Abstract
Genetically modified Metarhizium spp represent a major new arsenal for combating insect pests and insect-borne diseases. However, for these tools to be used safely and effectively, we need a much better understanding of their evolutionary potential and invasion ecology. In order to model natural as well as anthropogenic dispersal scenarios, we investigated evolutionary processes in a green fluorescent protein tagged strain of Metarhizium robertsii following transfer from a semitropical to a temperate soil community. Adaptive changes occurred over four years despite recurrent genetic bottlenecks and lack of recombination with locally well adapted strains. By coupling microarray-based functional analysis with DNA hybridizations we determined that expression of cell wall and stress response genes evolved at an accelerated rate in multiple replicates, whereas virulence determinants, transposons, and chromosome structure were unaltered. The mutable genes were enriched for TATA boxes possibly because they are larger mutational targets. In further field trials, we showed that the new mutations increased the fitness of M. robertsii in the new range by enhancing saprophytic associations, and these benefits were maintained in subsequent years. Consistent with selection being habitat rather than host specific, populations of an avirulent mutant cycled with seasons similarly to the wild type, whereas a mutant unable to adhere to plant roots showed a linear decrease in population. Our results provide a mechanistic basis for understanding postrelease adaptations, show that agents can be selected that lack gene flow and virulence evolution, and describe a means of genetically containing transgenic strains by disrupting the Mad2 gene.
Keywords: adaptive evolution, biocontrol, transgenic microbes, entomopathogenic fungi, soil fungi
Predicting the consequences of different types of human intervention has become an increasingly important challenge in light of habitat fragmentation, climate change, invasive species, and genetically modified introductions (1). Genetically modified (GM) Metarhizium strains represent a major new arsenal for combating agricultural pests, insect vectors of disease, or malaria parasites within mosquitoes (2, 3). However, the current predictive data base for risk assessment of genetically engineered microbes is very small. There is little information available on survival of individual genotypes (clones) of microbes in nature, nor is there experimentally derived information on gene transfer risks between strains. Lack of knowledge about the fate of genotypes at the population and ecosystem levels creates an inherent uncertainty about the efficacy, survivability, and environmental risk posed by any biocontrol agent. Even for classical biocontrol agents to be used safely and effectively there is a need to fill a vast gap in our knowledge regarding the potential consequences of their evolving (4). Some of the outstanding evolutionary questions address fundamental yet poorly understood issues, including: What roles do different kinds of mutants play in adaptation? When organisms adapt to new environments, do they do so because of changes in few genes or many? Are the same genes involved in independent cases of adaptation to the same environment? To date these questions have been addressed by long term culture of model organisms under conditions in which they were not originally suited (5). This approach has provided an unprecedented view of natural selection in action, but might involve unnaturally harsh selective pressures and the relevance to natural environments is unclear (5). Although there are also many examples of rapid adaptation of parasites in natural populations (6), it has been difficult to disentangle the effects of new mutations from preexisting genetic variation, and the beneficial mutations are rarely identified.
Metarhizium spp. are typically mitosporic, haploid soil fungi. Soil fungi have a central role in ecosystem functioning as pathogens, decomposers, and plant symbionts, but their natural selection and invasion ecology has received scant attention (7–9). It can however be reasonably assumed that genotypic plasticity and natural or anthropogenic dispersal of fungi has contributed to their abundance and diversity (10). Metarhizium spp may exemplify these processes, as they are very common (sometimes exceeding 106 propagules per gram of soil) (11), and contain numerous intraspecific variants that range from small areas to continents, and exhibit different insect host preferences (12). Recent studies have shown that some species, including Metarhizium robertsii (formerly known as M. anisopliae var. anisopliae) also colonize the rhizosphere (the layer of soil influenced by root metabolism) of specific plant types (13–15). This result was unexpected, and given the abundance of Metarhizium could have profound implications for plant biology. Potentially, a better understanding of these interactions could lead to the engineering of Metarhizium strains into comprehensive symbionts for plant growth enhancement (16).
In spite of risk assessment concerns, in practice many deliberately introduced pest pathogens and plant symbiotic fungi are reported to have had little invasive success, and this has been attributed to the absence of genetic variation and a failure to recombine with well adapted local populations (9). However, it is also possible that establishment and spread of inconspicuous introductions could have been missed because they were not tagged with a marker. In order to model natural or human dispersal scenarios beginning with a clonal founding population, we set out to identify evolutionary processes in a green fluorescent protein (GFP) expressing M. robertsii strain Mr2575 (Mr2575-GFP) (originally isolated in humid semitropical South Carolina), after its introduction into a temperate grassland habitat in Maryland. Mr2575 is the type strain of M. robertsii (17), and is frequently employed as a model for studies on host pathogen interactions and genetic engineering (18).
Results
Metarhizium robertsii Mr2575-GFP Persisted in the New Range.
