Abstract
Down-regulation of the enzyme hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase (HCT) in thale cress (Arabidopsis thaliana) and alfalfa (Medicago sativa) leads to strongly reduced lignin levels, reduced recalcitrance of cell walls to sugar release, but severe stunting of the plants. Levels of the stress hormone salicylic acid (SA) are inversely proportional to lignin levels and growth in a series of transgenic alfalfa plants in which lignin biosynthesis has been perturbed at different biosynthetic steps. Reduction of SA levels by genetically blocking its formation or causing its removal restores growth in HCT–down-regulated Arabidopsis, although the plants maintain reduced lignin levels. SA-mediated growth inhibition may occur via interference with gibberellic acid signaling or responsiveness. Our data place SA as a central component in growth signaling pathways that either sense flux into the monolignol pathway or respond to secondary cell-wall integrity, and indicate that it is possible to engineer plants with highly reduced cell-wall recalcitrance without negatively impacting growth.
Keywords: biofuel crops, defense signaling, lignin modification, Medicago truncatula
The phenylpropanoid-derived polymer lignin cross-links plant secondary cell walls to provide mechanical strength and hydrophobicity to the vascular system as well as contributes to defense against biotic stress (1). Reducing lignin levels by genetic manipulation improves both forage digestibility and processing of lignocellulosic biomass for liquid biofuel production (2). However, transgenic plants down-regulated in the hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase (HCT) enzyme, which have a strong reduction in lignin levels, show severe defects in growth (3–5). Similar, although less severe, effects are observed in plants down-regulated in some, but not all, of the other enzymes of the monolignol pathway (5–10).
The dwarf phenotype of HCT–down-regulated Arabidopsis plants can be alleviated by restoring lignin accumulation through expression of a Selaginella enzyme that bypasses the reactions catalyzed by HCT (4), suggesting that structural alterations to the partially lignified secondary cell walls are in some way linked to the growth defects. Such structural alterations include increased extractability of pectic polysaccharides (11). It is not clear whether it is possible to strongly reduce lignin content in plant cell walls without causing structural changes that will lead to deleterious effects.
HCT–down-regulated alfalfa plants contain increased levels of the stress hormone salicylic acid (SA) and SA-inducible pathogenesis-related (PR) protein transcripts (11). PR gene expression can be induced by cell-wall pectic fragments via processes involving SA (12, 13), and SA can impact plant growth and development through largely unknown mechanisms (14, 15). A previous report that lignin reduction in HCT–down-regulated plants results from flavonoid-mediated inhibition of auxin transport has recently been refuted (4).
Levels of SA correlate with the extent of lignin reduction in a series of transgenic alfalfa lines expressing antisense or RNAi constructs independently targeting seven enzymes of the monolignol pathway (16). The potential relationship between SA levels and lignin is intriguing because SA is biosynthetically related to lignin (17). Two major routes have been proposed for SA biosynthesis in plants. The initial pathway via isochorismate, derived from the shikimic acid pathway, is supported genetically (18), but enzymes that convert isochorismate to SA have yet to be shown in plants. Similarly, the previously favored pathway via cinnamate is believed to involve a benzoate 2-hydroxylase (19), but no gene encoding this enzyme has been shown in plants. Theoretically, back-up of flux into the lignin pathway could result in increased production of SA via either route. Alternatively, SA accumulation might be a result of activation of endogenous defense responses by elicitor-active polysaccharides released from improperly lignified cell walls (11).
We show here that removal of SA by genetic approaches relieves growth inhibition but maintains low lignin levels in HCT–down-regulated Arabidopsis thaliana. Thus, there is no a priori reason why plants cannot be engineered to contain highly processable cell walls yet also produce abundant biomass. The effects of SA are associated with alterations in gibberellin (GA) responsiveness and signaling.
Results
Correlations Between SA Levels and Growth in a Population of Lignin-Modified Plants.
