1, Hoxa1, and Btg2 genes. In MCF7 cells, knockdown of CUL-3 or RBX1 decreased the RA-induced activation of the three genes (Fig. 5 A–D) as assessed by quantitative RT-PCR. Knockdown of the other cullins, CUL-1 and CUL-2, had no significant effects (Fig. 5 A–D). Overexpression of CUL-3 also had no effect, indicating that CUL-3 is not in limiting amounts in MCF7 cells (Fig. S3). Altogether these results corroborate the importance of a CRL assembled with CUL-3 in the expression of RARα-target genes.
Fig. 5.
CUL-3 participates in the transcription of RA-target genes. (A–C) Silencing of CUL-3 or RBX1 decreases the RA-induced expression of the Cyp26A1, Hoxa1, and Btg2 genes as monitored by quantitative RT-PCR. Values, expressed as fold induction relative to untreated cells, correspond to a representative experiment among three or are the mean ± SD of three different experiments. (D) Knockdown efficiency was controlled by immunoblotting. (E) CAT activity in COS-1 cells transfected with the SRC-3WT or SRC-3(S860A) vectors along with RARα and the DR5-tk-CAT reporter gene and RA-treated for 6 h. Results are the mean ± SD of three experiments.
Next, because SRC-3 ubiquitination by the CUL-3 complex depends on the prior phosphorylation of SRC-3, the relevance of SRC-3 phosphorylation at S860 for the transcription of RA target genes was investigated in COS-1 cells transfected with RARα and a DR5-tk-CAT reporter gene. CAT activity increased in response to RA and overexpression of SRC-3 WT but not of SRC-3(S860A) enhanced this effect (Fig. 5E), corroborating that SRC-3 phosphorylation facilitates transcription (6).
SRC-3 Ubiquitination and Degradation Occur out of Chromatin.
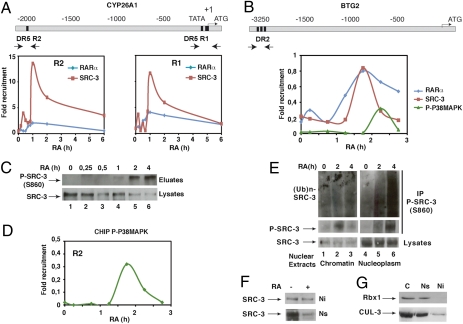
To substantiate the role of the phosphorylation-dependent ubiquitination of SRC-3 in transcription, we correlated the phosphorylation and ubiquitination state of SRC-3 to the recruitment of the coactivator to RARα-target promoters in chromatin immunoprecipitation (ChIP) experiments. In MCF7 cells, SRC-3 was rapidly recruited concomitantly with RARα, to the RA response element (RARE) located in the promoter of the Btg2 gene, and to both the proximal and distal RAREs located in the promoter of the Cyp26A1 gene, with a peak at 1 h after RA addition (Fig. 6 A and B) and a decrease at 2 h. The decrease occurred when active phosphorylated p38MAPK was recruited (Fig. 6 B and D) and when SRC-3 was phosphorylated (Fig. 6C), ubiquitinated, and degraded (Fig. 1), raising the hypothesis that these latter processes might play a role in clearing out SRC-3 from the promoters.
Fig. 6.
RA-induced SRC-3 ubiquitination and degradation occur out of chromatin. (A) Kinetic ChIP experiments performed with RA-treated MCF7 cells and showing the recruitment of SRC-3 and RARα to the R1 and R2 regions of the Cyp26A1 gene promoter. Values are expressed as fold enrichment relative to untreated cells and are the mean of three distinct experiments. (B) Recruitment of RARα, SRC-3, and active p38MAPK to the Btg2 gene promoter. (C) Kinetics of SRC-3 phosphorylation at S860 after phosphoprotein affinity purification and immunoblotting with the phospho-antibodies. (D) Recruitment of active phosphor-p38MAPK to the R2 region of the Cyp26A1 promoter. (E) Soluble nucleoplasmic (Ns) and insoluble chromatin (Ni) extracts were immunoprecipitated with the phospho-SRC-3 antibodies and immunoblotted with SRC-3 or ubiquitin antibodies. (F) Ni and Ns extracts were analyzed for SRC-3 degradation by immunoblotting. (G) Cytoplasmic (C), Ns, and Ni extracts were compared for the presence of RBX1 and CUL-3 by immunoblotting.
