Abstract
Integrin-mediated adhesions are critical for stem cell differentiation, cancer metastasis, and the immune response [Hynes RO (2009) Science 326:1216–1219]. However, the mechanisms of early adhesion formation remain unclear, especially the effects of lateral clustering of integrins and the role of the Src family kinases. Using mobile Arg–Gly–Asp (RGD) peptide ligands on lipid bilayers with nano-fabricated physical barriers [Salaita K, et al. (2010) Science 327:1380–1385], we observe surprising long-range lateral movements of ligated integrins during the process of cell spreading. Initially, RGD-activated integrin clusters stimulate actin polymerization that radiates from the clusters. Myosin II contraction of actin from adjacent clusters produces contractile pairs that move toward each other against barriers. Force generated by myosin II stimulates a Src kinase-dependent lamellipodial extension and outward movement of clusters. Subsequent retraction by myosin II causes inward movement of clusters. The final cell spread area increases with the density of periodic barriers. Early integrin clustering recruits adhesion proteins, talin, paxillin, and FAK, irrespective of force generation. However, recruitment of vinculin is only observed upon contraction. Thus, we suggest that integrin activation and early clustering are independent of lateral forces. Clustering activates Src-dependent actin polymerization from clusters. Myosin contraction of clusters to lines stimulates active spreading with outward forces from actin polymerization followed by a second wave of contraction. Many of these early mechanical steps are not evident in cells spreading on immobilized matrices perhaps because of the low forces involved. These observations can provide new targets to control integrin-dependent adhesion and motility.
Keywords: cell adhesion, integrin reorganization, supported membranes
The integrins, a heterodimeric transmembrane receptor family, regulate many cellular processes including growth, differentiation, and death (1–3). Both chemical recognition and mechanical sensing are important factors in the outside-in integrin signaling pathway (4–6). Chemically, ligation of integrin receptors, such as fibronectin binding to α5β1 and αvβ3, results in conformational changes of the external domains and cytoplasmic tails causing the recruitment of various signaling molecules and adaptor proteins during focal adhesion formation (7, 8). Mechanically, matrix rigidity and local matrix density regulate cell spreading, growth, and differentiation. Rigidity sensing depends upon local contractile force generation by the actin cytoskeleton on ligated integrin receptors (9–11). The spacing of Arg–Gly–Asp (RGD) peptide matrix ligands on 5 nm gold dots needs to be 60 nm or less for cell spreading (12). Recent findings with a constant overall density of RGD ligands, show that local clusters of four or more RGD ligands spaced by 60 nm are needed for sustained spreading, defining a minimal adhesion unit of four ligated integrins (13, 14). Nevertheless, because the ligands on solid substrates are physically fixed and lack long-ranged lateral mobility, it is not possible to know how spatial-temporal redistributions of integrins are involved in multimolecular assembly processes or when actomyosin forces are applied.
To investigate spatial reorganization of activated integrins and the effect of force, we spread cells on planar supported lipid bilayer membranes with lipid-attached cyclic RGD tripeptide ligands (Fig. 1A). Supported lipid membranes on nano-patterned substrates have been diversely used to study biophysical signaling events at cell-membrane junctions, such as immunological synapses and ephrin-mediated cell adhesions (15, 16). Two-dimensional fluidity of membrane-bound RGD, with a diffusion coefficient of about 2.5 μm2/s, enabled redistribution of ligated integrins and associated proteins in live cells (Fig. S1A). Using nano-imprint techniques, chromium lines of 100 nm in width and 5 nm in height were fabricated on the glass substrates and served as physical barriers to lateral movement (17, 18). When lipid membranes were partitioned by nano-fabricated lines, cytoskeletal forces could be generated on RGD-ligated integrin clusters. The metal lines were spaced at variable distances enabling local confinement of the RGD ligands and bound integrins within the plasma membrane plus associated cortical cytoskeletal components (19). Biotinylated RGD peptides were stably attached to biotinylated lipids via Cascade Blue labeled neutravidin, at a surface density approximately 1,750 ± 400 molecules/μm2 (16, 20). The planar sandwich of lipids with mobile ligands and live cells enabled sensitive high-resolution fluorescence microscopy of the dynamics of integrins and associated proteins.
