Abstract
Computational design of novel protein–protein interfaces is a test of our understanding of protein interactions and has the potential to allow modification of cellular physiology. Methods for designing high-affinity interactions that adopt a predetermined binding mode have proved elusive, suggesting the need for new strategies that simplify the design process. A solvent-exposed backbone on a β-strand is thought of as “sticky” and β-strand pairing stabilizes many naturally occurring protein complexes. Here, we computationally redesign a monomeric protein to form a symmetric homodimer by pairing exposed β-strands to form an intermolecular β-sheet. A crystal structure of the designed complex closely matches the computational model (rmsd = 1.0 Å). This work demonstrates that β-strand pairing can be used to computationally design new interactions with high accuracy.
Keywords: computational protein design, protein design, protein interface design, Rosetta
Protein–protein interactions and assemblies are essential for a wide array of cellular processes. The ability to rationally design unique protein interactions could provide scaffolds for functional reactions and new reagents for perturbing and monitoring cellular processes. Computational approaches for interface design have advanced rapidly in recent years and have allowed interactions to be engineered for increased affinity or altered specificity (1, 2). One long-standing goal is the creation of unique interactions. Thus far, most computational designs of new interactions have involved either the pairing of α-helices (3–6) or binding of an α-helix to an open groove on a target (7–10). Other methodologies have focused on grafting side-chain interactions from a known interaction onto another scaffold (7, 11, 12). There have been two examples of structurally confirmed unique computational interface designs (6, 7), however these sample a limited set of modes by which proteins can interact. New methods of constructing an interface are necessary to mimic the ways nature forms protein–protein interactions (13).
There are many examples of naturally occurring protein heterodimers, homodimers, and larger complexes where β-strands from each chain associate to form an intermolecular β-sheet (14); β-strand pairing has also been observed in evolved antibody-antigen interactions (15) and monobody-target interfaces selected from phage display libraries (16). It has been proposed that β-strand pairing is so favorable that naturally occurring proteins often use negative design to avoid edge-to-edge association. In one study, 75 monomeric β-sheet proteins were visually examined to see if they contained structural features that would be predicted to disfavor β-sheet formation across their edge strands (17). In almost every case, one or more negative design elements were present including prolines, strategically placed charges, very short edge strands, loop coverage, and irregular edge strands. The propensity of exposed β-strands to pair is reinforced by observations of intermolecular β-sheet formation at crystal contacts of crystallization chaperones (18, 19) and designed proteins (20, 21) with exposed strands. In addition to providing affinity, β-strand interactions are geometrically constrained (22), which could provide a stable building block for designing interactions with a predetermined binding orientation. The intrinsic preference of β-strands to interact suggests that they may serve as a good anchor point for de novo interface design.
Formation of symmetric homodimers is one of the most common ways that proteins interact (23). Symmetric oligomerization provides increased stability, strict control over the number of protein units in the assembly, and low-energy structures (24). A survey of secondary structure at interfaces found that strand pairing represents 8.8% of contacts in homodimers (14). Paired β-strands at a homodimer interface are typically antiparallel (14) and longer than noninterface forming exposed strands (25). Protecting elements, typical of exposed strands in monomeric proteins, are less prevalent at β-strand mediated protein interfaces (17, 25).
There have been few successful rational designs of β-strand mediated protein interactions. Peptides that form β-strand mimetics are therapeutically used to inhibit proteases or protein–protein interactions (26, 27). One approach targeted amyloid fibrils by computationally designing a peptide to form a terminating β-strand on a growing fibril (28). Another study took the sequence of the β-strand of a known β-strand mediated homodimer and embedded it in a cyclic peptide (29). A crystal structure of the peptide showed it formed an antiparallel β-strand paired dimer as predicted (30). However, there have been no structurally verified computational designs of a unique protein–protein interaction between two domains where the interface contains interactions between β-strands.