Therefore, we investigated whether phosphorylation, ubiquitination, and degradation concern SRC-3 associated to chromatin or out of chromatin. After preparation of highly purified intact nuclei from MCF7 cells, insoluble chromatin and soluble nucleoplasm were separated and subjected to immunoprecipitation with our phosphospecific antibodies. In chromatin, SRC-3 was only transiently phosphorylated 2 h after RA addition (Fig. 6E) when its promoter occupancy decreased. However, no ubiquitination (Fig. 6E) and no degradation could be detected (Fig. 6F). In contrast, in soluble nucleoplasm, SRC-3 was more abundant and became markedly phosphorylated, ubiquitinated, and degraded in response to RA (Fig. 6 E and F). Collectively, these results suggest that ubiquitination and degradation of phosphorylated SRC-3 occur out of chromatin. Indeed, we demonstrated that S860 phosphorylation induces the dissociation of SRC-3 from RARα (6). Moreover, CUL-3 and RBX1 were detected in nucleoplasmic extracts (Fig. 6G) and not in chromatin.
SRC-3 Is Not Phosphorylated nor Degraded in erbB-2 Positive Cells.
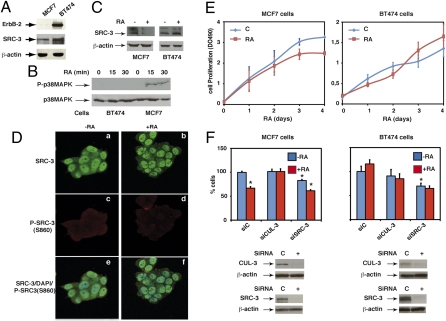
Given the importance of SRC-3 phosphorylation/degradation in the dynamics of RARα target genes transcription, we examined whether this process is affected in erbB-2 positive breast cancer cells that are characterized by aberrant kinase pathways downstream of erbB-2 (17). In these cells, exemplified by the BT474 cell line, SRC-3 is overexpressed (Fig. 7A) (18) and the nongenomic effects of RA, i.e., the activation of the p38MAPK pathway, were abrogated (Fig. 7B) (14). Consequently, SRC-3 was not phosphorylated at S860 (Fig. 7D, c and d) nor degraded (Fig. 7 C and D, a and b) and the classical RA target genes (Cyp26A1, Btg2, and Hoxa1) were not regulated by RA (Fig. S4). Similar observations were made with another erbB-2 positive cell line, MDA-MB361 (Fig. S5 A–C). Collectively, these results corroborate the importance of the p38MAPK pathway and the subsequent phosphorylation, ubiquitination, and degradation of SRC-3 in the RA response.
Fig. 7.
SRC-3 is not phosphorylated nor degraded in erbB-2 positive breast cancer cells. (A–C) Immunoblots showing that in BT474 cells, erbB-2, and SRC-3 are overexpressed, p38MAPK is not activated, and SRC-3 not degraded. (D) In BT474 cells, SRC-3 is not phosphorylated at S860 in confocal microscopy experiments performed as in Fig. 4A. (E) RA decreases the proliferation rate of MCF7 cells but not of BT-474 cells. (F) MCF7 (Left) and BT474 (Right) cells, RA-treated or not, were analyzed for proliferation after knockdown of CUL-3 and SCRC-3. Knockdown efficiency was checked by immunoblotting. Results are the mean ± SD of two distinct experiments performed in quadruplate. Statistically significant differences are indicated (*P < 0.05, control versus RA or siRNA).
SRC-3 Degradation by a CUL-3–Based E3 Ligase Contributes to the Antiproliferative Effect of RA.