Fig. 1.
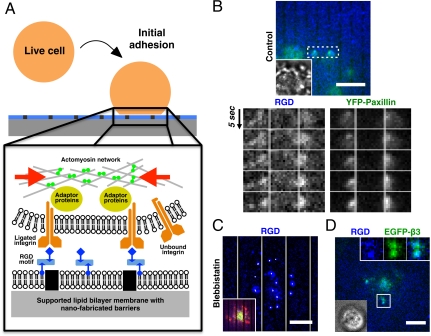
Formation of contractile pair assembly during initial adhesion. (A) Experimental schematic of live cells over nano-patterned supported lipid bilayer membranes. RGD peptides were chemically linked to lipids in supported bilayer membranes, and then bound to integrin receptors on the plasma membrane. (B) Contractile pair assembly of ligated RGD-integrin clusters across nano-patterned metal lines during initial adhesion. RGD-integrin clusters and associated YFP-paxillin laterally moved towards each other and piled up against the physical barrier of metal lines (two right boxes, 2 μm gap spacing between lines). (C) Preincubation with 50 μM blebbistatin blocked contractile movement of RGD-integrin clusters. White trajectories over each RGD cluster were 90 s time-projection of cluster positions. Inset overlay: Lifeact-Ruby (red) and EGFP-myosin light chain (green). (D) Colocalization of RGD and EGFP-integrin-β3 during early cell adhesion. Inset overlay: bright-field image. Membrane-bound RGD peptides attached to Cascade Blue neutravidin were utilized as reporters for activated integrins. (Scale bars, 5 μm).
Results and Discussion
Formation of Integrin Clusters and Actin Remodeling.
As cells contacted RGD-functionalized supported lipid membranes, integrin binding to RGD caused the formation of submicron-sized clusters (Fig. S2A). The fluorescently labeled mobile RGD complexes on the supported membrane localized with EGFP-integrin-β3 clusters and were used as reporters for ligated integrins (Fig. 1D, Fig. S2B). The RGD-integrin clusters were circularly shaped and fluorescence intensity linearly increased with time in the initial 200 s of attachment (Fig. S2A). Various cytoplasmic adaptor proteins, such as paxillin, were recruited at ligated integrin cluster sites in a force-independent manner during initial adhesion cluster formations, because their associations were not inhibited by either blebbistatin or latrunculin A (Fig. S3). Early association of paxillin and other adhesion proteins with activated integrin clusters was observed in a variety of adherent cell types, such as HeLa epithelial cells, MDCK epithelial cells, and mouse embryonic fibroblasts. As negative controls, neutravidin alone on supported bilayers, EDTA-containing media (Fig. S4), or soluble RGD in the media (1) blocked cell adhesion. The nearly circular shape of these multimolecule clusters supported the hypothesis that the initial growth was by diffusion and capture of ligated integrins.
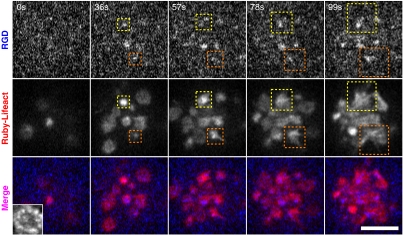
Concurrent with the early nucleation of RGD-integrin clusters, there was actin network remodeling and locally stimulated actin polymerization (21–23) (Fig. 2). As clusters formed, actin filaments (Ruby-Lifeact staining) were enriched over RGD-integrin cluster sites and then filaments grew from RGD-integrin clusters (Movie S1). Often ribbons of actin filaments appeared to emanate from the clusters and extended outward over time (Fig. 2, dashed boxes). Thus, the formation of clusters appeared to stimulate actin polymerization.
Fig. 2.
Stimulation of actin polymerization at newly formed RGD-integrin clusters during the initial contact of cell and RGD-membrane (blue). Actin filaments, labeled by Ruby-Lifeact (red), were enriched in the beginning of RGD-integrin cluster formation (0 to 36 s). Integrin-mediated actin network remodeling then took place and resulted in active polymerization of actin around RGD-integrin clusters (dashed box, 57 to 99 s). (Scale bars, 5 μm).