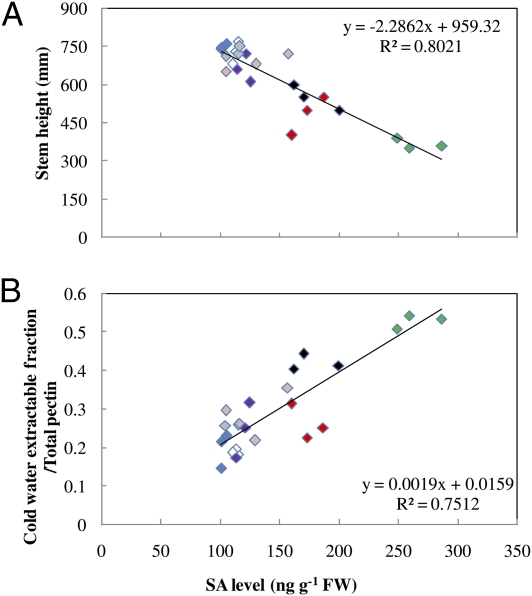
To investigate whether SA levels were directly related to growth phenotypes, we examined a series of transgenic alfalfa lines in which lignin biosynthesis had been perturbed via antisense or RNAi-mediated down-regulation (2). The collection of plants analyzed consisted of three WT controls and sets of three independent transgenic events down-regulated independently at each of six successive steps of the monolignol pathway (24 plants grown in parallel) (Fig. 1). Lignin levels (thioacidolysis yields) in the first six internodes were from WT levels (130 nmol⋅g−1 dry weight) to as low as 30 nmol⋅g−1 dry weight and inversely correlated with SA levels (R2 = 0.796) (16). Stem height was inversely correlated with endogenous SA levels across this population of plants (Fig. 1A). In addition, a positive correlation was observed between SA levels and the amount of cell-wall pectic material that was extractable by cold water from alcohol-insoluble cell-wall residues prepared from the aerial portions of the plants (Fig. 1B). Altogether, these data indicate that the previous observations of elevated SA levels and water-soluble pectic elicitors of PR protein induction in HCT–down-regulated alfalfa (11) are not specific to HCT down-regulation but reflect the extent of lignin reduction independent of the lignin pathway enzyme targeted.
Fig. 1.
SA levels, growth, and cold-water–extractable pectin in a population of transgenic alfalfa plants with lignin levels down-regulated at different steps in the monolignol pathway. (A) Stem height. (B) Pectin extractable from cell walls by cold water. The plant lines are color-coded depending on the gene target: red, 4-coumarate CoA ligase; black, coumaroyl shikimate 3′-hydroxylase; green, HCT; white, caffeoyl CoA 3-O-methyltransferase; violet, CCR; pink, cinnamyl alcohol dehydrogenase; gray, caffeic acid 3-O-methyltransferase; blue, WT. P < 0.0001. Stems with six internodes were harvested before flowering. For each transgenic line, three biological replicates were taken, each consisting of a pool of three stems from different plants.
Blocking SA Production Restores Growth to HCT–Down-Regulated A. thaliana Plants.
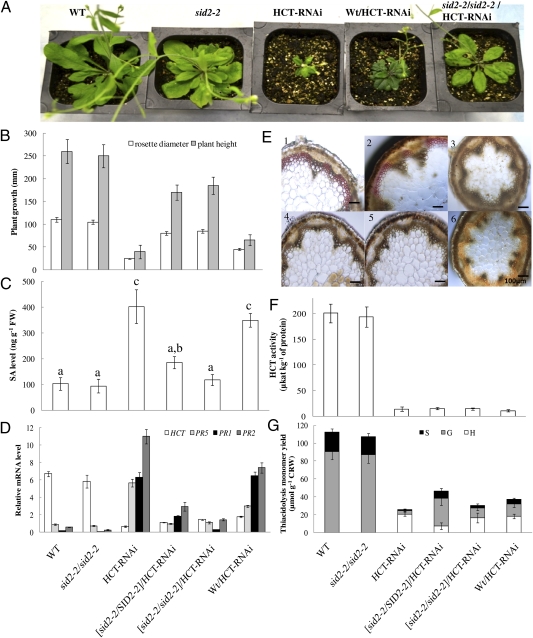
We used A. thaliana to genetically separate reduced lignin from elevated SA levels because mutants of Arabidopsis in which SA biosynthesis is blocked are available. As in alfalfa (11), Arabidopsis HCT-RNAi lines are severely dwarfed compared with WT plants (Fig. 2A) (3) and exhibit increased SA levels and expression of the PR-1, PR-2, and PR-5 defense genes (Fig. S1 A and B). SA and PR transcript levels are also elevated in the cinnamoyl CoA reductase 1 (ccr1) mutant of Arabidopsis (Fig. S2), suggesting that, as in alfalfa, elevated SA production is not restricted to plants with lignin modification targeted only to HCT.