We next asked whether SRC-3 levels and/or SRC-3 degradation directly influence cell growth. MCF7 cells respond to RA through a decrease in their proliferation rate (Fig. 7E, Left). Knockdown of CUL-3 did not affect MCF7 cells growth but abrogated the decrease observed after RA addition (Fig. 7F, Left). In contrast, the erbB-2 positive BT474 and MDA-MB361 cell lines were both resistant to the antiproliferative action of RA (Fig. 7E, Right, and Fig. S5D) (17) and knockdown of CUL-3 had no effect, either in the presence or absence of RA (Fig. 7F, Right, and Fig. S5D). Collectively, these observations highlight the importance of SRC-3 phosphorylation and turnover in the antiproliferative effect of RA. Of note is that knockdown of SRC-3 decreased significantly the proliferation rate of the three cell lines (Fig. 7F and Fig. S5D), suggesting that a reduction in SRC-3 levels inhibits cell growth. However, knockdown of SRC-3 did not restore the RA sensitivity of the BT474 and MDA-MB361 cells, in terms of cell proliferation (Fig. 7F and Fig. S5D).
Discussion
SRC-3 is a model coactivator of nuclear receptors for studying the influence of posttraductional modifications. Indeed, SRC-3 has been shown to be phosphorylated at several residues by different kinases in response to different signaling pathways (2, 5). Moreover, our laboratory demonstrated that in response to RA, SRC-3 is phosphorylated by p38MAPK and subsequently degraded by the proteasome (6). Here we identified S860 as the residue that is phosphorylated in response to RA and that promotes SRC-3 ubiquitination and degradation. We also expanded the repertoire of SRC-3 E3 ligases by characterizing a CUL-3–based complex as the E3 ubiquitin ligase involved in the ubiquitination/degradation of SRC-3 in a RA and phospho-dependent manner.
Classically, CUL-3–based complexes recognize their substrate through an adaptor containing a Bric-a Brac/Tramtrack/Broad (BTB) domain (19). The human genome encodes 190 BTB proteins (20), which were not included in our screen, but one can speculate that SRC-3 ubiquitination involves a CRL3 with a BTB protein that recognizes phosphorylated motifs (10, 11), as described for CUL-1 complexes (4, 21), or a nearby domain created by S860 phosphorylation through an allosteric mechanism (22). However, one cannot exclude that CUL-3 interacts directly with the serine-rich domain of SRC-3 containing phosphorylated S860, independently of any adaptor (12).
It is worth noting that SRC-3 can be ubiquitinated by other CRLs such as CUL-1-skp1-Fbw7α, in response to other signal kinase pathways and to estrogens via other phosphorylated domains (4). When this manuscript was submitted, another study was issued, reporting also a role for a CRL3 complex in SRC-3 ubiquitination and proteolysis (23). However, in this study, CUL-3 was recruited through the BTB protein SPOP (speckle-typePOZ protein) to a different motif that was phosphorylated by a different kinase. Thus, different signals and/or phosphorylation of distinct serine residues can select distinct E3 ligases to regulate SRC-3 ubiquitination/degradation and, thus, SRC-3 levels.
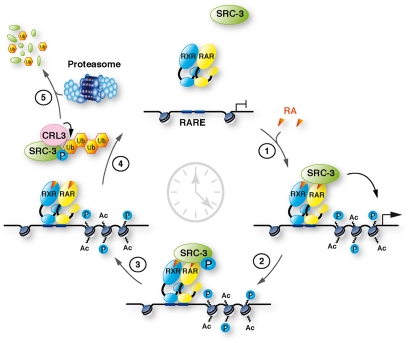
The other interesting point of this study is that ubiquitination and degradation of SRC-3 occur out of chromatin and that these processes, together with CUL-3, are required for the expression of RA target genes. Therefore, we proposed a model (Fig. 8) in which the RA-induced phosphorylation of DNA-bound SRC-3 at S860 is the signal that promotes the dissociation of SRC-3 from chromatin (6) and SRC-3 interaction with CUL-3 complexes out of chromatin. Then SRC-3 is ubiquitinated and degraded. Thus, one can suggest that ubiquitination and degradation cooperate with phosphorylation to clear SRC-3 out of the promoters according to the model proposed for another CUL-3 target (10) so that other coregulators can come and participate to transcription (3), in line with the dynamics of transcription. Corroborating this model, in erbB-2 positive breast cancer cell lines, where the p38MAPK pathway is not activated by RA, SRC-3 is not phosphorylated nor degraded and most of the classical RA target genes are not regulated.