Contractile Movement Triggers Outward Translocation of Ligated Integrin Clusters.
After clusters formed, they often moved laterally in interesting patterns. Unlike conventional centripetal retrograde movement, adjacent integrin complexes moved toward each other initially. In the case of supported membranes with nano-fabricated metal lines, ligated clusters separated by metal lines moved laterally towards each other and then piled up against the metal lines (Fig. 1B, Movie S2). To avoid nonspecific interactions, bovine serum albumin or casein was used to passivate metal surfaces. The thin metal lines not only effectively blocked lateral movements of integrin-RGD clusters, but also defined where forces could be generated by passively opposing transport forces exerted on integrin clusters. Both actin and myosin were present between contractile RGD clusters (Fig. S5). Pretreatment with blebbistatin, a specific myosin-II inhibitor effectively blocked contractile movement of ligated integrin complexes (Fig. 1C). Because bipolar myosin was normally excluded from early integrin adhesions (24), we postulated that cluster formation stimulated local actin assembly and myosin was then recruited to pull clusters together.
Contraction to Nano-Fabricated Barriers Activated Spreading.
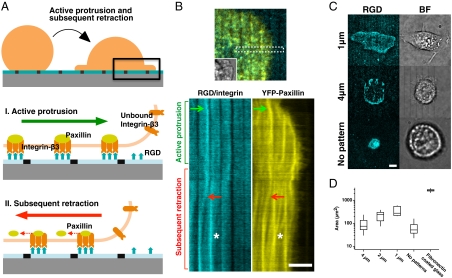
When initial local contraction occurred across physical barriers, cells were activated to spread via lamellipodial extension in over 80% of the cells observed (Fig. 3 A, I and B) whereas only 30% of cells on continuous bilayers were able to spread. During active lamellipodial extension, we observed unreported outward movements of ligated integrin clusters. As cells spread over supported membranes with RGD ligands, ligated RGD-integrin complexes moved outward and concentrated at the inner side of prefabricated metal lines that disrupted the supported bilayers (Fig. 3B, Movie S3). Previous studies have not reported this outward movement during cell spreading, perhaps because of the physical restraint of matrix ligands, as well as limited sensitivity of current force measurement methodologies.
Fig. 3.
Active cell spreading and subsequent retraction on nano-patterned RGD-membranes. (A) Schematic diagrams of sequential movements of ligated integrin complexes in supported bilayers to nano-fabricated lines. (B) Lateral movement of ligated integrin complexes. Inset overlay: bright-field image. Kymographs (two lower boxes) of both RGD-integrin and YFP-paxillin as they moved outwards and then inwards. 2 μm gap spacing between lines. (C) Cell adhesion area was modulated by spacing between nano-patterned metal barriers. Larger spacing between lines resulted in a smaller spread area after 60 min of adhesion. (D) Adhesion area (log scale) on substrates with different gap distance and fibronectin-coated glass. Boxes, 1st and 3rd quartiles; whiskers, 10th and 90th percentiles; total 84 cells. (Scale bars, 5 μm).
Subsequently, the spreading of cells was followed by the activation of myosin-II mediated retraction that generated high forces at adhesion clusters (25). With lipid-attached mobile RGD ligands, the activation of retraction often resulted in large-scale inward translocation of integrin complexes, due to the lack of resistance (Fig. 3 A, II). As the retraction started, many smaller ligated integrin clusters from the lamellipodia, along with temporarily stationary large clusters, rapidly moved centripetally, as fast as 30 nm/s. On the patterned substrates, retraction caused ligated integrin-RGD complexes to move away from the inner side and then concentrated on the outer side of nano-patterned parallel chromium lines (Fig. 3B). Adaptor proteins, such as paxillin, paused temporarily at the lines with laterally confined integrin-RGD clusters but broke away and continued to move inwards while integrins stayed at the lines (Fig. 3B white aster). Because of the ability of myosin-II to develop high forces during the retraction phase (5), typically in the range of 5–10 nN/μm2, the cytoplasmic early adhesion complexes containing paxillin may be physically separated from ligated integrins (Fig. S6A).
