Abstract
Bardet-Biedl syndrome (BBS) is a heterogeneous disorder characterized by obesity, retinopathy, polydactyly, and congenital anomalies. The incidence of hypertension and diabetes are also increased in BBS patients. Mutation of 16 genes independently causes BBS, and seven BBS proteins form the BBSome that promotes ciliary membrane elongation. BBS3 (ARL6), an ADP ribosylation factor-like small GTPase, is not part of the BBSome complex. The in vivo function of BBS3 is largely unknown. Here we developed a Bbs3 knockout model and demonstrate that Bbs3−/− mice develop BBS-associated phenotypes, including retinal degeneration, male infertility, and increased body fat. Interestingly, Bbs3−/− mice develop some unique phenotypes not seen in other BBS knockout models: no overt obesity, severe hydrocephalus, and elevated blood pressure (shared by some but not all BBS gene knockout mice). We found that endogenous BBS3 and the BBSome physically interact and depend on each other for their ciliary localization. This finding explains the phenotypic similarity between Bbs3−/− mice and BBSome subunit knockout mice. Loss of Bbs3 does not affect BBSome formation but disrupts normal localization of melanin concentrating hormone receptor 1 to ciliary membranes and affects retrograde transport of Smoothened inside cilia. We also show that the endogenous BBSome and BBS3 associate with membranes and the membrane association of the BBSome and BBS3 are not interdependent. Differences between BBS mouse models suggest nonoverlapping functions to individual BBS protein.
The cilium is a microtubule-based organelle present on the cell surface that plays multiple roles during development and in adult tissue homeostasis. Loss of cilia or ciliary malfunction is involved in a wide range of human diseases, named ciliopathies, including primary ciliary dyskinesia, polycystic kidney disease, nephronophthisis, Joubert syndome, Senior-Loken syndrome, Meckel-Gruber syndrome, oro-facial-digital syndrome, Alstrom syndrome, and Bardet Biedl syndrome (BBS).
BBS (OMIM 209900) is an autosomal recessive, pleiotropic disorder characterized by obesity, retinal degeneration, polydactyly, renal abnormalities, hypogenitalism, and cognitive impairment (1). In addition, BBS is associated with an increased susceptibility to hypertension, diabetes mellitus, and heart defects (2). To date, 16 BBS genes have been identified (3–15). Seven BBS proteins (BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, and BBS9) form a complex (the BBSome) necessary for ciliary membrane biogenesis (16). This complex localizes to nonmembranous centriolar satellites in the cytoplasm as well as the ciliary membrane. BBS6, BBS10, and BBS12 have homology with the type II chaperonin superfamily and are required for BBSome assembly (17).
Analysis of several BBS mutant mice, including Bbs1M390R knockin, Bbs2−/−, Bbs4−/−, and Bbs6−/− reveals similarities to the human phenotypes, including blindness, obesity, renal abnormalities, and neurological deficits (18–22). In addition, the absence of normal Bbs1, Bbs2, Bbs4, and Bbs6 protein results in failure to form flagella during spermatogenesis. However, these mutant mice are able to form other motile cilia, such as tracheal cilia (23) and primary cilia, including the connecting cilia of photoreceptors. The mutant mice initially form photoreceptors, but subsequently undergo a progressive retinal degeneration as mice age (24). Mislocalization of rhodopsin to the inner segment has been shown to occur in BBS mutant photoreceptors, suggesting that BBS proteins are involved in ciliary transport (21). BBS mutant mice develop obesity because of hyperphagia and decreased calorie expenditure, have elevated leptin levels, and develop leptin resistance (25, 26). The ADP ribosylation factor (ARF) and ARF-like (ARL) GTPases are well characterized to function in membrane-trafficking pathways. However, few mouse models have been generated for these small GTPases to evaluate their in vivo functions. BBS3/ARL6 is a member of the Ras superfamily of small GTP-binding proteins. Studies from Caenorhabditis elegans showed that BBS3 undergoes intraflagellar transport (IFT) (6, 27). Jin et al. showed that the BBSome directly interacts with the ciliary localization signal of SST3R and serve as coat proteins for ciliary membrane proteins (28). BBS3 is required for the ciliary localization of the BBSome. The in vivo function of BBS3 is not fully characterized. To gain insight into the function of BBS3, a BBS protein that is not part of the BBSome and does not have chaperone homology, we generated Bbs3 knockout mice. Evaluation of Bbs3−/− mice revealed common BBS associated phenotypes and Bbs3 unique phenotypes.
Results
Generation of Bbs3 Knockout Mice.
To investigate the in vivo function of the Bbs3 gene, we targeted the Bbs3 gene in mice by replacing exon 6 and 7 with a neomycin cassette. This targeting construct was aimed at creating a frame-shift that would knockout both known Bbs3 mRNA isoforms (29) (Fig. S1A). The absence of Bbs3 mRNA was verified by RT-PCR analysis using RNA isolated from Bbs3−/− mouse testes and a primer specific for the knockout (Fig. S1B). Western blotting (Fig. S1C) confirmed the absence of Bbs3 protein.
Bbs3−/− Mice Exhibit Retinal Degeneration, Loss of Sperm Flagella, and Severe Hydrocephalus.