Here, we redesign a monomeric protein to form a symmetric homodimer via an intermolecular β-sheet. To design a β-strand mediated homodimer, we first identified structures in the protein database with exposed β-strands that could self-associate by β-strand pairing. We then used symmetric docking and sequence optimization (31) to create favorable interactions surrounding the interacting strands. Four designs were experimentally characterized and one was found to adopt the structure of the computational model.
Results
Scaffold Search Protocol.
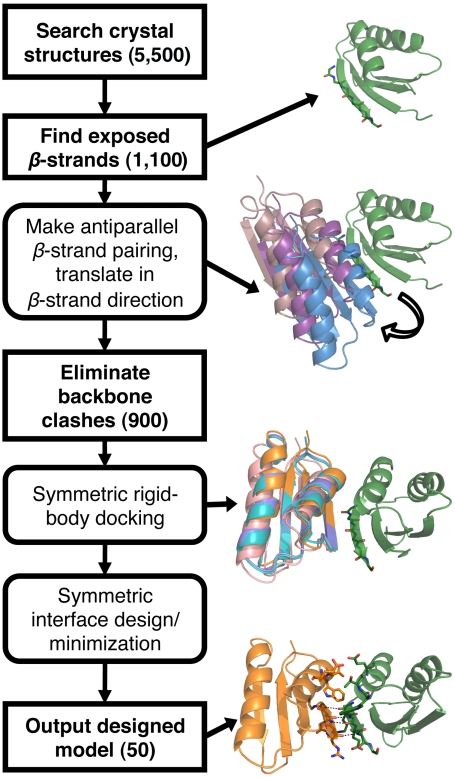
To find proteins with surface-exposed β-strands, we performed a computational search on a set of 5,500 protein crystal structures with resolution better than 2.2 Å to find proteins with a surface-exposed β-strands (Fig. 1). A strand was defined as exposed if there was a continuous stretch of five or more residues in which every second residue did not form backbone–backbone hydrogen bonds and were not occluded from solvent (see Materials and Methods). This criterion yielded 1,500 exposed β-strands on 1,100 unique proteins. We then tested each exposed β-strand for its potential to form the basis of a homodimer interface. A copy of the entire chain of each protein with an exposed strand was rotated and translated to an ideal backbone–backbone hydrogen bonding distance to the original protein chain (Fig. 1). The copied chain was then translated along the exposed strand in steps of 7 Å to identify alternate conformations that had no clashing backbone atoms. After this step, there were 2,800 potential alignments of 900 different proteins.
Fig. 1.
Search and design protocol for a symmetric β-strand mediated homodimer. Method used to search for, then design, scaffold proteins to create a symmetric homodimer (see full details in Materials and Methods). Numbers in parentheses represent the total number of unique input structures used in each step. Individual steps are illustrated by the structures generated during each step using the protein Atx1 (Protein Data Bank ID 1CC8).
To narrow down the list further, we performed a brief design and minimization protocol to determine which of our potential homodimers gave favorable binding energies and interface sizes after design (see Materials and Methods). This step reduced the overall number of targets to 200. From the final set of designable alignments, we removed all proteins that had not previously been expressed in Escherichia coli, were natural oligomers, had crystal contacts that resulted in an intermolecular β-sheet, were over 500 amino acids in length, or whose interacting β-strands were not part of globular domains. These steps generated 50 possible starting points for design of a homodimer.
Design Protocol.
Each design simulation consisted of one round of symmetric protein docking (32) followed by five successive rounds of symmetric sequence optimization and minimization of side-chain and backbone residues at the homodimer interface (see Materials and Methods). For some protein scaffolds it was necessary to build alanine into all positions at the interface to obtain a docked structure that formed hydrogen bonds between the β-strands. We applied a stringent filter to eliminate designs that were unlikely to produce the desired experimental results. First, from each run, we selected only designs in the top 10% in total score, ΔGbinding, and β-strand hydrogen-bond energy. These selections were further filtered for interface energy density (ΔGbinding/ΔSASA, where SASA represents the solvent-accessible surface area), number of buried polar atoms failing to form hydrogen bonds, and packing quality (RosettaHoles score, ref. 33).