Barrier lines may activate further spreading during the extensile phase and then stabilize cells in the spread state during the retraction phase. When we tested cell spreading on surfaces with different densities of prefabricated metal lines, we found that line density correlated with cell contact area after 30 min of spreading, and a larger gap spacing between the 100 nm lines resulted in a smaller spread area (Fig. 3C). With 1 μm-pitch lines, cells spread to a relatively large area (800 μm2) that was still only about 30% of the spread area on fibronectin-coated glass (2,500 μm2) (Fig. 3D). With the lower density 4 μm-pitch lines, cells retracted to a very small contact area (50–100 μm2). On continuous supported bilayers, cell contact area decreased dramatically, and ligated integrin complexes moved to the center of the contact area (Fig. 3C, lower boxes). The cells on continuous RGD-supported membranes eventually formed a contractile ring that lifted up the rest of the cell in a nearly spherical morphology surrounded by mesh-like cortical actomysin network and a small contact footprint (Fig. S6B, Movie S4). Thus, the density of force-bearing sites positively correlated with the spread area (Fig. 3D). Because integrin adhesions were strengthened by force, the cells formed adhesive contacts at the lines that kept the cells spread on the substrate.
Src-Family Kinases Regulate Lateral Movement of Integrin Clusters.
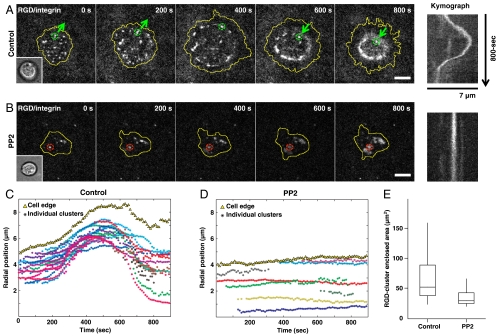
On the lined substrates, outward movements were limited and left many unanswered questions about how outward forces were generated. To better understand the process of outward movement, we analyzed the cluster movements in continuous bilayers when spreading was activated (Fig. 4A). Initially, lateral contractile movements of ligated clusters were also observed (Fig. S7A). Outward spreading was activated in 30% of cells possibly by forces generated at local bilayer imperfections during local contractile movements of integrin clusters. Upon activation of spreading, there was a remarkable long-ranged outward movement of the complexes behind the leading edge of the spreading cell (Fig. 4A, Movie S5). At early times, ligated integrin complexes and the cell leading edge both moved outwards with a similar radial velocity, as fast as 20 nm/s (Fig. 4C). Within 11 cells that actively spread on continuous RGD-supported membranes, the average number of contractile and outward-moving clusters per cell were 4.2 ± 1.7 (standard deviation, SD) and 19.5 ± 5 (SD), respectively. Actin polymerization around RGD-integrin clusters was observed as clusters moved outward (Fig. S7B). There was a relatively constant spatial separation, approximately 800 nm to 1 μm, between the cell edge and the peripheral integrin clusters during the initial outward movement. However, outward movement of integrin clusters ceased before lamellipodial extension and clusters were temporarily fixed in space. Because there was no biased outward movement of bulk membranes during the early adhesion phase (Fig. S1B), we suggest that actin polymerization powered the outward movement of clusters. At later times, the edge polymerization appeared to dominate, perhaps because of increased membrane tension.
Fig. 4.
Outward movement of integrin clusters follows protrusion of cell edges. (A) Long-ranged outward then inward translocation of ligated integrin complexes. (B) Preincubation of 10 μM PP2, a Src-kinase inhibitor, blocked outward movement and cell spreading. Inset overlay: bright-field images, respectively. Kymographs: lateral movement of marked clusters, respectively. (C, D) Radial positions of each cluster’s trajectory (colored aster) and averaged cell edges (yellow triangle) of (A, 27 clusters) and (B, 8 clusters). (E) Cell contact area under control condition and PP2 treatment. Boxes, 1st and 3rd quartiles; whiskers, 10th and 90th percentiles; total 50 cells. (Scale bars, 5 μm).