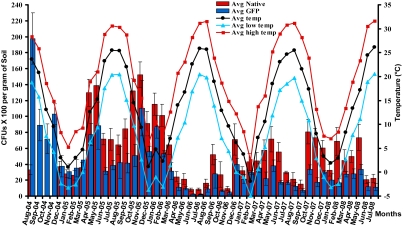
An assumption about genetically modified organisms is that they will be eliminated from the environment due to the metabolic burden of expressing a foreign gene (19). Following the initial application of Mr2575-GFP, there was a 94% decline in population size, measured as colony forming units (CFUs) in soil samples, but several intermittent recoveries occurred over four years (Fig. 1). These population cycles coincided with temperature extremes. Although Mr2575 belongs to a genotypic class mostly found in semitropical parts of the United States (12), which usually have milder winters and longer summers than Maryland, temperatures > 32 °C repressed population size more than temperatures < 0 °C (Mann-Whitney test, p < 0.001). The more days in a particular month with temperatures above 32 °C, the lower the CFU counts for that month (p < 0.001). While the introduced Mr2575-GFP initially suppressed the native Metarhizium strains, suggesting a maximum carrying capacity for the soil, the native populations had recovered four months post application, and the introduced strain comprised ∼30% of the total population throughout the rest of the study. Subsequently, native and Mr2575-GFP strains cycled in tandem (Fig. 1), indicating that their population sizes are controlled by the same biotic and abiotic factors.
Fig. 1.
Metarhizium robertsii Mr2575-GFP in turf plots cycled with seasonal changes. Soil samples were collected over four years from turf grass plots. The seasonal changes in population counts of the introduced strain were also shown by the native Metarhizium strains, indicating that they were controlled by the same biotic and abiotic factors. Error bars indicate SD, n = 15.
Metarhizium robertsii Mr2575-GFP Exhibits Adaptive Evolution in Gene Expression.
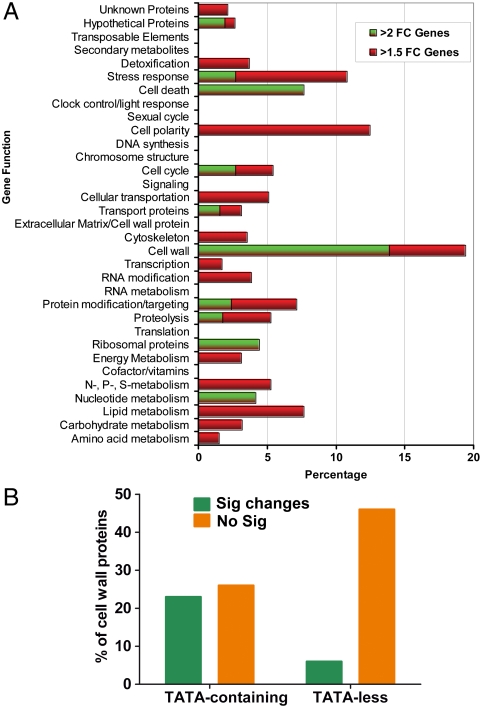
Measuring changes in gene expression with microarrays provides a very sensitive assay for mutant accumulation as mutations in many different loci can affect abundance of any one transcript and a single mutation in a regulatory locus can affect dozens of genes (20). We used cDNA microarrays composed of genes differentially expressed in soil or insect hosts (21), to inventory the accumulation of mutations in five Mr2575-GFP isolates (Re01-Re05) collected after four years in separate turf plots; i.e., in replicate populations. We found that 3.6% of the arrayed genes had significant statistical differences (≥1.5 fold change, Student’s T-test, p < 0.05) in expression across isolates, with 1.3% showing differences higher than twofold relative to the introduced strain. The average percentage of mutated genes per isolate was 0.72%. Significantly, genes coding for traits with obvious ecosystem effects such as the secreted products and pathogenicity determinants (including 45 genes for proteases), that are listed in refs. 21–23 were unaltered in regulation (Fig. 2A), in spite of approximately 60% of the arrayed genome encoding secreted products (22). Genes for most metabolic pathways were also highly conserved whereas > 12% of genes encoding cell wall proteins, cell polarity, and stress response proteins were mutable; a highly significant overrepresentation in each case (Fisher’s exact test, p < 0.0005) (Fig. 2A). We used quantitative reverse transcription-polymerase chain reaction (qRT-PCR) (24) to validate the microarray results for five cell wall proteins, one stress response protein, one hypothetical protein, and one transporter protein in Re01-Re05, and five additional isolates (Re06-Re10) (Tables S1 and S2). The same cell wall and stress response genes showed a ≥1.5-fold change (p < 0.01) in expression in at least 4 of the 10 isolates, whereas the transporter and hypothetical genes were each altered in just two of the isolates (Table S2). We next investigated whether promoter properties might influence the evolvability of gene expression. In studies with model systems, the minority of genes with TATA boxes were more likely than TATA-less genes to accumulate gene expression mutations owing to their larger cis- and trans-mutational target sizes (25). Consistent with this, mutable genes in the recovered isolates are significantly enriched for TATA boxes (Fisher’s exact test, p = 0.02). Thus, while 49% of genes encoding cell wall proteins harbor a TATA box, 47% of these had significantly (p < 0.05) altered gene expression in one or more isolates, as compared to only 11% of TATA-less genes (Fig. 2B). TATA boxes do not increase the likelihood that mutations will be adaptive as ∼70% of genes encoding secreted proteases have TATA boxes, but none were altered in regulation, suggesting that mutations in these genes had been selected against.