Histological analysis of Bbs3−/− eyes demonstrates degeneration of the photoreceptor cells associated with a lack of the outer nuclear layer and the absence of photoreceptor inner and outer segments (Fig. 1 A and B). The inner retina of these animals appears to be intact, with normal inner nuclear and ganglion cell layers. Bbs3−/− males failed to produce offspring. To investigate the male infertility, we examined the seminiferous tubules from Bbs3−/− mice. H&E staining of 4-mo-old Bbs3−/− testes revealed a lack of sperm flagella (Fig. 1 C and D), a finding consistent with other BBS mutant mice (18–21).
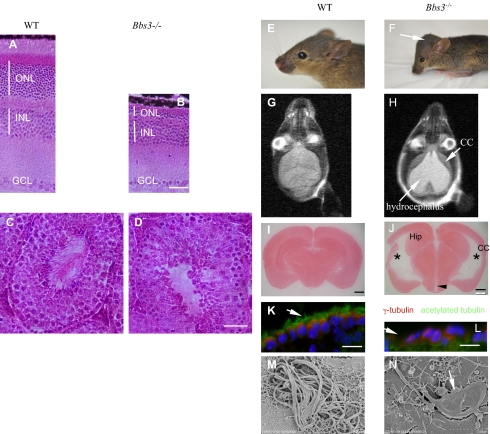
Fig. 1.
Photoreceptor cell loss, absent sperm flagella, and severe hydrocephalus in Bbs3−/− mice. H&E-stained WT (A) and mutant (B) eyes show loss of photoreceptor inner and outer segments, as well as degeneration of the outer nuclear layer. Scale bar, 100 μm. H&E-stained 4-mo-old WT (C) and Bbs3−/− (D) seminiferous tubules show a lack of spermatozoa flagella. Scale bar, 100 μm. (E and F) Pictures of 3-wk-old WT and Bbs3−/− brains. Arrow points to the domed cranium in a Bbs3−/− mouse. (G and H) MRI images of WT and Bbs3−/− brains. The long arrow indicates a hydrocephalic region and the short arrow points to thinning of the cerebral cortex. (I and J) Neutral-red–stained 60- to 100-μm thick coronal brain sections from WT (I) and mutant mice (J) show enlarged lateral ventricles (*), an enlarged dorsal third ventricle (arrowhead), reduced hippocampus (Hip), and thinning of the cerebral cortex (CC). Scale bar, 1 mm. Immunofluorescent staining of brain ventricle ependymal cells using antiacetylated tubulin (green) and anti–γ-tubulin antibody (red) reveals the lack of cilia and shortened cilia (arrow) in the ependymal cells of Bbs3−/− mice (L) compared to WT (K). Scale bar, 20 μm. (M and N) Scanning electron microscopy showed cilia morphological abnormalities in cultured ependymal cells from Bbs3−/− mice. Shortened and paddle-shaped cilia (arrow) are seen in Bbs3−/− ependymal cell cultures.
Bbs3−/− mice have early craniofacial abnormalities, with the appearance of a domed cranium and splaying of the cranial sutures compared with their WT littermates (Fig. 1 E and F). This domed cranium is not seen in other BBS gene knockout mice. Brain MRI of Bbs3−/− mice showed severe hydrocephalus (Fig. 1 G and H) and was confirmed by H&E staining of coronal sections of Bbs3−/− mouse brains (Fig. 1 I and J). We also observed thinning of the cerebral cortex and a reduction in the size of the hippocampus and corpus striatum.
Motile cilia of the ependymal cell layer lining the cerebral ventricles were examined by immunofluorescent staining with an antiacetylated tubulin antibody as a cilia marker protein and γ-tubulin as basal body marker. Compared with WT, Bbs3−/− mice showed a reduced number and misshaping of ependymal cell cilia (Fig. 1 K and L). Cultured ependymal cells from Bbs3−/− mice have paddle-shaped cilia with dilatations along the terminal portion of the cilia (Fig. 1 M and N). The Bbs3−/− mice also possess altered cilia beat mechanics, with chaotic directional beating of the cilia compared with the laminar beating pattern of the WT cells. This finding suggests that altered ciliary beating in ependymal cilia may contribute to the development of hydrocephalus in Bbs3−/− mice. (See Movies S1 and S2)
To examine primary cilia in vivo, we stained tissue sections from the kidney, eye, and pancreas with acetylated tubulin and did not find differences in cilia numbers or length between WT and Bbs3−/− mice (Fig. S2). Primary cilia also form in cultured kidney epithelial cells and fibroblasts derived from Bbs3−/− mice.
Bbs3−/− Mice Have Increased Fat Mass Without Overt Obesity and Develop Increased Arterial Pressure.
Obesity is one of the cardinal features of BBS patients. To determine whether Bbs3−/− mice develop obesity as found in BBS human patients and other BBS mutant mice, we weighed Bbs3−/− and their WT littermate controls. Surprisingly, Bbs3−/− mice have minimally elevated body weight (Fig. 2A); however, fat mass analysis by MRI and by weighing individual fat depots revealed that Bbs3−/− mice have increased fat mass (Fig. 2 B and C). There is a trend for higher leptin levels in Bbs3−/− mice relative to WT controls, but the differences are not statistically significant (P = 0.09). Food intake measurements show that Bbs3−/− mice are not hyperphagic, unlike other BBS knockout mice.
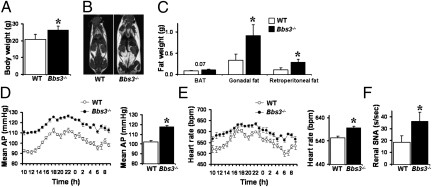
Fig. 2.