Previously, it was shown that ligated αvβ3 integrin receptors promote Rac1GTP-mediated cell spreading by activating focal adhesion kinase (FAK) and Src-family kinases (SFK) such as Fyn (26). If activation of cell spreading was driven by a similar process, then localized Fyn activation could support the outward translocation of ligated integrin complexes by directly promoting Rac1GTP-mediated cell spreading (Fig. S7A). When 10 μM of 4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine (PP2) was added to inhibit SFKs, both initial contractile movement and long-ranged outward movement of ligated integrin complexes were inhibited (Fig. 4 B and D). In addition, the robust polymerization of actin from the clusters was blocked. However, PP2 treatment did not block formation of integrin-RGD clusters. Adhesion area of PP2 treated cells, defined by area enclosed by RGD clusters, was merely 34.3 ± 14.3 μm2 after 30-min of spreading (Fig. 4E). Thus, we suggest that SFKs were needed to stimulate the force-dependent steps that resulted in activation of spreading.
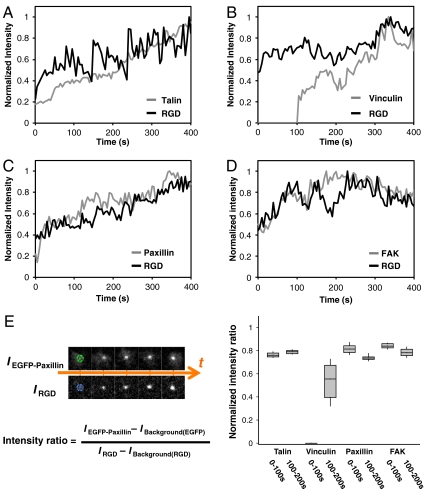
Integrin Clustering Recruits Talin, Paxillin, and FAK Without Contraction, but Vinculin Recruitment Requires Contraction.
The outside-in activation of integrins by both RGD binding and cytoplasmic association of adhesion components was thought to be a force-dependent process (27, 28). However, we found that talin, paxillin, and FAK were promptly associated with ligated integrin clusters during the early adhesion phase (Fig. 5 A, C, D, Fig. S8). By measuring the time dependence of the relative fluorescence intensities of GFP-tagged adhesion proteins and Cascade Blue neutravidin-RGD in newly formed clusters, we found that talin, paxillin, and FAK were quickly recruited to submicrometer RGD-integrin clusters, and the fluorescence intensity steadily increased during the early adhesion formation. The intensity ratio between GFP-tagged adhesion proteins (talin, paxillin, and FAK) and Cascade Blue neutravidin-RGD at each cluster also indicated steady growth within the initial 200 s. However, vinculin association was delayed until inward movement of ligated integrin-RGD clusters began (Fig. 5B, Fig. S9). The time-dependent recruitment of vinculin potentially resulted from an increased unfolding of talin as a result of contractile tension on the integrin clusters. This observation is further evidence that contraction can generate greater force than outward spreading (29, 30).
Fig. 5.
Temporal and spatial localization of adaptor proteins at activated integrin clusters. Time-dependent normalized intensity of (A) EGFP-Talin, (B) EGFP-Vinculin, (C) YFP-Paxillin, (D) EGFP-FAK relative to membrane-bound RGD-Cascade Blue at individual newly-formed clusters. Talin, paxillin, and FAK were promptly recruited with newly-formed RGD-integrin clusters. Vinculin recruitment was delayed till inward movement of RGD-integrin clusters (also see Fig. S9). (E) Normalized intensity ratio between the adaptor proteins and RGD-Cascade Blue within the initial 200 s of cluster formation. Boxes, 1st and 3rd quartiles; whiskers, 10th and 90th percentiles; a total of 29 clusters were measured over 200 s.