Fig. 8.
Working model for the role of SRC-3 phosphorylation, ubiquitination, and degradation in RA target genes transcription. In response to RA, RARα and SRC-3 are recruited to target genes promoters to initiate transcription (1). Then SRC-3 becomes phosphorylated at S860, dissociates from RARα and DNA (2), and interacts with CUL-3 complexes that promote its ubiquitination (3) and degradation by the proteasome (4).
Finally, our knockdown experiments indicate that SRC-3 participates to cell growth and that SRC-3 degradation via a CUL-3 complex is involved in the antiproliferative action of RA. Such results highlight the importance of SRC-3 phosphorylation and turnover in the RA response. Thus, one can predict that cancers characterized by aberrant signaling pathways (24) would be RA resistant. In line with this hypothesis, erbB-2 positive breast cancer cell lines, in which SRC-3 is not degraded in response to RA, are resistant to the antiproliferative effect of RA. Note that in such cells, overexpression of CUL-3 markedly inhibited cell growth (Fig. S5E), most probably through the ability of the CUL-3–based ubiquitin ligases, which are known to function as breast cancer tumor suppressors, to target several signaling proteins for ubiquitination and degradation (25).
Remarkably, SRC-3 is a coactivator not only for RARs, but also for several nuclear receptors such as the estrogen receptor (3). Because several RAR regulated genes cross-talk with estrogen signaling (26, 27), one can speculate that the RA-induced degradation of SRC-3 would influence RA target genes at the cost of estrogen-ER function. With estrogen being the predominant hormone involved in the proliferation of breast cancer cells, SRC-3 segradation might be part of the RA rationale in the treatment of breast cancer with functional signaling pathways (28). Reciprocally, according to our results, one cannot exclude that the degradation of SRC-3 that occurs in response to estrogens (4) might potentiate the antiproliferative action of RA.
In conclusion, our work highlights the importance of phosphorylation and ubiquitination processes in the regulation of RA target genes through the control of SRC-3 turnover. It also reveals that RA resistance may be correlated, at least in part, to the deregulation of these processes.
Materials and Methods
Plasmids, reagents, antibodies, and cell lines are described in SI Experimental Procedures.
Complete details for the RNAi E3 Ubiquitine Ligase screen, immunoblotting, immunoprecipitation, chromatin immunoprecipitation, qRT-PCR, immunofluorescence analysis, and proximity ligation assay (PLA) are also described in the SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Dr. W. Krek for the cullin vectors; Dr. B. W. O'Malley for the phosphorylation defective SRC-3 mutants; M. Oulad Abdelghani (Institut de Génétique et de Biologie Moléculaire et Cellulaire; IGBMC) for the mouse monoclonal antibodies; J. M. Garnier (IGBMC) for constructs; Amélie Weiss and Laure Froidevaux from the high-throughput screening facility (IGBMC), the cell culture facilities; and Regis Lutzing for help. This work was supported by funds from Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Agence Nationale pour la Recherche Grants ANR-05-BLAN-0390-02 and ANR-09-BLAN-0297-01, Association pour la Recherche sur le Cancer Grant ARC-07-1-3169, Fondation pour la Recherche Médicale (FRM) Grant DEQ20090515423), and Institut National du Cancer Grants INCa-PL09-194 and PL07-96099. C.F. and E.S. were supported by the Ministère de l'Enseignement Supérieur et de la Recherche and A.P. by FRM and the Lady TATA Memorial Trust. Association pour la Recherche sur le Cancer supported G.P.-B. and C.F.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.A. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102572108/-/DCSupplemental.