Fig. 2.
Blocking SA accumulation restores growth to HCT-RNAi Arabidopsis. (A) Fifteen-week-old plants grown in soil. (B) Rosette diameters and plant heights for the genotypes in A. (C) SA levels. Values with different letters are significantly different (P < 0.05). (D) Transcript levels of the HCT, PR5, PR1, and PR2 genes as determined by qRT-PCR. (E) Maüle staining of stem cross-sections. Note loss of red staining in interfascicular fibers in all lines expressing HCT-RNAi. 1, WT; 2, sid2-2 null mutant; 3, HCT-RNAi; 4, HCT-RNAi with heterozygous sid2-2; 5, HCT-RNAi with homozygous sid2-2; 6, WT × HCT-RNAi. (F) Extractable HCT enzyme activity. (G) Lignin content and composition as determined by thioacidolysis. H, hydroxyphenyl unit; G, guaiacyl unit; S, syringyl unit. Results are means ± SD of three biological replicates (each a pool of three plants of the same genotype).
SA biosynthesis may occur through either the isochorismate synthase (ICS) route or via benzoic acid derived from cinnamate (17). The fast-neutron–generated sid2-2 mutant of Arabidopsis contains an exon deletion in the gene encoding ICS (20) that is implicated in SA biosynthesis (18). Homozygous sid2-2 plants have growth, basal SA levels, and HCT and PR transcript levels similar to those in WT plants (Fig. 2 A–D). To determine the impact of blocking SA biosynthesis through loss of function of ICS on the growth of HCT–down-regulated plants, homozygous sid2-2 mutant plants were crossed with a homozygous T5 HCT-RNAi plant containing a WT SID2-2 allele. A control cross between WT and HCT-RNAi plants was performed in parallel. In the F1 population, heterozygous HCT-RNAi × WT plants exhibited reduced plant height similar to that in the HCT-RNAi dwarf parents, whereas heterozygous sid2-2 × HCT-RNAi progeny showed a growth phenotype intermediate between that of the parents. The heterozygous sid2-2 × hemizygous HCT-RNAi plants from the F1 population were crossed again with the sid2-2 null mutant. Among a population of 40 progeny, four genotypes were found at approximately equal frequency: sid2-2/SID2-2, sid2-2/sid2-2, sid2-2/SID2-2 with HCT-RNAi, and sid2-2/sid2-2 with HCT-RNAi. The HCT-RNAi × WT progeny retained their dwarf growth habit (Fig. 2 A and B), high SA levels (Fig. 2C), and elevated PR transcript levels (Fig. 2D). In contrast, SA and PR transcript levels were reduced and growth was considerably restored in HCT-RNAi lines either homozygous or heterozygous for the sid2 mutation (Fig. 2 A–D). The SA levels of the sid2-2 heterozygotes were intermediate, between those of the HCT-RNAi and sid2-2 or WT values (Fig. 2C). Heterozygous sid2 mutants in the WT background do not exhibit reduced SA levels after fungal infection (21); the apparent lack of haploinsufficiency with regard to the SA phenotype in the present crosses may be the result of the different HCT expression levels in infected plants in a WT background (such as in ref. 19) and the present uninfected plants with an HCT-RNAi background.
Extractable HCT enzymatic activity, staining of syringyl lignin with Maüle reagent (1), and lignin monomer thioacidolysis yields were strongly reduced in all lines expressing the HCT-RNAi construct, including those homozygous for the sid2 mutant with restored growth (Fig. 2 E–G). The lignin composition of the HCT-RNAi lines in the sid2-2/sid2-2 background was typical of plants with reduced HCT expression, namely elevated hydroxyphenyl lignin units, and strongly reduced levels of guaiacyl and syringyl units (22). Thus, HCT–down-regulated Arabidopsis plants in which SA biosynthesis is blocked no longer display a dwarf phenotype, despite their strongly reduced lignin levels and drastically altered lignin composition.
Conversion of SA to Catechol Also Restores Growth to HCT–Down-Regulated A. thaliana Plants.