Fig. 2.
Microarray analysis of five isolates recovered four years after field application compared with the original introduced strain Mr2575-GFP. (A) Percentage of genes in each functional group with a significant > 1.5-fold change in gene expression in at least one of the five isolates. The cell wall, cell polarity, and stress response categories contained the highest proportions of genes with > 1.5 fold changes in expression. (B) Cell wall genes with significantly (p < 0.05) altered expression (Sig changes) are enriched for TATA boxes compared to genes with unaltered expression (No sig). FC, fold change.
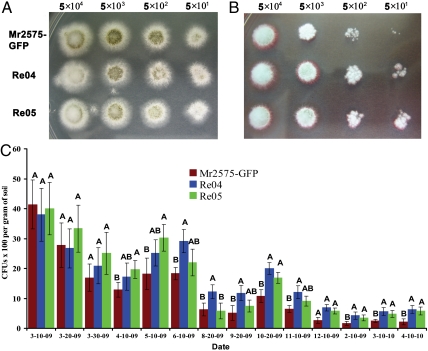
Re04 and Re05 have several parallel mutations in cell wall and stress response genes. To determine if the microarray results are predictive of phenotypic differences, we conducted a cell wall rigidity assay using Congo red which interferes with the construction of the cell wall. Both Re04 and Re05 exhibited increased tolerance to Congo red as shown by more rapid colony growth relative to the introduced strain (Fig. 3 A and B), indicative of more rigid and impermeable cell walls (26).
Fig. 3.
The evolved isolates Re04 and Re05 possessed more rigid and impermeable cell walls than the parent strain (Mr2575-GFP), and were better adapted to field conditions. To determine susceptibility of Mr2575-GFP, Re04, and Re05 to Congo red, 5 μL of each spore suspension (range 5 × 10 conidia/mL to 5 × 104 conidia/mL) was spotted onto PDA (A) or PDA complemented with Congo red (300 μg/mL) (B). The sensitivity to Congo Red (CR) is determined by comparing the extent of colony formation. To measure population levels, Re04, Re05 and the introduced strain Mr2575-GFP were collected from turf plots and CFUs were counted (C). Error bars indicate SD, n = 15.
Field Evolved Isolates Rescued Four Years After Release of Mr2575-GFP Are Better Adapted to the New Range.
A state of local adaptation can fluctuate in time as well as space (4). To determine if the accumulated mutations have fitness benefits that were maintained in subsequent years, Re04 and Re05 were reintroduced to the field and their survival compared with their parent strain Mr2575-GFP (Fig. 3C). In the first month following application (March 10 to April 10, 2009), Mr2575-GFP declined significantly more than Re04 and Re05, whereas throughout summer (June to August), Re05 declined the most. We hypothesized that Re05 may show less heat tolerance as a result of reduced expression of heat-shock protein HSP70 (Fig. S1A). As predicted, Re05 showed a significantly reduced germination rate following heat-shock (6 h at 42 °C) compared to other isolates (Student’s T-test, p < 0.03) whereas growth rates under optimal conditions were the same (Fig. S1 B and C). A possible explanation for the altered expression of HSP70 is that the responsible mutation is pleiotropic and beneficial effects offset deleterious effects. During the cooler fall season, Re05 recovered more rapidly than the other isolates, and for the rest of the year trended with Re04. Both Re04 and Re05 ended the year at significantly higher levels than Mr2575-GFP (p = 0.002). Bioassays showed no significant differences between Re04, Re05, and Mr2575-GFP in their virulence against Galleria mellonella caterpillars (Log-rank test, p = 0.42) (Fig. S1D). Unaltered virulence against caterpillars is consistent with the better fitness of field evolved isolates not being dependent on insects although it cannot exclude a change in virulence against one or more native species of field insect.
Populations of M. robertsii Are Principally Maintained by Rhizospheric Associations.