Bbs3−/− mice have increased body weight and arterial pressure. (A–C) Comparison of (A) body weight and fat mass as assessed by MRI (B) and weight of individual fat depots (C) between Bbs3−/− mice and WT littermate controls. Although Bbs3−/− mice have increased white adipose tissues like gonadal fat and retroperitoneal fat, they do not have increased brown adipose tissue (BAT). (D–F) Comparison of (D) mean arterial pressure (AP), (E) heart rate, and (F) renal SNA between Bbs3−/− mice and WT littermate controls. *P < 0.05 vs. WT controls.
Some BBS mutant mice, such as Bbs4−/− and Bbs6−/− mice, have increased arterial pressure, but others, including Bbs2−/− mice, do not. To determine if Bbs3−/− mice develop hypertension, we used radio-telemetry to measure arterial pressures. The Bbs3−/− mice have higher 24-h arterial pressure and heart rates compared with WT controls (Fig. 2E). Consistent with the hemodynamic changes, Bbs3−/− mice have higher renal sympathetic nerve activity (SNA) when multiple fiber recordings were performed on the kidneys of anesthetized mice (Fig. 2F).
BBS3 and the BBSome Are Interdependent for Ciliary Localization but Pericentriolar Satellite Localization of the BBSome Is BBS3-Independent.
Given that BBS3 is not an integral part of the BBSome, yet loss of Bbs3 leads to overlapping phenotypes with mice with loss of BBSome subunits, we investigated the effect of Bbs3 loss on BBSome protein levels and on BBSome formation. Loss of Bbs3 does not affect BBSome subunit protein levels or BBSome formation (Fig. S3). However, loss of Bbs3 affects the ciliary localization of the BBSome and vice versa, as shown by using RNAi against BBS8, BBS9, and BBS3 (Fig S4). Unlike pericentriolar material 1 and CEP290, which stably localize to the pericentriolar satellites (Fig. S5C), the endogenous BBSome shows dynamic changes, moving from the cytosol to the pericentriolar satellites to cilia when serum was withdrawn to induce cilia formation (Fig. S5A). This observation is dependent on normal microtubule structure (Fig. S5B), but is independent of BBS3 (Fig. S4). BBS3 is shown to localize to cilia but not to pericentriolar satellites. Basal body staining of BBS3 appears to be nonspecific, because this signal is not decreased by depleting BBS3 using RNAi (Fig. S4).
N-Terminal Hydrophobic Amino Acids Regulate BBS3–BBSome, but Human Homozygous Point Mutations Disrupt This Interaction.
To determine how BBS3 and the BBSome depend on each other for cilia localization, we examined the biochemical interactions between BBS3 and the BBSome. Using purified protein, Jin et al. showed that BBS3 physically interacts with the BBSome when they used BBS3 with the N-terminal 16 amino acids deleted (28). However, pair-wise interactions of BBS3 with individual BBSome subunits by cotransfection into human 293T cells fail to detect any direct interaction between BBS3 and individual BBSome subunits. Examination of the amino acid sequence and secondary structure analysis of BBS3 protein reveals that the N terminus contains several hydrophobic amino acids and forms an α-helix. This structure is shared by other ARF proteins, such as ARF1, but not by Rab proteins, including Rab8a, and is predicted to help ARF proteins to be recruited to vesicle membranes (30). We reasoned that the BBS3 N-terminal sequences might regulate the interaction between BBS3 and the BBSome. To test this hypothesis, we deleted the N-terminal 11 amino acids, added an N-terminal Flag tag, and transfected this modified BBS3 construct into 293T cells. We pulled down the N-terminal–deleted BBS3 with Flag beads and detected the existence of endogenous BBSome proteins. All BBSome subunits were present in this pull down, but were not detected in full-length BBS3 or negative control (Flag-pVHL) pull down (Fig. 3A). Mutation of the six hydrophobic amino acids in the first 11 amino acids to alanine give similar results to those observed with the BBS3 N-terminal deletion mutant (Fig. 3B). Mutation of the predicted myristoylation site G2 to alanine did not have any effect, indicating that BBS3 function is not dependant on myristoylation.
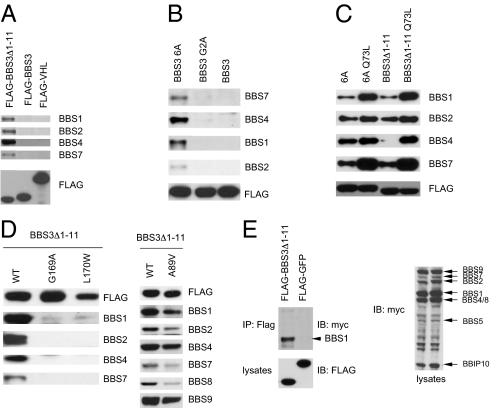
Fig. 3.
BBS3 N-terminal hydrophobic amino acids regulate BBS3–BBSome interaction, but human homozygous point mutations disrupt this interaction. (A) The N-terminal hydrophobic amino acids regulated the interaction between BBS3 and the endogenous BBSome. Shown are Western blots of Flag pull-down of 293T cells transfected with Flag-tagged full-length BBS3, BBS3 without the first 11 amino acids, and VHL as a negative control. (B) Mutation of the hydrophobic amino acids to Alanine in the first 11 amino acids of the N terminus of BBS3 increased the interaction between BBS3 and endogenous BBSome. (C) The GTP-bound form of BBS3 regulates the interaction between BBS3 and the BBSome. (D) Mutated BBS3 with mutations found in BBS patients lose the ability to interact with the BBSome but a mutation found in nonsyndromic retinal pigmentosa patients (A89V) only partially decreases the interaction between BBS3 and the BBSome. (E) BBS3 interacts with the BBSome through the BBS1 subunit. Shown are coimmunoprecipitation assays of 293T cells transfected with Flag-tagged BBS3Δ1–11 and myc-tagged BBSome subunits.