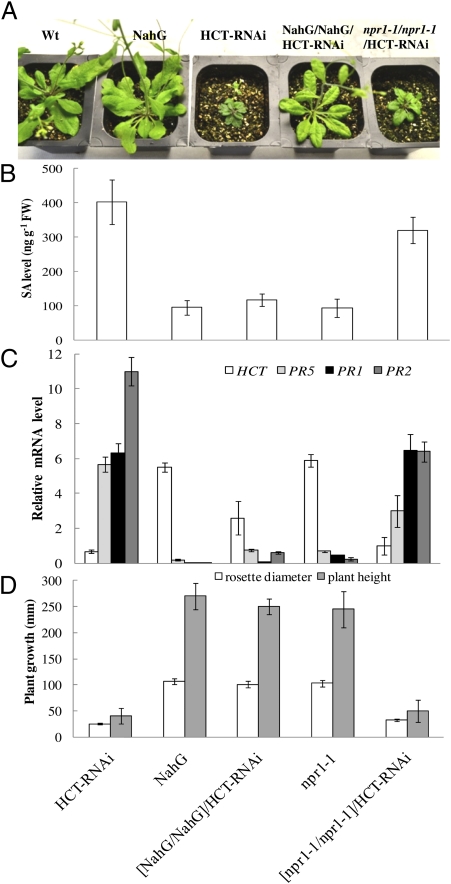
Because of the potential of multiple pathways for SA biosynthesis, we sought additional genetic confirmation of the role of SA in the growth phenotype of HCT-RNAi plants. The HCT-RNAi line was therefore crossed with plants expressing the bacterial NahG gene, which encodes a salicylate hydroxylase that removes SA by converting it to catechol (23). NahG Arabidopsis plants have WT growth habit (Fig. 3 A and D), and introduction of the NahG transgene into HCT-RNAi plants restored growth to WT levels and reduced SA and PR transcript levels (Fig. 3 A–D) in a manner similar to the introduction of the sid2 mutation. The extractable HCT activity of the individual NahG/HCT-RNAi progeny plants analyzed varied considerably, as did the overall lignin level, both of which, however, were always lower than those of the NahG parent line (Fig. S3 A and B). In addition, all NahG:HCT-RNAi lines showed the hydroxyphenyl lignin signature characteristic of HCT down-regulation (Fig. S3B). The elevation of HCT expression in many of the lines from this cross is likely the result of partial silencing of the 35S promoter derived HCT-RNAi by the additional copy of the 35S promoter driving the NahG gene. Importantly, however, HCT expression and lignin levels in some progeny lines were as low as in the HCT-RNAi parent line, but growth was normal (Fig. S3 B and C).
Fig. 3.
Removal of SA by expression of NahG restores growth to HCT-RNAi Arabidopsis, but interference with SA signaling does not. Plants are F2 progeny of the crosses NahG × HCT-RNAi and npr1-1 × HCT-RNAi. (A) Plant phenotypes. (B) SA levels. (C) Transcript levels of HCT, PR5, PR1, and PR2 as determined by qRT-PCR. (D) Rosette diameters and plant height. Results are means ± SD of three biological replicates (each a pool of three plants of the same genotype).
Reduction of SA-Mediated Defense Signaling Does Not Restore Growth to Low-Lignin Arabidopsis Plants.
To test whether PR expression, distinct from SA production, was causing the growth defects in HCT-RNAi plants, we crossed HCT-RNAi plants with plants harboring a mutation in NPR1, a gene involved in signal transduction between SA and PR expression (24). HCT-RNAi lines homozygous for the npr1 mutation retained elevated SA levels (Fig. 3B) and exhibited a partial reduction (∼50%) of PR5 and PR2 transcript levels, although PR1 transcripts were unaltered (Fig. 3C). The growth of these plants remained severely stunted (Fig. 3 A and D). These results are consistent with the fact that PR genes can be regulated by an alternative NPR1-independent pathway (25, 26); for example, constitutive PR gene expression in the cpr6-1 dominant mutation in Arabidopsis requires elevated SA levels but not NPR1 function (25).
Restoring Growth by Removal of SA Is Not Mediated by Altered Flavonoid Levels.