Conclusion
The fluid RGD ligands enable observation of the early mechanical steps in integrin-dependent adhesion and spreading. Although there is some question about the value of studying focal adhesions, in general and adhesions to a lipid bilayer in particular, because they may not form normally in vivo (31), more recent studies have found adhesions in three-dimensional (3D) matrice (32) and the premise that such phenomena reflect physiologically relevant processes has generally proven true. We suggest that the adhesions formed on the bilayers do reflect associations that normally form in vivo in certain cell states and that the elucidation of the steps involved in adhesion formation on bilayers provides a testable hypothesis for the steps involved in formation of other adhesions. Based upon these studies we suggest that, first, integrins triggered by extracellular ligand binding recruit other integrins and various adaptor proteins to form submicrometer clusters by means of lateral diffusion and capture. Second, assembly of actin filaments occurs at ligated integrin complexes, and those filaments are then aggregated by myosin contraction, leading to even larger cluster formation (33) (Fig. 1A). Third, actin-dependent spreading is triggered by mechanical stimulation of SFK-dependent phosphorylation events, resulting in activation of guanine nucleotide exchange factors (GEFs) and a presumed rise in Rac1-GTP levels (26, 34). Fourth, during SFK-stimulated actin-dependent spreading, mechanically linked integrin clusters serve as local anchoring points for outward polymerization of actin that drives ligated integrin clusters outwards (Fig. 3 A, I). Clusters spread outward at the rate of edge extension initially, but stop moving before the cell edge stops spreading, causing an increased spatial separation of the edge from the clusters. Fifth, myosin-II mediated retraction is activated (35), and the integrin clusters are rapidly drawn inward (Fig. 3 A, II). If there is local resistance to retraction, then adhesions will mature at the sites of greatest force.
Immobile-ligand surfaces, such as fibronectin-coated glass, have been widely used to study integrin-mediated cell adhesion and migration. The fixed-ligand configuration supports regular cell-matrix adhesion ex vivo, but the forces associated with early cell adhesion formation have not been determined. Instead of measuring force directly, which is usually difficult in early phases of cell adhesion, we utilized mobile ligands and high-resolution microscopy to investigate lateral redistribution of RGD-integrin clusters in plasma membranes. The lateral movement of clusters, which is easier to detect with higher accuracy, can be driven by actin polymerization from clusters and subsequent myosin contraction. The physical barriers to RGD ligand movement enable force generation needed for some signaling processes. Future identification of the driving force for outward translocation of mobile integrin-RGD will improve our understanding of the integrin-cytoskeletal linkage. The overall behavior of cells in our studies is generally consistent with earlier models of cell spreading (24, 36). However, the additional steps revealed by mobile RGD-supported membranes, including contractile pair assembly and outward translocation of integrins, yield an important understanding of the basic mechanisms that underlie cell spreading on rigid matrices as well as mobile adhesion molecules. We further postulate that similar steps occur during fibroblast migration and lamellipodial extension to new matrix sites. Knowing these additional steps will help to target approaches to understand the details of the adhesion process related to growth, differentiation, and death (4, 37).
Materials and Methods
Additional information on fusion protein constructs, supported membrane preparation, substrate fabrication, and data analysis is available in SI Text.
Cell Culture and Fluorescent Fusion Proteins.
HeLa epithelial cells, MDCK epithelial cells, RPTP+/+ fibroblasts were grown in DMEM media supplemented with 10% (Vol/Vol) fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 20 mM Hepes in 37 °C incubators with 5% CO2. These cell types behaved similarly on RGD-membranes. Unless otherwise noted, HeLa cells were utilized to characterize lateral reorganizations of ligated integrin complexes. Fig. 4, Fig S6 are examples of MDCK cells. Electroporation was utilized to transiently transfect fusion constructs into cells, by the manufacturer’s protocol. Cells were harvested after 18 to 24 h of transfection. To avoid nonspecific interaction from serum, cells were then resuspended in serum-free DMEM media in a 37 °C incubator with 5% CO2 for 30 min before imaging.
Nano-Patterned Glass Substrate.