Because BBS3 is a small GTPase, it is cycled between a GTP-bound form and a GDP-bound form. We tested whether GTP or GDP binding affects BBS3–BBSome interaction. We generated a T31N mutation, which locks BBS3 in a GDP-bound form, and a Q73L mutation, which locks BBS3 in a GTP-bound form. The T31N mutant protein is not stable when transfected into cells. The protein level is very low and it fails to interact with the endogenous BBSome. The Q73L mutation increases the interaction between the N-terminal deletion or N-terminal alanine mutant forms of BBS3 and the endogenous BBSome (Fig. 3C). These results demonstrate that both the N-terminal hydrophobic amino acids and the GTP-bound form of BBS3 facilitate the interaction between BBS3 and the BBSome. We also show that mutations found in human BBS patients disrupt BBS3–BBSome interaction. A BBS3 mutation (A89V) found in nonsyndromic retinal pigmentosa patients decreases but do not totally eliminate the interaction between BBS3 and the BBSome (Fig. 3D). These results highlight the importance of BBS3–BBSome interactions. To determine which BBSome subunit mediates this interaction, we cotransfected the Flag-tagged N-terminal–deleted BBS3 with myc-tagged BBSome subunits into 293T cells. BBS1 showed the strongest interaction with BBS3; other BBSome subunits have weak or absent interaction with BBS3 (Fig. 3E). This result demonstrates that BBS1 is the BBSome subunit that likely directly interacts with BBS3, consistent with results using in vitro purified proteins (28).
Both Endogenous BBS3 and the BBSome Associate with Membranes.
The requirement of BBS3 for the ciliary localization of the BBSome could explain why loss of Bbs3 leads to phenotypes similar to those caused by the loss of BBSome subunits. Of note, we observed that Bbs3−/− mice have some unique phenotypes compared with mice mutant in BBSome subunits, including severe hydrocephalus, increased fat without frank obesity, and hypertension. This finding prompted us to examine the differences between BBS3 and the BBSome. As demonstrated in Fig. S4, the localization of BBS3 and the BBSome overlap inside the cilium, but the pericentriolar satellite localization of the BBSome is not shared by BBS3 and this localization is BBS3-independent. Because BBS3 is a small GTPase, and small GTPases can bind to vesicle membranes, we asked whether endogenous BBS3 also binds to membranes. We fractionated WT testes total lysates by mechanically disrupting the tissues without adding detergent to avoid disruption of the membranes. We then used differential ultracentrifugation to separate the nucleus and the cytosol from the membrane fraction. The membrane fraction was then loaded on to a sucrose gradient. Both endogenous Bbs3 and the BBSome are associated with membranes, demonstrated by Western blot analysis of different sucrose gradient fractions (Fig. 4A). Most Bbs3 is in the cytosol, whereas only a fraction of Bbs3 is associated with membranes. In contrast, a large portion of the endogenous BBSome is associated with membranes. Membrane fractionation using testes from Bbs4−/− mice demonstrated that loss of Bbs4 did not alter the overall membrane association of Bbs3 (Fig. 4B). Similarly, membrane fractionation using testes from Bbs3−/− mice demonstrate that loss of Bbs3 did not alter the overall membrane association of BBSome proteins (Fig. 4C). Together with data from Figs. S4 and S6, these results indicate that there are several pools of Bbs3 and the BBSome inside the cells: the cytosolic pool, the general membrane pool, and the cilia pool.
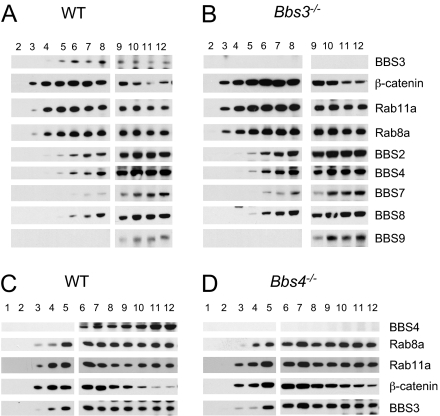
Fig. 4.
Both endogenous BBS3 and the BBSome associate with membranes. Shown are Western blots of sucrose gradient fractions from mechanically disrupted testicular tissue. Both endogenous Bbs3 and the BBSome are associated with membranes (A). Loss of Bbs3 does not affect the general membrane association of the BBSome (B). Compared with WT (C), loss of Bbs4 does not affect the general membrane association of Bbs3 (D).
Bbs3 Is Required for Proper Ciliary Localization of Melanin Concentrating Hormone Receptor 1.