Arabidopsis HCT-RNAi plants accumulate high levels of flavonoids because of metabolic spillover from the lignin to the flavonoid pathway, which shares 4-coumaroyl CoA, the substrate for HCT, as a common early intermediate (3, 4). Flavonoid levels were reduced somewhat in the sid2-2 × HCT-RNAi plants, but they still contained significantly higher levels of flavonoids than WT plants did (Fig. S4), confirming the previous conclusion that elevated flavonoid accumulation is not the primary reason for the growth defect in HCT-RNAi plants (4).
SA-Mediated Growth Effects in Low-Lignin Plants May Operate via GA Signaling.
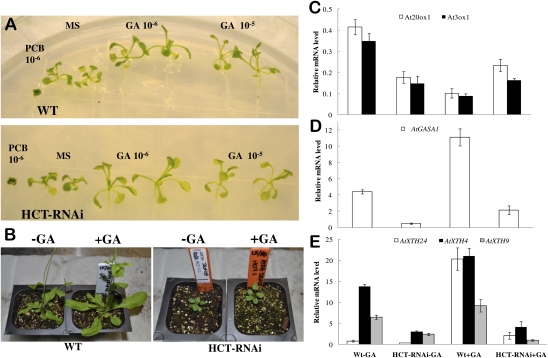
HCT-RNAi alfalfa plants show reduced GA levels and perception (11). To test whether the dwarf phenotype of Arabidopsis HCT-RNAi lines with elevated levels of SA is mediated via altered GA sensing or signaling, we first examined the effects of exogenous addition of GA4 to Arabidopsis seedlings germinated on Petri plates for 10 d. The GA biosynthesis inhibitor paclobutrazol caused total inhibition of growth of both WT and HCT-RNAi lines (Fig. 4A). Both WT and HCT-RNAi seedlings showed similar growth enhancement at GA concentrations of 10−6 M and above (Fig. 4A). However, at 8 wk postgermination, GA induced similar increases in rosette diameter and inflorescence length in WT, NahG, and sid2-2 plants but not in HCT-RNAi lines (Fig. 4B). GA did, however, increase rosette diameter and petiole length in lines in which the HCT-RNAi was expressed in the sid2-2 or NahG background, and inflorescence length also increased in NahG × HCT-RNAi lines (Table S1). Thus, GA responsiveness is lost in HCT-RNAi lines at later stages of vegetative development when lignification is occurring but can be restored by genetic manipulation to block accumulation of SA.
Fig. 4.
Mature HCT-RNAi Arabidopsis plants have reduced responsiveness to GA. (A) Response of 12-d-old WT and HCT-RNAi seedlings to supplementation of the medium with the GA biosynthesis inhibitor paclobutrazol (PCB; 10−6 M) or GA4 at 10−6 or 10−5 M. (B) Response of 8-wk-old plants to 10−5 M GA4. Pictures were taken after 2 wk of twice-per-week GA treatment. (C–E) Transcript levels (by qRT-PCR) of GA biosynthesis and response genes in 8-wk-old Arabidopsis plants with and without GA application (8 d postapplication). Means and SDs from three biological replicates are given relative to the Arabidopsis serine/threonine phosphatase (AtPP2A) transcript levels. (C) GA20 and GA3 oxidase. (D) GASA1. (E) XTH4, XTH9, and XTH24.
To test whether the reduction in growth of HCT-RNAi lines is associated with SA-mediated impairment of GA biosynthesis/signaling, we designed PCR primers for the Arabidopsis GA3 and GA20 oxidases (GA biosynthesis, repressed by exogenous GA application), gibberellic acid-stimulated transcript protein homolog 1 (GASA1), and xyloglucan endotransglycosylase/hydrolase (XTH) genes XTH4, XTH9, XTH24, and XTH32 (GA-induced) (Table S2), orthologs of which are down-regulated in HCT–down-regulated alfalfa (11). Quantitative RT-PCR (qRT-PCR) analysis indicated that these genes were repressed in the HCT-RNAi Arabidopsis line compared with WT, even after exogenous GA application (Fig. 4 C–E). However, the GA responsiveness of these genes (repression for GA20 and GA3 oxidases and induction for GASA1 and XTH genes) was restored in HCT-RNAi lines in the sid2-2 and NahG backgrounds after exogenous GA application (Fig. S5 A–C), with the notable exception of XTH9 in sid2 lines (Fig. S5C).