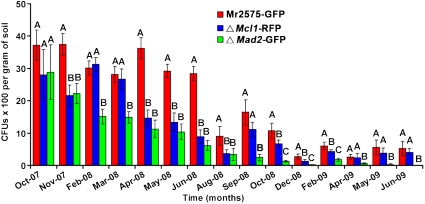
To determine whether populations of Metarhizium are principally maintained by rhizospheric associations or in insects, we compared survival of Mr2575-GFP vs. two mutant strains, ΔMcl1-RFP [expressing red fluorescent protein (RFP); disrupted in an immune evasion gene and with low infectivity to insects] (27) and ΔMad2-GFP (expressing GFP; disrupted in a gene required for adhesion to roots) (28) (Fig. S2). Mcl1 and Mad2 are specifically up-regulated by insect blood and root exudates, respectively, and lack pleiotropic effects in a panel of tests (27, 28). Scarab beetle larvae (Phyllophaga spp.), comprised approximately 80% of insects infected by green fluorescent Mr2575-GFP and ΔMad2-GFP in the field plots, with the balance being made up largely of other beetles (SI Text). We also identified ants and hemipteran bugs infected with GFP-expressing Mr2575-GFP and ΔMad2-GFP during the first month postrelease, when spore levels were very high in the application zones. Throughout the course of the study we did not find any insects infected with RFP-expressing fungus, confirming that ΔMcl1-RFP is avirulent. The ΔMcl1-RFP population cycled with seasons in the same manner as Mr2575-GFP (Spearman correlation coefficient ρ = 0.99, p = 1.6 × 10-7), albeit CFU counts were lower (Fig. 4). Between October 2007 and March 2008, ΔMad2-GFP declined in parallel with Mr2575-GFP (ρ = 0.90, p < 0.001), but correlation was less between March and May 2008 (ρ = 0.8, p = 0.1) and between June 2008 to June 2009 (ρ = 0.58, p = 0.2) because ΔMad2-GFP exhibited a continuing significant (Mann-Whitney test, p < 0.01) linear decrease in population levels without intermittent increases. In contrast to the wild type and ΔMcl1-RFP, ΔMad2-GFP was not detected in the field eighteen months post release.
Fig. 4.
Rhizosphere competence plays an important role in maintaining Metarhizium’s population. Mr2575-GFP, the avirulent mutant ΔMcl1-RFP and rhizosphere incompetent mutant ΔMad2-GFP were recovered from turf plots. Different letters indicate a significant difference (p < 0.05) in counts of CFUs. Error bars indicate SD, n = 15.
M. robertsii Mr2575 Is Exclusively Clonal.
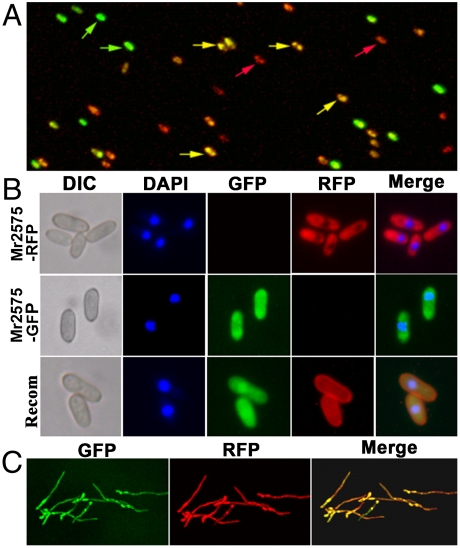
Mutation is the ultimate source of genetic variation, but genetic and fitness changes could also result from recombination, whereas local adaptation will be favored by low gene flow (4). To assay for recombination, we released a double-labeled strain Mr2575-GFP/RFP in which gfp and rfp are on two different chromosomes. The strain stability and possible dissemination of genetic material through hybridization was investigated by examining whether Mr2575-GFP/RFP retained both marker elements in their original state. During two years of continuous screening (n = 126,820 colonies), we did not find any colonies with only GFP or RFP (Table S3). We also released Mr2575-GFP and Mr2575-RFP together to measure intrastrain recombination events and found that within one month, 1.2% of CFUs were unstable monokaryon diploids (Table S3). In the laboratory, these diploids gave rise to parasexual segregants during the first parameiosis (Fig. S3). Although parasexuality can enhance adaptation (29), we did not detect segregants in the field, suggesting that they have reduced fitness.