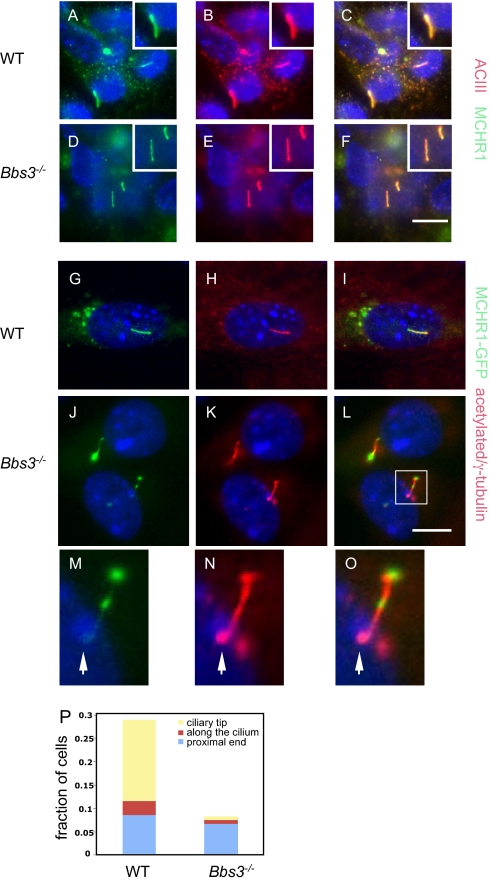
Functions of the BBSome include ciliary membrane biogenesis and ciliary membrane protein targeting. The membrane receptor melanin concentrating hormone receptor 1 (MCHR1) has been shown to localize to the ciliary membrane in brain neurons and in cultured mouse inner medullary collecting duct cells (31). MCHR1 ciliary membrane localization is lost in Bbs2−/− and Bbs4−/− brain neurons. To determine if Bbs3 is required for MCHR1 ciliary localization, we cultured neurons from P0 mouse brain from WT and Bbs3−/− mice. We stained cultured neurons with anti-ACIII antibody and anti-MCHR1 antibody. Consistent with published results, MCHR1 localizes to neuronal cilia in WT neurons (Fig. 5 A–C). Loss of Bbs3 does not affect ciliary localization of MCHR1 in Bbs3−/− neurons (Fig. 5 D–F). When cultured Bbs3−/− kidney cells were infected with adenovirus-expressing GFP-MCHR1, MCHR1 localized to the ciliary membrane (Fig. 5 G–L), confirming that Bbs3 is not required for ciliary localization of MCHR1. However, of those cilia that express GFP-MCHR1, about 27% of them show MCHR1-containing bulges along the ciliary membrane in Bbs3−/− cells, but only ∼8% of the cilia in WT cells have MCHR1-containing bulges. Among those ciliary bulges in the Bbs3−/− cells, ∼56% of them localize to the proximal end of the cilia, as demonstrated by their colocalization close to γ-tubulin staining (Fig. 5 M–O), ∼29% of them localize to the tip of the cilia, and ∼15% localize along the length of the cilia (Fig. 5P). This finding is consistent with previous findings that Bbs gene knockdown in zebrafish leads to retrograde melanosome transport delay in addition to Kupffer vesicle defects (32).
Fig. 5.
BBS3 is required for proper ciliary localization of MCHR1. Cultured neurons from WT (A–C) and Bbs3−/− (D–F) mice were stained with anti-ACIII antibody (red) as a neuronal cilia marker and anti-MCHR1 antibody (green). (G–L) Cultured kidney epithelial cells from WT (G–I) and Bbs3−/− (J–L) mice were infected with adenovirus expressing MCHR1-GFP (green). Cilia were labeled with antiacetylated tubulin and γ-tubulin (red). (M–O) Enlarged (3×) images of MCHR1 containing bulges along Bbs3−/− cilia. (P) Quantitation of improper localization of MCHR1 on the ciliary membranes. Scale bars, 20 μm.
Bbs3 Is Required for Proper Smoothened Ciliary Localization.
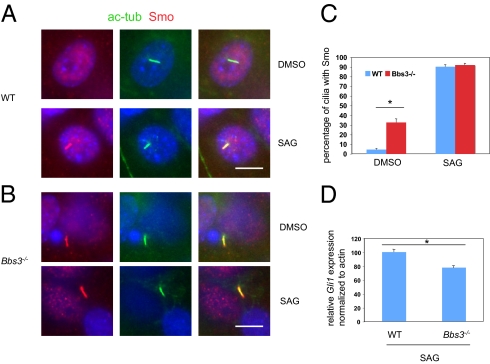
The relevance of MCHR1 to BBS phenotypes is not known. Therefore, we attempted to identify the physiologically relevant endogenous ciliary membrane proteins that are regulated by BBS3. One of the cardinal features of BBS patients is polydacyly. Most polydactyly phenotypes typically have sonic hedgehog (Shh) pathway defects. Recent studies from our laboratory have identified that Smoothened (Smo), one of the ciliary membrane proteins that is required for Shh pathway activation, is a cargo of the BBSome and loss of the BBSome results in Smo accumulation inside cilia and decreased Shh response. We asked if Smo ciliary localization is also regulated by BBS3. We stained mouse embryonic fibroblast (MEF) cells from WT and Bbs3−/− mice with antiacetylated tubulin and anti-Smo. Smo was accumulated inside cilia in 30–40% of Bbs3−/− MEF cells, even in the absence of Shh pathway stimulation with SAG (a Smo-binding Shh pathway agonist) treatment (Fig. 6), compared with less than 5% in WT MEF cells. SAG treatment further increased the percentage of cilia positive for Smo in Bbs3−/− MEF cells (Fig. 6C), suggesting that Bbs3−/− MEF cells can still respond to ligand treatment. To determine if loss of Bbs3 affects Shh pathway activation, we determined gli1 (an indicators of Shh pathway activation) mRNA levels by quantitative PCR. Loss of Bbs3 leads to a 22.3% decrease of Shh pathway activation (Fig. 6D). This modest decrease in Shh pathway activation explains the lack of polydactyly in Bbs3−/− mice. Our results are consistent that with that of loss of BBSome subunits and further demonstrated that BBS3 and the BBSome are interdependent in transporting ciliary membrane proteins.