Discussion
Down-regulation of lignin pathway genes in transgenic plants often leads to reduced biomass associated with altered growth and vascular morphology. This phenomenon has been observed in plants with partial knockdown of enzyme activities through expression of antisense or RNAi constructs (5, 7) and is more apparent in genetic knockouts, which are often highly dwarfed (4, 27, 28). If these growth effects are primarily caused by altered vascular morphology and function, it will be difficult to reverse-engineer low-lignin plants for improved growth. This concept was questioned when it was proposed that metabolic spillover from the lignin to the flavonoid pathway was the cause of the reduced growth phenotype in HCT–down-regulated Arabidopsis plants via flavonoid-mediated inhibition of auxin transport (3). However, although down-regulation of HCT clearly does result in increased flavonoid levels (3, 11), reduced growth in HCT–down-regulated alfalfa plants is not associated with alterations in auxin transport (11); flavonoids are elevated in some, but not all, reduced lignin lines with altered growth (2). Furthermore, reexamination of the original experiments linking flavonoids to growth effects in Arabidopsis (4), as well as the present analyses of flavonoid levels in Arabidopsis plants with low lignin but restored growth, strongly support the argument that flavonoids are not causally linked to growth defects in lignin-modified plants but that SA levels are.
Two pathways have been postulated for SA biosynthesis in plants: from cinnamate via benzoate (19) or from shikimate via isochorismate (18). The fact that the elevated SA levels present in HCT-RNAi plants were reduced to WT levels in HCT-RNAi plants homozygous for the sid2 mutation suggests that lignin down-regulation induces SA formation primarily through the isochorismate pathway, which operates in Arabidopsis during responses to pathogens (18). The correlation between SA levels and the proportion of cold-water–extractable pectin to total pectin (Fig. 1B) in multiple transgenic reduced lignin alfalfa lines is consistent with the hypothesis that release of pectic elicitors from underlignified secondary cell walls in HCT-RNAi lines induces SA, and thereby defense responses, in the same manner as would occur when pathogens degrade cell walls during ingress (11, 12) rather than that the SA accumulates simply as a result of metabolic spillover from the monolignol pathway. A program of secondary wall formation with limited lignification can result in pectin that is more extractable (11), and pectic oligosaccharides are well known as elicitors of plant defense responses (12, 29, 30).
HCT-RNAi alfalfa (11) and Arabidopsis (the present work) are nonresponsive to GA as a growth enhancer when beyond the early seedling stage. GA-induced growth is restored upon reduction of the SA pool in Arabidopsis, suggesting that SA may mediate its growth-reducing effects through GA signaling. Our data do not support an alternative model in which reduced growth is largely a result of the diversion of energy into defense responses (e.g., PR protein expression). Although SA is increasingly being shown to impact plant growth (15), there has been little study of its potential involvement in elongation growth. Our work therefore provides a unique system for studying the poorly understood cross-talk between plant defense and growth control mechanisms (31, 32). In conclusion, we have shown that SA-mediated events are central to the orchestration of the reduced growth response of HCT-RNAi plants. This finding has clear implications for the engineering of improved bioenergy crops with optimal agronomic performance.
Materials and Methods
Plant Material.
Arabidopsis HCT-RNAi lines were obtained from Purdue University and were as previously described (4). Plants used for crossing were the mutant alleles sid2-2 (18, 20) and npr1-1 (24) along with plants expressing the NahG transgene (33); all were in the A. thaliana Columbia-0 ecotype background. The HCT-RNAi transgene was detected by RT-PCR using the primer pair reported previously (4). Mutants carrying the npr1-1 mutation were identified with the codominant amplified polymorphic sequences (CAPS) markers described in ref. 24. Arabidopsis ccr1 (AT1G15950.1) mutant seeds were obtained from the Arabidopsis Biological Resource Center (line SALK_123689). Primer pairs for plant genotyping were as follows: left primer, 5′-GTGTCGTAGAGGCTTTGCTTG-3′; right primer, 5′-TTGTGGAAATATTTCCGGTTG-3′; and LBb1.3, 5′-ATTTTGCCGATTTCGGAAC-3′. Alfalfa antisense lines analyzed were as described previously (5, 34, 35). For the specific lines used, see SI Materials and Methods.
GA Application.
Treatments with GA4 were as described in SI Materials and Methods.