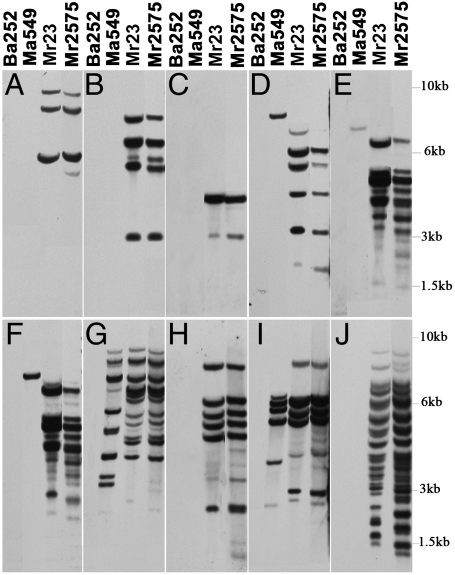
Insects are hot spots for recombination events for some Metarhizium strains (30). To confirm that Mr2575 has a very low capacity for gene introgression, we looked for recombinants in insects coinfected with different strains labeled with either GFP or RFP (Table S4). We found that twenty-four percent of spores harvested from insects infected with Mr2575-GFP and Mr2575-RFP were unstable diploids (Table S5, Fig. 5). However, no recombinants were observed when Mr2575-RFP and M. robertsii strain Mr23-GFP [from North Carolina, and in the same genotypic class as Mr2575 (12)] attacked the same insect, and several other combinations of strains were also incompatible (Table S5). Changes in genome structure and population genetic variability in fungi are often attributed to transposable elements (31). However, the distribution of transposons in Mr2575 and Mr23 are very similar (Fig. 6), suggesting that chromosomal changes and recombination events have not played a part in adaptation because Mr2575 and Mr23 were somatically compatible. Analysis of the complete genome of Mr23 revealed that it is probably exclusively asexual and clonal (23), and the current study provides experimental evidence for clonality. Of the ten selected Mr2575 transposable elements, five could not be detected and three were in single copy in M. anisopliae strain Ma549, suggesting these transposons had been active in the common ancestor of Mr23 and Mr2575 after it diverged from M. anisopliae. Isolates recovered from the field four years after application displayed no distribution pattern variation in these transposons (Fig. S4). These results suggest that new beneficial mutations are the only cause of fitness increases in Re04 and Re05, and that there is little risk of introgression of new mutations or transgenes into native strains of fungi.
Fig. 5.
Intrastrain recombinants are unstable diploids. (A) Confocal fluorescent image of conidia harvested from cadavers of Galleria mellonella coinfected by Mr2575-GFP and Mr2575-RFP. Green and red arrows indicate conidia expressing GFP or RFP only. Yellow conidia are recombinants expressing both GFP and RFP. (B) Parental strains (Mr2575-GFP and Mr2575-RFP) and their recombinants are uninucleate, confirming that recombinants are diploid. DNA was stained with DAPI (blue, 4′,6-diamidino-2-phenylindole). (C) Recombinant mycelia expressing both GFP and RFP appear yellow when GFP and RFP fields are merged. Scale bar indicates 10 μm.
Fig. 6.
Genetic relatedness revealed by transposable elements in Southern blot analysis. Beauveria bassiana Bb252, Metarhizium anisopliae Ma549, Metarhizium robertsii Mr23 and Mr2575 genomic DNA were hybridized with DIG-11-dUTP-labeled probes corresponding to the complete ORF sequences of Mr2575 transposable elements: transposase CN808808 (A), transposase AJ272685 (B), pol polyprotein AJ274203 (C), gag-like polyprotein AJ274338 (D), polyprotein AJ272783 (E) polyprotein AJ274240 (F), reverse transcriptase CN809546 (G), transposase-like protein AJ273429 (H), transposase CN808708 (I), restless-like transposase AJ274202 824J). Molecular size markers are indicated on the right.
Discussion
Most of the existing data on how fungi adapt to their environments has been obtained from highly artificial experiments (32). The present work on M. robertsii is of interest both fundamentally and practically with respect to several understudied areas of fungal evolution and invasion biology, and provides a potential model for assessing fungal evolution in natural communities. We show that contrary to expectations (4, 9), adaptive changes in a clonal lineage can occur within a few years, even in the absence of preexisting genetic variation and recombination, and that they occur despite recurrent genetic bottlenecks.
Biocontrol agents are usually selected from regions of the native range that environmentally match the release area reasonably well, but the importance of finer scale adaptation of biocontrol agents is unclear (4). We reasoned that dispersal ∼450 miles north of its origin would not subject Mr2575 to harsh selective pressures, and it would model common usages of biocontrol agents, as well as many natural dispersal scenarios. The recurring drops in population levels caused by thermal stresses are consistent with laboratory determinations of permissive temperatures for sporulation, germination, growth, and infection processes (33). However, many grasses go semidormant in temperatures > 32 °C, with reduced photosynthetic rates, root formation, and the death of older roots (34). Reduced productivity of plants could detrimentally impact population structures of insects and M. robertsii. In contrast to ΔMad2, the ΔMcl1 mutant cycles with the wild type suggesting that population levels depend on temperature-dependent interactions with plants rather than insects.