Fig. 6.
BBS3 is required for proper Smo ciliary localization and modulates Shh pathway activation. Cultured MEF cells from WT (A) and Bbs3−/− (B) mice were stained with antiacetylated tubulin (green) as a marker for cilia and anti-Smo (red). (C) Quantitation of percentage of cilia with Smo staining when treated with DMSO or SAG. (D) Quantitative PCR for Gli1 from cultured Bbs3−/− MEF cells stimulated with SAG showed a 22.3% decrease compared with WT MEF cells when normalized to actin. *P < 0.03. (Scale bars, 10 μm.)
Discussion
Phenotypic Similarities and Differences Between Bbs3−/− Mice and Other BBS Mutant Mice.
BBS patients share similar features, regardless of the gene mutated, although differences between individual patients are well documented. Studies in model organisms like zebrafish also show that knockdown of different BBS genes result in common phenotypes, including abnormal Kuppfer vesicle formation, cilia abnormalities, and retrograde melanosome transport defects (32). Therefore, we anticipated that Bbs3−/− mice would be like other BBS mutant mice, developing retinal degeneration, obesity, ventriculomegaly, and male infertility. Indeed, phenotypic characterization of Bbs3−/− mice revealed that they developed retinal degeneration and are male-infertile because of the absence of sperm flagella. Consistent with these phenotypic similarities, our biochemical analysis demonstrates that BBS3 interacts with the BBSome and is required for the ciliary localization of the BBSome, confirming in vitro data published recently (28). Furthermore, we showed that mutations found in BBS3 patients disrupt BBS3–BBSome interaction, but mutations found in nonsyndromic retinitis pigmentosa only partially decrease the interaction, highlighting the importance of BBS–BBSome interactions. Interestingly, the Bbs3−/− mice develop more severe and earlier hydrocephalus than do other BBS mutants. Unlike other BBS mutants, which develop ventriculomegaly around 2–3 wk of age, the hydrocephalus can be detected as early as postnatal day 5 in Bbs3−/− mice. These mice go on to develop cranial abnormalities, with splayed cranial sutures and domed cranial vaults. This finding suggests the accumulation of cerebrospinal fluid early in the neonatal period.
The alteration of motility of ependymal cell cilia is most likely because of the accumulation of materials inside the bulges, although we cannot rule out the possibility of subtle defective axonemal structures. The accumulation of materials that appear vesicle-like has been shown in trachea motile cilia and motile cilia of ependymal cells of other BBS mutants (18, 23). The movement of motile cilia without bulges appears to be normal. Unlike the severe defects observed in motile cilia of the ventricle ependymal layer and spermatozoa flagella, trachea motile cilia appear grossly normal, although a small fraction of them have abnormal cilia morphology, as is seen in other BBS mutants (23). The ciliogenesis of trachea motile cilia may differ from other motile cilia, as a similar situation is observed in Tg737orpk mice. Understanding ciliogenesis differences between these motile cilia is of interest, and BBS mouse models will provide valuable tools for future studies.
Interestingly, unlike other BBS mutants that develop obesity, Bbs3−/− mice do not have increased body weight, although they do have increased body fat. One possible explanation is that the severe hydrocephalus developed in Bbs3−/− mice affect their food intake, or that the severe hydrocephalus compresses other brain regions regulating body weight. However, when we knocked-out the Bbs1 gene in GFAP-positive cells by crossing GFAP-cre mice with Bbs1 conditional knockout mice, they develop severe hydrocephalus with a domed cranium but still develop obesity, indicating severe hydrocephalus does not affect obesity development. Another possibility is that BBS3 has an additional BBSome-independent function.
A BBSome-independent function for BBS3 is also suggested by the fact that Bbs3−/− mice developed hypertension. The hypertension phenotype is shared by Bbs4−/− and Bbs6−/− mice, but it is not found in Bbs2−/− and Bbs1M390R knockin mice (18, 25). This finding indicates that some BBS proteins have other functions in addition to the BBSome function, or that the knockout of some components of the BBSome results in a BBSome subcomplex that retains partial function capable of regulating blood pressure.
A BBSome-independent function for BBS3 is also suggested by studies showing that overexpression of WT BBS3 increases response to canonical Wnt signals, but loss of BBSome function, including knockdown of BBS1 and BBS4, leads to an increased Wnt response. In contrast, overexpression of the BBSome component BBS4 does not have any effect on canonical Wnt pathway activity (33, 34).
N-Terminal BBS3 Amino Acids Regulate the Interaction Between BBS3 and the BBSome.