Plant Growth Measurement and Histochemical Analysis.
Plant height was measured as described in SI Materials and Methods. Maüle staining for lignin was performed as previously described (7) in cross-sections from the base of the mature stems.
Determination of SA Levels.
SA levels were determined by using the biosensor organism Acinetobacter sp. ADPWH_lux as described previously (36, 37) and in SI Materials and Methods.
Assay of HCT Activity and Determination of Lignin and Flavonoid Levels.
HCT activity was determined as described in SI Materials and Methods. Lignin content and composition was determined by thioacidolysis (38). Soluble phenolic compounds were extracted by using the protocol described in ref. 4 and as further described in SI Materials and Methods.
Determination of Pectic Compounds.
Extraction of pectic materials from alcohol-insoluble cell-wall residues was determined as described previously (11). The proportion of pectic material released by cold-water extraction was determined as a proportion of total pectin.
Measurement of Transcript Levels by qRT-PCR.
qRT-PCR analysis was performed as described previously (11). Gene-specific primers are listed in Table S2.
Statistical Analysis.
Statistical treatment of data was performed by ANOVA using Fisher's least significant difference procedure for multiple-comparison tests (Statgraphics Plus, version 5.1 for Windows). Significance of correlations was obtained by using the online calculator for Pearson correlation P values at http://www.danielsoper.com/statcalc/calc44.aspx.
Supplementary Material
Acknowledgments
We thank Dr. Clint Chapple for Arabidopsis HCT-RNAi lines, Dr. Hui Wang for Acinetobacter sp. ADPWH_lux, David Human for assistance with liquid chromatography/MS data analysis, and Drs. Rujin Chen and Rao Uppalapati for critical reading of the manuscript. This work was supported by the Oklahoma Bioenergy Center and the Bioenergy Sciences Center (which is supported by the Office of Biological and Environmental Research in the Department of Energy Office of Science).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117873108/-/DCSupplemental.
References
- 1.Lewis NG, Yamamoto E. Lignin: Occurrence, biogenesis and biodegradation. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:455–496. doi: 10.1146/annurev.pp.41.060190.002323. [DOI] [PubMed] [Google Scholar]
- 2.Chen F, Dixon RA. Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol. 2007;25:759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- 3.Besseau S, et al. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell. 2007;19:148–162. doi: 10.1105/tpc.106.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Bonawitz ND, Weng JK, Chapple C. The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell. 2010;22:1620–1632. doi: 10.1105/tpc.110.074161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shadle G, et al. Down-regulation of hydroxycinnamoyl CoA: Shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry. 2007;68:1521–1529. doi: 10.1016/j.phytochem.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 6.Jackson LA, et al. Improving saccharification efficiency of alfalfa stems through modification of the terminal stages of monolignol biosynthesis. BioEnergy Res. 2008;1:180–192. [Google Scholar]
- 7.Nakashima J, Chen F, Jackson L, Shadle G, Dixon RA. Multi-site genetic modification of monolignol biosynthesis in alfalfa (Medicago sativa): Effects on lignin composition in specific cell types. New Phytol. 2008;179:738–750. doi: 10.1111/j.1469-8137.2008.02502.x. [DOI] [PubMed] [Google Scholar]
- 8.Ranjeva R, Refeno G, Boudet AM, Marmé D. Activation of plant quinate:NAD 3-oxidoreductase by Ca and calmodulin. Proc Natl Acad Sci USA. 1983;80:5222–5224. doi: 10.1073/pnas.80.17.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chabannes M, et al. In situ analysis of lignins in transgenic tobacco reveals a differential impact of individual transformations on the spatial patterns of lignin deposition at the cellular and subcellular levels. Plant J. 2001;28:271–282. doi: 10.1046/j.1365-313x.2001.01159.x. [DOI] [PubMed] [Google Scholar]
- 10.Chabannes M, et al. Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 2001;28:257–270. doi: 10.1046/j.1365-313x.2001.01140.x. [DOI] [PubMed] [Google Scholar]
- 11.Gallego-Giraldo L, Jikumaru Y, Kamiya Y, Tang Y, Dixon RA. Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.) New Phytol. 2011;190:627–639. doi: 10.1111/j.1469-8137.2010.03621.x. [DOI] [PubMed] [Google Scholar]