Nano-imprint lithography was utilized to fabricate the physical barriers on glass substrates. A silicon-based imprint mold was made by electron-beam lithography and anisotropic etching processes. Patterns were then transferred from the mold to the glass by high pressure stamping the imprint mold onto the polymer-coated cover glass and curing the polymer. After demolding, oxygen plasma etching was used to extend imprinted trenches vertically to the surface of the cover glass. A thin chromium metal layer was deposited by thermal evaporation and then the chromium outside the lines was selectively removed by resist lift-off processing. Typically, metal lines were 100 nm in width and 5 nm in height with a gap distance ranging from 500 nm to 4 μm.
Membrane Functionalization.
The lipids composed of 0.4 mol% of 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) (16∶0 biotinyl-Cap-PE) and 99.6 mol% of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) (both from Avanti Polar Lipids) were used to form supported lipid bilayer membranes. Detailed preparation methods were previously described (17, 18). Before RGD functionalization, supported lipid membranes and the nano-patterned substrates were blocked by incubation with 10 ∼ 50 μg/mL of BSA or casein for 30 min. Cascade Blue Neutravidin (Invitrogen) serves as the link between biotinyl-Cap-PE and biotinylated RGD peptide (20). Excess neutravidin was removed before the addition of biotinylated RGD peptide. Based on quantitative fluorescence calibration (16), the surface density of biotinylated RGD linked by neutravidin on 0.4 mol% biotin-lipid membranes was approximately 1,750 ± 400 molecules/μm2.
Inhibition Chemicals.
Blebbistatin, latrunculin A, and PP2 were purchased from Sigma-Aldrich. Chemicals were first dissolved in dimethyl sulfoxide (DMSO) with a stock concentration 1,000-times higher than the final concentration. Final concentrations were: blebbistatin 50 μM, latrunculin A 200 nM, PP2 10 μM. Before applying to cells, chemicals were diluted 1,000-times into DMEM media.
Microscopy and Data Analyses.
Fluorescent images of live cells were taken by a spinning-disk confocal inverted microscope (PerkinElmer UltraVIEW VoX), with 100x oil immersion lens (1.40 NA, UPlanSApo 100x) and back-thinned electron multiplier CCD camera (C9100-13, Hamamatsu Photonics). An environmental chamber (37 °C and 5% CO2) was attached to the microscope body for long-term time-lapse imaging. Acquired images were analyzed by ImageJ (NIH), Imaris (Bitplane AG), and Matlab (MathWorks). Within individual cells, clusters of ligated RGD-integrins and associated adaptor proteins were identified by Imaris under identical particle-searching criteria, including center position, area, intensity, and time point. These parameters for each cluster were then processed by a home-written Matlab code. Kymographs were generated by ImageJ. The normalized intensity ratios between RGD-integrins (IRGD) and associated adaptor proteins (IEGFP-protein) were calculated by the following procedure. Along each cluster trajectory, we first measured the fluorescence intensity with background subtraction at each cluster area, as Fig. 5 A–D. The series of intensity ratio (IEGFP-protein/IRGD) per unit area at each time point were then normalized within each trajectory. To compare the intensity ratio, the first time point of a newly detected integrin cluster was assigned to time zero. Normalized intensity ratios between each colocalized cluster of RGD-integrin and adaptor proteins were then plotted together, as Fig. 5E.
Supplementary Material
Acknowledgments.
The authors acknowledge helpful discussions with Dr. Alexander Bershadsky, Dr. Naila Alieva, and Dr. Jay T. Groves. C.H.Y. acknowledges support from National Science Council of Taiwan, Grant NSC98-2917-I-564-165.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109485108/-/DCSupplemental.