The N-terminal sequence of BBS3 contains several hydrophobic amino acids and forms an α-helix structure. The α-helix domain appears to regulate the interaction between BBS3 and the BBSome. Similar structures are shared by other ARFs (30). Membrane binding, nucleotide affinity, and nucleotide hydrolysis by ARFs are all sensitive to the presence and sequence composition of N-terminal amino acids. The structure and position of this region has only been determined for ARFs bound to GDP and is presumed to change dramatically upon binding GTP. We anticipate that the N terminus of BBS3 will behave like ARF1. When the BBS3 crystal structure was solved, the N-terminal 15 amino acids were removed, precluding the determination of the effect of the N-terminal region on the 3D structure (34). Nevertheless, it is likely that deletion of the N terminus or binding to membranes through the N-terminal hydrophobic amino acids changes the structure of BBS3, therefore regulating the interaction between BBS3 and the BBSome. Binding to GTP may change the positioning of the N-terminal α-helix and facilitate BBS3–BBSome interaction.
BBS3 and Cilia Membrane Protein Trafficking and BBSome Cargo Selection.
One of the functions of the BBSome is to transport ciliary membrane proteins to the cilium. One type of seven transmembrane G protein-coupled receptors (GPCRs), which contain the conserved motif of AX[S/A]XQ in the intracellular loop serving as the ciliary localization signal, are localized to cilia (35): these include SST3R and MCHR1. BBS proteins are required for the ciliary localization of these GPCRs. Jin et al. showed that the BBSome directly interact with the ciliary localization signal of SST3R and serve as coat proteins for ciliary membrane proteins (28). However, our studies showed that Bbs3 is not required for the ciliary localization of MCHR1. One possible explanation for this difference may relate to differences in the neuron type between our study and others. We cultured the neurons from the whole brain sans olfactory bulb and cerebellum, but other studies used hippocampal neurons. Alternatively, the discrepancy may reflect functional differences between BBS3 and other BBS proteins. Consistent with this possibility, expression of MCHR1 in cultured kidney epithelial cells from Bbs3−/− mice showed ciliary localization of MCHR1. However, MCHR1 was often associated with bulges along the cilium in Bbs3−/− cells. These bulges are similar to those found in the tracheal and ependymal cilia of BBS mutant mice (18, 23), and reminiscent of bulges found in IFT complex A mutants (36, 37). Similarly, endogenous Smo accumulates inside cilia without ligand stimulation. Interestingly, we do not find accumulation of the IFT complex B protein IFT88, unlike that observed in other IFT mutants, such as Tg737orpk or Dync2h1. This finding is consistent with the hypothesis that the BBSome is specifically involved in membrane protein trafficking. These data indicate that BBS proteins function in retrograde trafficking, consistent with studies in zebrafish (32).
Other BBS3 and BBSome Functions.
Biochemical analysis demonstrates that both BBS3 and the BBSome are associated with membranes. The membrane association is important, given that both BBS3 and the BBSome localize to cilia, and the BBSome is required for cilia membrane biogenesis. BBS3 and the BBSome depend on each other for ciliary localization. However, loss of Bbs3 does not affect the membrane association of the BBSome and loss of the BBSome does not affect the membrane association of Bbs3. This finding indicates that the membrane association of BBS3 and the BBSome occur before they enter the cilia. Indeed, BBSome staining is diffuse in the cytosol before serum withdrawal, concentrates over time at pericentriolar satellites, and eventually BBSomes are transported to cilia (Fig. S5). Depletion of BBS3 in cells affect BBSome ciliary localization but not the pericentriolar satellite localization. In fact, sucrose gradient analysis of lysates of hTERT-immortalized retinal pigment epithelial (RPE1) cells cultured under conditions in which cilia do not form (10% serum and nonconfluence) (Fig. S6) and lysates of 293T cells, which do not form cilia under normal culture conditions, reveal that both the BBSome and BBS3 still associate with membranes. These data indicate that both BBS3 and the BBSome may have general membrane-associated functions in addition to the known functions of ciliary membrane biogenesis and cilia membrane protein sorting. Such functions likely include trafficking to other membrane compartments, for example the trafficking of the leptin receptor to the plasma membrane (26).
Materials and Methods
Hemodynamic and Sympathetic Measurements.
Arterial pressure and heart rate were recorded in conscious mice, as described previously (25). Briefly, mice were anesthetized with (91 mg/kg) and xylazine (9.1 mg/kg) and the catheter was inserted in the carotid artery. Animals were allowed to recover for several days before arterial pressure was recorded continuously in the conscious unrestrained state for 7 d.
To measure direct multifiber renal SNA, a nerve fascicle to the left kidney of anesthetized WT and Bbs3−/− mice was carefully isolated. A bipolar platinum-iridium electrode (Cooner Wire) was suspended under the nerve and secured with silicone gel (Kwik-Cast; World Precision Instruments). The nerve signal was amplified and filtered as described previously (25). Reagents and procedures are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Dr. Maxence Nachury for anti-ARL6 antibody. This work was supported by National Institutes of Health Grants R01EY110298 and R01EY017168 (to V.C.S.) and HL084207 (to K.R.). V.C.S. and E.M.S. are investigators of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.W.-B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113220108/-/DCSupplemental.