- 12.Roco A, Castaneda P, Pérez LM. Oligosaccharides released by pectinase treatment of citrus limon seedlings are elicitors of the plant response. Phytochemistry. 1993;33:1301–1306. [Google Scholar]
- 13.Delaney TP, et al. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 14.Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant Mol Biol. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 15.Rivas-San Vicente M, Plasencia J. Salicylic acid beyond defence: Its role in plant growth and development. J Exp Bot. 2011;62:3321–3338. doi: 10.1093/jxb/err031. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y, Chen F, Gallego-Giraldo L, Dixon RA, Voit EO. Integrative analysis of transgenic alfalfa (Medicago sativa L.) suggests new metabolic control mechanisms for monolignol biosynthesis. PLoS Comp Biol. 2011;7:e1002047. doi: 10.1371/journal.pcbi.1002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Zheng Z, Huang J, Lai Z, Fan B. Biosynthesis of salicylic acid in plants. Plant Signal Behav. 2009;4:493–496. doi: 10.4161/psb.4.6.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 19.León J, Shulaev V, Yalpani N, Lawton MA, Raskin I. Benzoic acid 2-hydroxylase, a soluble oxygenase from tobacco, catalyzes salicylic acid biosynthesis. Proc Natl Acad Sci USA. 1995;92:10413–10417. doi: 10.1073/pnas.92.22.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dewdney J, et al. Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 2000;24:205–218. doi: 10.1046/j.1365-313x.2000.00870.x. [DOI] [PubMed] [Google Scholar]
- 21.Nawrath C, Métraux JP. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann L, et al. Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell. 2004;16:1446–1465. doi: 10.1105/tpc.020297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaffney T, et al. Requirement of salicylic acid for the induction of systemic acquired resistance. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- 24.Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 25.Clarke JD, Liu Y, Klessig DF, Dong X. Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell. 1998;10:557–569. doi: 10.1105/tpc.10.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah J, Kachroo P, Nandi A, Klessig DF. A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 2001;25:563–574. doi: 10.1046/j.1365-313x.2001.00992.x. [DOI] [PubMed] [Google Scholar]
- 27.Franke R, et al. The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 2002;30:33–45. doi: 10.1046/j.1365-313x.2002.01266.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhou R, et al. Distinct cinnamoyl CoA reductases involved in parallel routes to lignin in Medicago truncatula. Proc Natl Acad Sci USA. 2010;107:17803–17808. doi: 10.1073/pnas.1012900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albersheim P, et al. Structure and function of complex carbohydrates active in regulating the interactions of plants and their pests. Rec Adv Phytochem. 1981;15:37–58. [Google Scholar]
- 30.Darvill AG, Albersheim P. Phytoalexins and their elicitors—A defense against microbial infection in plants. Annu Rev Plant Physiol. 1984;35:243–275. [Google Scholar]
- 31.Robert-Seilaniantz A, Navarro L, Bari R, Jones JDG. Pathological hormone imbalances. Curr Opin Plant Biol. 2007;10:372–379. doi: 10.1016/j.pbi.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Achard P, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 33.Ryals JA, et al. Systemic acquired resistance. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen F, et al. Multi-site genetic modulation of monolignol biosynthesis suggests new routes for formation of syringyl lignin and wall-bound ferulic acid in alfalfa (Medicago sativa L.) Plant J. 2006;48:113–124. doi: 10.1111/j.1365-313X.2006.02857.x. [DOI] [PubMed] [Google Scholar]
- 35.Reddy MS, et al. Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.) Proc Natl Acad Sci USA. 2005;102:16573–16578. doi: 10.1073/pnas.0505749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang WE, et al. Chromosomally located gene fusions constructed in Acinetobacter sp. ADP1 for the detection of salicylate. Environ Microbiol. 2005;7:1339–1348. doi: 10.1111/j.1462-5822.2005.00821.x. [DOI] [PubMed] [Google Scholar]
- 37.Huang WE, et al. Quantitative in situ assay of salicylic acid in tobacco leaves using a genetically modified biosensor strain of Acinetobacter sp. ADP1. Plant J. 2006;46:1073–1083. doi: 10.1111/j.1365-313X.2006.02758.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Q, et al. An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J. 2010;63:100–114. doi: 10.1111/j.1365-313X.2010.04223.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.