References
- 1.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans R, et al. Integrins in immunity. J Cell Sci. 2009;122:215–225. doi: 10.1242/jcs.019117. [DOI] [PubMed] [Google Scholar]
- 3.Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Bio. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 5.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DuFort CC, Paszek MJ, Weaver VM. Balancing forces: architectural control of mechanotransduction. Nat Rev Mol Cell Biol. 2011;12:308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo B-H, Springer TA. Integrin structures and conformational signaling. Biol. 2006;18:579–586. doi: 10.1016/j.ceb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moser M, Legate KR, Zent R, Fassler R. The tail of integrins, talin, and kindlins. Science. 2009;324:895–899. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 9.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson CM, et al. Emergent patterns of growth controlled by multicellular form and mechanics. Proc Nat'l Acad Sci USA. 2005;102:11594–11599. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giannone G, Sheetz MP. Substrate rigidity and force define form through tyrosine phosphatase and kinase pathways. Trends Cell Biol. 2006;16:213–223. doi: 10.1016/j.tcb.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Arnold M, et al. Induction of cell polarization and migration by a gradient of nanoscale variations in adhesive ligand spacing. Nano Lett. 2008;8:2063–2069. doi: 10.1021/nl801483w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schvartzman M, et al. Nanolithographic control of the spatial organization of cellular adhesion receptors at the single-molecule level. Nano Lett. 2011;11:1306–1312. doi: 10.1021/nl104378f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature. 2003;424:334–337. doi: 10.1038/nature01805. [DOI] [PubMed] [Google Scholar]
- 15.Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310:1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 16.Salaita K, et al. Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science. 2010;327:1380–1385. doi: 10.1126/science.1181729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin W-C, Yu C-H, Triffo S, Groves JT. Supported membrane formation, characterization, functionalization, and patterning for application in biological science and technology. Current Protocols in Chemical Biology. 2010;2:235–269. doi: 10.1002/9780470559277.ch100131. [DOI] [PubMed] [Google Scholar]
- 18.Yu CH, Groves JT. Engineering supported membranes for cell biology. Med Biol Eng Comput. 2010;48:955–963. doi: 10.1007/s11517-010-0634-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu CH, Wu HJ, Kaizuka Y, Vale RD, Groves JT. Altered actin centripetal retrograde flow in physically restricted immunological synapses. PloS one. 2010;5:e11878. doi: 10.1371/journal.pone.0011878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nair PM, Salaita K, Petit RS, Groves JT. Using patterned supported lipid membranes to investigate the role of receptor organization in intercellular signaling. Nat Protoc. 2011;6:523–539. doi: 10.1038/nprot.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endlich N, Otey CA, Kriz W, Endlich K. Movement of stress fibers away from focal adhesions identifies focal adhesions as sites of stress fiber assembly in stationary cells. Cell Motil Cytoskel. 2007;64:966–976. doi: 10.1002/cm.20237. [DOI] [PubMed] [Google Scholar]
- 22.Rossier OM, et al. Force generated by actomyosin contraction builds bridges between adhesive contacts. EMBO J. 2010;29:1055–1068. doi: 10.1038/emboj.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiesner S, Legate KR, Fässler R. Integrin-actin interactions. Cell Mol Life Sci. 2005;62:1081–1099. doi: 10.1007/s00018-005-4522-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giannone G, et al. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128:561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, et al. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat Cell Biol. 2008;10:1062–1068. doi: 10.1038/ncb1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kostic A, Sheetz MP. Fibronectin rigidity response through Fyn and p130Cas recruitment to the leading edge. Mol Biol Cell. 2006;17:2684–2695. doi: 10.1091/mbc.E05-12-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- 28.Ye F, et al. Recreation of the terminal events in physiological integrin activation. J Cell Biol. 2010;188:157–173. doi: 10.1083/jcb.200908045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.del Rio A, et al. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grashoff C, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraley SI, et al. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat Cell Biol. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kubow KE, Horwitz AR. Reducing background fluorescence reveals adhesions in 3D matrices. Nat Cell Biol. 2011;13:3–5. doi: 10.1038/ncb0111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shemesh T, Geiger B, Bershadsky AD, Kozlov MM. Focal adhesions as mechanosensors: a physical mechanism. Proc Nat'l Acad Sci USA. 2005;102:12383–12388. doi: 10.1073/pnas.0500254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Na S, et al. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc Nat'l Acad Sci USA. 2008;105:6626–6631. doi: 10.1073/pnas.0711704105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai Y, et al. Nonmuscle myosin IIA-dependent force inhibits cell spreading and drives F-actin flow. Biophys J. 2006;91:3907–3920. doi: 10.1529/biophysj.106.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, et al. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat Cell Biol. 2008;10:1062–1068. doi: 10.1038/ncb1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Discher DE, Mooney DJ, Zandstra PW. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.