References
- 1.Rochette-Egly C, Germain P. Dynamic and combinatorial control of gene expression by nuclear retinoic acid receptors. Nucl Recept Signal. 2009;7:e005. doi: 10.1621/nrs.07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han SJ, Lonard DM, O'Malley BW. Multi-modulation of nuclear receptor coactivators through posttranslational modifications. Trends Endocrinol Metab. 2009;20:8–15. doi: 10.1016/j.tem.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Malley BW, Qin J, Lanz RB. Cracking the coregulator codes. Curr Opin Cell Biol. 2008;20:310–315. doi: 10.1016/j.ceb.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu RC, Feng Q, Lonard DM, O'Malley BW. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 2007;129:1125–1140. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 5.Wu RC, et al. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol Cell. 2004;15:937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 6.Giannì M, et al. P38MAPK-dependent phosphorylation and degradation of SRC-3/AIB1 and RARalpha-mediated transcription. EMBO J. 2006;25:739–751. doi: 10.1038/sj.emboj.7600981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferry C, et al. SUG-1 plays proteolytic and non-proteolytic roles in the control of retinoic acid target genes via its interaction with SRC-3. J Biol Chem. 2009;284:8127–8135. doi: 10.1074/jbc.M808815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mani A, et al. E6AP mediates regulated proteasomal degradation of the nuclear receptor coactivator amplified in breast cancer 1 in immortalized cells. Cancer Res. 2006;66:8680–8686. doi: 10.1158/0008-5472.CAN-06-0557. [DOI] [PubMed] [Google Scholar]
- 9.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 10.Spoel SH, et al. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell. 2009;137:860–872. doi: 10.1016/j.cell.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q, et al. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc Natl Acad Sci USA. 2009;106:21191–21196. doi: 10.1073/pnas.0912008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xing H, Hong Y, Sarge KD. PEST sequences mediate heat shock factor 2 turnover by interacting with the Cul3 subunit of the Cul3-RING ubiquitin ligase. Cell Stress Chaperones. 2010;15:301–308. doi: 10.1007/s12192-009-0144-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruck N, et al. A coordinated phosphorylation cascade initiated by p38MAPK/MSK1 directs RARalpha to target promoters. EMBO J. 2009;28:34–47. doi: 10.1038/emboj.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piskunov A, Rochette-Egly C. A retinoic acid receptor RARα pool present in membrane lipid rafts forms complexes with G protein αQ to activate p38MAPK. Oncogene. 2011 doi: 10.1038/onc.2011.499. in press. [DOI] [PubMed] [Google Scholar]
- 15.Söderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 16.Welcker M, Larimore EA, Frappier L, Clurman BE. Nucleolar targeting of the fbw7 ubiquitin ligase by a pseudosubstrate and glycogen synthase kinase 3. Mol Cell Biol. 2011;31:1214–1224. doi: 10.1128/MCB.01347-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tari AM, Lim SJ, Hung MC, Esteva FJ, Lopez-Berestein G. Her2/neu induces all-trans retinoic acid (ATRA) resistance in breast cancer cells. Oncogene. 2002;21:5224–5232. doi: 10.1038/sj.onc.1205660. [DOI] [PubMed] [Google Scholar]
- 18.Xu J, Wu RC, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Heuvel S. Protein degradation: CUL-3 and BTB—partners in proteolysis. Curr Biol. 2004;14:R59–R61. [PubMed] [Google Scholar]
- 20.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Privé GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 22.Samarut E, et al. Evolution of nuclear retinoic acid receptor alpha (RARα) phosphorylation sites. Serine gain provides fine-tuned regulation. Mol Biol Evol. 2011;28:2125–2137. doi: 10.1093/molbev/msr035. [DOI] [PubMed] [Google Scholar]
- 23.Li C, et al. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30:4350–4364. doi: 10.1038/onc.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 25.Emanuele MJ, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross-Innes CS, et al. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev. 2010;24:171–182. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darro F, et al. Growth inhibition of human in vitro and mouse in vitro and in vivo mammary tumor models by retinoids in comparison with tamoxifen and the RU-486 anti-progestagen. Breast Cancer Res Treat. 1998;51:39–55. doi: 10.1023/a:1006098124087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.