References
- 1.Harnett JD, et al. The spectrum of renal disease in Laurence-Moon-Biedl syndrome. N Engl J Med. 1988;319:615–618. doi: 10.1056/NEJM198809083191005. [DOI] [PubMed] [Google Scholar]
- 2.Green JS, et al. The cardinal manifestations of Bardet-Biedl syndrome, a form of Laurence-Moon-Biedl syndrome. N Engl J Med. 1989;321:1002–1009. doi: 10.1056/NEJM198910123211503. [DOI] [PubMed] [Google Scholar]
- 3.Badano JL, et al. Identification of a novel Bardet-Biedl syndrome protein, BBS7, that shares structural features with BBS1 and BBS2. Am J Hum Genet. 2003;72:650–658. doi: 10.1086/368204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang AP, et al. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11) Proc Natl Acad Sci USA. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang AP, et al. Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3) Am J Hum Genet. 2004;75:475–484. doi: 10.1086/423903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan Y, et al. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat Genet. 2004;36:989–993. doi: 10.1038/ng1414. [DOI] [PubMed] [Google Scholar]
- 7.Leitch CC, et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 8.Li JB, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 9.Mykytyn K, et al. Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet. 2001;28:188–191. doi: 10.1038/88925. [DOI] [PubMed] [Google Scholar]
- 10.Mykytyn K, et al. Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat Genet. 2002;31:435–438. doi: 10.1038/ng935. [DOI] [PubMed] [Google Scholar]
- 11.Nishimura DY, et al. Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedl syndrome (BBS2) Hum Mol Genet. 2001;10:865–874. doi: 10.1093/hmg/10.8.865. [DOI] [PubMed] [Google Scholar]
- 12.Nishimura DY, et al. Comparative genomics and gene expression analysis identifies BBS9, a new Bardet-Biedl syndrome gene. Am J Hum Genet. 2005;77:1021–1033. doi: 10.1086/498323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoetzel C, et al. BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat Genet. 2006;38:521–524. doi: 10.1038/ng1771. [DOI] [PubMed] [Google Scholar]
- 14.Stoetzel C, et al. Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. Am J Hum Genet. 2007;80:1–11. doi: 10.1086/510256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SK, et al. Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science. 2010;329:1337–1340. doi: 10.1126/science.1191184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 17.Seo S, et al. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc Natl Acad Sci USA. 2010;107:1488–1493. doi: 10.1073/pnas.0910268107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis RE, et al. A knockin mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc Natl Acad Sci USA. 2007;104:19422–19427. doi: 10.1073/pnas.0708571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fath MA, et al. Mkks-null mice have a phenotype resembling Bardet-Biedl syndrome. Hum Mol Genet. 2005;14:1109–1118. doi: 10.1093/hmg/ddi123. [DOI] [PubMed] [Google Scholar]
- 20.Mykytyn K, et al. Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc Natl Acad Sci USA. 2004;101:8664–8669. doi: 10.1073/pnas.0402354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimura DY, et al. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc Natl Acad Sci USA. 2004;101:16588–16593. doi: 10.1073/pnas.0405496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulaga HM, et al. Loss of BBS proteins causes anosmia in humans and defects in olfactory cilia structure and function in the mouse. Nat Genet. 2004;36:994–998. doi: 10.1038/ng1418. [DOI] [PubMed] [Google Scholar]
- 23.Shah AS, et al. Loss of Bardet-Biedl syndrome proteins alters the morphology and function of motile cilia in airway epithelia. Proc Natl Acad Sci USA. 2008;105:3380–3385. doi: 10.1073/pnas.0712327105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swiderski RE, et al. Gene expression analysis of photoreceptor cell loss in bbs4-knockout mice reveals an early stress gene response and photoreceptor cell damage. Invest Ophthalmol Vis Sci. 2007;48:3329–3340. doi: 10.1167/iovs.06-1477. [DOI] [PubMed] [Google Scholar]
- 25.Rahmouni K, et al. Leptin resistance contributes to obesity and hypertension in mouse models of Bardet-Biedl syndrome. J Clin Invest. 2008;118:1458–1467. doi: 10.1172/JCI32357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seo S, et al. Requirement of Bardet-Biedl syndrome proteins for leptin receptor signaling. Hum Mol Genet. 2009;18:1323–1331. doi: 10.1093/hmg/ddp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- 28.Jin H, et al. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pretorius PR, et al. Identification and functional analysis of the vision-specific BBS3 (ARL6) long isoform. PLoS Genet. 2010;6:e1000884. doi: 10.1371/journal.pgen.1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amor JC, et al. Structures of yeast ARF2 and ARL1: Distinct roles for the N terminus in the structure and function of ARF family GTPases. J Biol Chem. 2001;276:42477–42484. doi: 10.1074/jbc.M106660200. [DOI] [PubMed] [Google Scholar]
- 31.Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci USA. 2008;105:4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yen HJ, et al. Bardet-Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer's vesicle cilia function. Hum Mol Genet. 2006;15:667–677. doi: 10.1093/hmg/ddi468. [DOI] [PubMed] [Google Scholar]
- 33.Gerdes JM, et al. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat Genet. 2007;39:1350–1360. doi: 10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
- 34.Wiens CJ, et al. Bardet-Biedl syndrome-associated small GTPase ARL6 (BBS3) functions at or near the ciliary gate and modulates Wnt signaling. J Biol Chem. 2010;285:16218–16230. doi: 10.1074/jbc.M109.070953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berbari NF, Johnson AD, Lewis JS, Askwith CC, Mykytyn K. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol Biol Cell. 2008;19:1540–1547. doi: 10.1091/mbc.E07-09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iomini C, Li L, Esparza JM, Dutcher SK. Retrograde intraflagellar transport mutants identify complex A proteins with multiple genetic interactions in Chlamydomonas reinhardtii. Genetics. 2009;183:885–896. doi: 10.1534/genetics.109.101915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran PV, et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet. 2008;40:403–410. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.