There is considerable controversy regarding the proportion of expression divergence in model systems that is attributable to natural selection rather than genetic drift (35). However, several lines of evidence suggest that the regulatory mutations in M. robertsii cell wall and stress response genes are the result of positive selection. First, parallel mutations in replicate populations is consistent with directional (positive) selection (36). Second, stress responses, cell polarity, and the cell wall, are functionally linked. The cell wall determines shape and environmental signals redirect cell wall deposition effecting cell polarity during stress responses (26). Third, these genes are likely to have a direct impact on fitness of individual conidia and germtubes. Stress responses are an essential component of survival (26), while the cell wall is the interphase with the environment, and plays a vital role in morphology, adhesion, resource acquisition, and resistance to toxins. The fact that the survival of diversified field isolates was increased suggests that microarrays can identify the mechanistic basis for adaptation to the Maryland field site.
In the absence of similar field-based evolution studies with other fungi we cannot say whether the mutation rate or functional categories of genes undergoing adaptive regulatory evolution in M. robertsii are typical. The genetic variation we observed suggests divergence is habitat rather than host specific, and this is consistent with the broad host range M. robertsii being commonest in grasslands, whereas several other broad host range Metarhizium species prefer woodlands (13). However, in light of extensive empirical support for the Red Queen hypothesis (37), the absence of changes in pathogenicity-related genes is not a result we would have predicted. TATA boxes are present in many genes encoding secreted products that have been implicated in pathogenicity. The failure to detect mutations in these genes suggests that mutations have been selected against. It is possible that purifying selection was intense because the Mr2575 phenotype is already well adapted for pathogenicity to multiple hosts. Mr2575 was isolated from the pecan weevil in South Carolina but principally infected scarab beetle larvae in Maryland. The ability to attack multiple host species may require conservation of a generic pathogenicity “tool kit,” and heterogeneous hosts could also constrain the ability to coevolve with just one host. We suggest that as hosts were heterogeneous M. robertsii instead locally adapted to more consistent components of the environment, such as grass roots. In spite of purifying selection of pathogenicity genes, the ability of the avirulent ΔMcl1-RFP to cycle with the seasons in a similar manner to Mr2575-GFP suggests that insects were not the major constraint on Mr2575-GFP populations. Although it has wild-type infectivity to insects (28), the ΔMad2-GFP mutant showed a linear decrease in population confirming that the ability to adhere to root surfaces plays an important part in maintaining population size and seasonal cycling, irrespective of the presence of insects. Metarhizium is thus in the fortuitous position of being able to maintain populations on plant roots while the additional lifestyle option of entomopathogenicity enables it to build up population levels above the carrying capacity of the rhizosphere, and possibly also escape competition from other microbes. There is not enough experimental data to conclude whether or not adaptation in a new environment is an important part of efficacious biological control (4). However, even in the absence of changes to virulence, if local adaption of Re04 and Re05 increases population size then this by itself may enhance biocontrol.
Little is known about the extended periods many plant, animal, and insect pathogens survive in soil in the absence of their hosts (38). As plant roots stabilize Mr2575-GFP populations it is feasible they play a similar role with other fungal pathogens. If so, this information would be useful for being able to predict and control outbreaks of commercially important pathogens. The data thus far from our field trials suggest that at least some Metarhizium strains pose little risk from gene flow or from evolution of host virulence in a new habitat. This knowledge alleviates science-based risk assessment concerns that are a potential obstacle to application of transgenic Metarhizium technologies. However, surveying native tropic interactions of biocontrol agents is clearly crucial in order to fully understand patterns of establishment, and also to identify sets of genes that can be used for monitoring adaptation. Furthermore, with knowledge of such genes, the precision and malleability of molecular techniques could allow the design of multiple pathogens with different strategies to be used for different ecosystems. While root colonization increases the potential of an introduced biocontrol agent to replicate and persist in the environment providing long term control, it will also increase the difficulty of eliminating an agent following unanticipated environmental effects. Fortunately, the ability to selectively block root colonization by knocking out the Mad2 gene provides an easy means of genetically containing the persistence of introduced or transgenic strains, if so required by regulatory agencies. This containment strategy could be immediately useful for transgenic strains which are not designed to recycle; e.g., Metarhizium strains expressing arthropod toxins to control vector borne diseases (2).
Materials and Methods
A detailed description of the methods is available in SI Text. Briefly, we used two strains of Metarhizium robertsii (Mr2575 and Mr23), two strains of M. anisopliae (Ma549 and Ma1080), and one strain of Beauveria bassiana (Bb252) (Table S4). Plasmid construction and fungal genetic transformation were performed as described in SI Text. Fungal release and field sample collection were performed as previously described (14); additional details are available in SI Text. We investigated genetic recombination by coinfecting insects and coinoculation of GFP and RFP-expressing strains in the field. Southern blot, real-time PCR, heat tolerance, cell wall rigidity assays, insect bioassay, and other procedures were performed as described in SI Text.
Microarray Analysis.
We used cDNA microarrays for array based mutation accumulation assays. The cDNA microarrays and RNA hybridizations were performed as described (22, 39). A loop design was used to compare gene differential expression between the original introduced strain (designated as Mr2575-GFP) and five field strains (designated as Re01, Re02, Re03, Re04, and Re05), and three microarray slides were used per comparison. Competitive hybridization of the second biological replicate was performed using a reverse dye-assignment to eliminate bias from dye incorporation. Microarray dataset and methods are available at the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo) under accession number GSE30175.
Statistical Analysis.
Comparisons of population differences were analyzed using Wilcoxon-Mann-Whitney tests. Survival curves for insects were analyzed using the Log-rank (Mantel-Cox) test. The correlation between changes in gene expression and TATA boxes was analyzed by Fisher’s exact test. Correlations in population sizes were analyzed using Spearman correlation analysis. Pair-wise comparisons of mRNA expression levels in qRT-PCR were analyzed using Student’s T-test. Except for microarray data, all statistics were performed using GraphPad Prism version 5.00 for Windows (GraphPad Software, www.graphpad.com). P-values of less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments.
This work was supported by the United States Department of Agriculture (USDA) Biotechnology Risk Assessment Research Grants Program (2006-03692 and 2009-05805)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE30175).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113824108/-/DCSupplemental.
References
- 1.Burdon JJ, Thrall PH. Coevolution of plants and their pathogens in natural habitats. Science. 2009;324:755–756. doi: 10.1126/science.1171663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C, St Leger RJ. A scorpion neurotoxin increases the potency of a fungal insecticide. Nat Biotechnol. 2007;25:1455–1456. doi: 10.1038/nbt1357. [DOI] [PubMed] [Google Scholar]
- 3.Fang WG, et al. Development of transgenic fungi that kill human malaria parasites in mosquitoes. Science. 2011;331:1074–1077. doi: 10.1126/science.1199115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hufbauer RA, Roderick GK. Microevolution in biological control: Mechanisms, patterns, and processes. Biol Control. 2005;35:227–239. [Google Scholar]
- 5.Ferea TL, Botstein D, Brown PO, Rosenzweig RF. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc Nat'l Acad Sci USA. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez P, Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- 7.van der Putten WH, Klironomos JN, Wardle DA. Microbial ecology of biological invasions. Isme J. 2007;1:28–37. doi: 10.1038/ismej.2007.9. [DOI] [PubMed] [Google Scholar]
- 8.Tirosh I, Reikhav S, Levy AA, Barkai N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science. 2009;324:659–662. doi: 10.1126/science.1169766. [DOI] [PubMed] [Google Scholar]
- 9.Desprez-Loustau ML, et al. The fungal dimension of biological invasions. Trends Ecol Evol. 2007;22:472–480. doi: 10.1016/j.tree.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Rouxel T, et al. Effector diversification within compartments of the Leptosphaeria maculans genome affected by repeat-induced point mutations. Nature Communications. 2011;22:202. doi: 10.1038/ncomms1189. doi: 10.1038/ncomms1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milner RJ. Selection and characterization of strains of Metarhizium anisopliae for control of soil insects in Australia. In: Lomer CJ, Prior C, editors. Biological Control of Locusts and Grasshoppers. Proc. Workshop Internat. Republic of Benin: Inst. Tropical Agricult., CAB International; 1992. pp. 200–207. [Google Scholar]
- 12.St Leger RJ, et al. Genetic differences in allozymes and in formation of infection structures among isolates of the entomopathogenic fungus Metarhizium anisopliae. J Invertebr Pathol. 1992;60:89–101. [Google Scholar]
- 13.Wyrebek M, Huber C, Sasan RK, Bidochka MJ. Three sympatrically occurring species of Metarhizium show plant rhizosphere specificity. Microbiology. 2011;157:2904–2911. doi: 10.1099/mic.0.051102-0. [DOI] [PubMed] [Google Scholar]
- 14.Hu G, St Leger RJ. Field studies using a recombinant mycoinsecticide (Metarhizium anisopliae) reveal that it is rhizosphere competent. Appl Environ Microb. 2002;68:6383–6387. doi: 10.1128/AEM.68.12.6383-6387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher JJ, Rehner SA, Bruck DJ. Diversity of rhizosphere associated entomopathogenic fungi of perennial herbs, shrubs and coniferous trees. J Invertebr Pathol. 2011;106:289–295. doi: 10.1016/j.jip.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Vega FE, et al. Fungal entomopathogens: new insights on their ecology. Fungal Ecol. 2009;2:149–159. [Google Scholar]
- 17.Bischoff JF, Rehner SA, Humber RA. A multilocus phylogeny of the Metarhizium anisop liaelineage. Mycologia. 2009;101:512–530. doi: 10.3852/07-202. [DOI] [PubMed] [Google Scholar]
- 18.St Leger RJ, Joshi L, Bidochka MJ, Roberts DW. Construction of an improved mycoinsecticide overexpressing a toxic protease. Proc Nat'l Acad Sci USA. 1996;93:6349–6354. doi: 10.1073/pnas.93.13.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velkov VV. Stress-induced evolution and the biosafety of genetically modified microorganisms released into the environment. J Bioscience. 2001;26:667–683. doi: 10.1007/BF02704764. [DOI] [PubMed] [Google Scholar]
- 20.Gibson G. Mutation accumulation of the transcriptome. Nat Genet. 2005;37:458–460. doi: 10.1038/ng0505-458. [DOI] [PubMed] [Google Scholar]
- 21.Wang CS, Hu G, St Leger RJ. Differential gene expression by Metarhizium anisopliae growing in root exudate and host (Manduca sexta) cuticle or hemolymph reveals mechanisms of physiological adaptation. Fungal Genet Biol. 2005;42:704–718. doi: 10.1016/j.fgb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Wang C, St Leger RJ. Developmental and transcriptional responses to host and nonhost cuticles by the specific locust pathogen Metarhizium anisopliae var. acridum. Eukaryot Cell. 2005;4:937–947. doi: 10.1128/EC.4.5.937-947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Q, et al. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 2011;7:e1001264. doi: 10.1371/journal.pgen.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S, Fang W, Wang C, St Leger RJ. Insertion of an esterase gene into a specific locust pathogen (Metarhizium acridum) enables it to infect caterpillars. PLoS Pathog. 2011;7:e1002097. doi: 10.1371/journal.ppat.1002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL. Genetic properties influencing the evolvability of gene expression. Science. 2007;317:118–121. doi: 10.1126/science.1140247. [DOI] [PubMed] [Google Scholar]
- 26.Ram AFJ, Klis FM. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat Protoc. 2006;1:2253–2256. doi: 10.1038/nprot.2006.397. [DOI] [PubMed] [Google Scholar]
- 27.Wang C, St Leger RJ. A collagenous protective coat enables Metarhizium anisopliae to evade insect immune responses. Proc Nat'l Acad Sci USA. 2006;103:6647–6652. doi: 10.1073/pnas.0601951103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang CS, St Leger RJ. The MAD1 adhesin of Metarhizium anisopliae links adhesion with blastospore production and virulence to insects, and the MAD2 adhesin enables attachment to plants. Eukaryot Cell. 2007;6:808–816. doi: 10.1128/EC.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoustra SE, Debets AJ, Slakhorst M, Hoekstra RF. Mitotic recombination accelerates adaptation in the fungus Aspergillus nidulans. PLoS Genet. 2007;3:e68. doi: 10.1371/journal.pgen.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leal-Bertioli SCM, Butt TM, Peberdt JF, Bertioli DJ. Genetic exchange in Metarhizium anisopliae strains coinfecting Phaedon cochleariae is revealed by molecular markers. Mycol Res. 2000;104:409–414. [Google Scholar]
- 31.Rep M, Charlotte van der Does H, Cornelissen BJC. Drifter, a novel, low copy hAT-like transposon in Fusarium oxysporum is activated during starvation. Fungal Genet Biol. 2005;42:546–553. doi: 10.1016/j.fgb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Frenkel O, et al. Ecological genetic divergence of the fungal pathogen Didymella rabiei on sympatric wild and domesticated Cicer spp. (Chickpea) Appl Environ Microbiol. 2010;76:30–39. doi: 10.1128/AEM.01181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Croos JNA, Bidochka MJ. Cold-induced protein in cold-active isolates of the insect-pathogenic fungus Metarhizium anisopliae. Mycol Res. 2001;105:868–873. [Google Scholar]
- 34.Huang BR, Liu XZ. Summer root decline: production and mortality for four cultivars of creeping bentgrass. Crop Sci. 2003;43:258–265. [Google Scholar]
- 35.Fay JC, Wittkopp PJ. Evaluating the role of natural selection in the evolution of gene regulation. Heredity. 2008;100:191–199. doi: 10.1038/sj.hdy.6801000. [DOI] [PubMed] [Google Scholar]
- 36.Orr HA. The probability of parallel evolution. Evolution. 2005;59:216–220. [PubMed] [Google Scholar]
- 37.Wayne ML, et al. Expression of defense genes in Drosophila evolves under a different selective regime from expression of other genes. Evolution. 2011;65:1068–1078. doi: 10.1111/j.1558-5646.2010.01197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerry BR. Rhizosphere interactions and the exploitation of microbial agents for the biological control of plant-parasitic nematodes. Annu Rev Phytopathol. 2000;38:423–441. doi: 10.1146/annurev.phyto.38.1.423. [DOI] [PubMed] [Google Scholar]
- 39.Pava-Ripoll M, et al. The rhizosphere-competent entomopathogen Metarhizium anisopliae expresses a specific subset of genes in plant root exudate. Microbiology. 2011;157:47–55. doi: 10.1099/mic.0.042200-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.