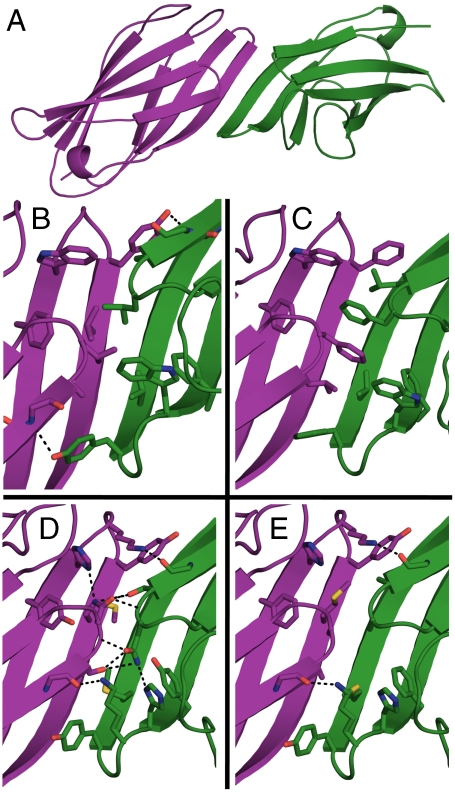
We selected four homodimer designs based on the γ-adaptin appendage domain (Protein Data Bank ID 2A7B, ref. 34). This scaffold protein was chosen because the designs of 2A7B scored favorably compared to other potential homodimers according to all metrics described above. Two of the four designs chosen had predominantly hydrophobic interfaces (βdimer1 and βdimer2), whereas the other two contained more polar interactions (βdimer3 and βdimer4) (Table 1), which allowed us to test our ability to design hydrogen bonding networks and hydrophobic packing interactions. All four designs exhibited a similar overall conformation and β-strand register to that of βdimer1 (Fig. 2A). The maximum Cα rmsd from βdimer2, βdimer3, and βdimer4 to βdimer1 is 1.5 Å. All four designs have a total of six main-chain hydrogen bonds between residues 104, 106, and 108 on one chain to residues 108, 106, and 104 on the other chain, respectively. One face of the intermolecular β-sheet is exposed to solvent, whereas the other is occluded by a loop formed by residues 10–12. The crystal structure 2A7B has no crystal lattice contacts along the exposed strand, suggesting that the wild-type sequence is not prone to form an intermolecular β-sheet. Wild-type γ-adaptin appendage domain is likely prevented from self-association by a salt bridge between residues K10 and D107 that would be buried at the designed homodimer interface. In the designs, K10 is mutated to alanine, leucine, or serine and D107 is mutated to serine or threonine. A common feature in all four designs is charge complementation on the solvent-accessible side of the interacting strands between residues 104 and 108 on opposite chains. For example, in βdimer1, residue 104 is a lysine and residue 108 is a glutamate. In βdimer3, residue 104 is an arginine and residue 108 is a glutamate. The buried side of the interface is dominated by either hydrophobic or polar interactions depending on the design (Fig. 2 B–E). A search of Protein Data Bank Protein Interfaces, Surfaces, and Assemblies (35) yielded no known interfaces bearing any similarity to the designs, suggesting that the designed complexes represent a unique configuration of a protein–protein interaction.
Table 1.
Computational evaluation of designed homodimer models
| Model | No. mutations | Etotal | ΔGbind | No. buried-unsatisfied | Polar interface area, % |
| Wild type | 0 | −561 | −13 | 8 | 48 |
| βdimer1 | 11 | −597 | −29 | 2 | 39 |
| βdimer2 | 7 | −593 | −30 | 0 | 31 |
| βdimer3 | 5 | −596 | −32 | 0 | 54 |
| βdimer4 | 9 | −593 | −27 | 0 | 46 |
Computational values used to select homodimer designs are shown compared to the wild-type protein represented as a homodimer forced into a conformation similar to the designs. “No. mutations” is the number of mutations to the wild type to generate the design, “Etotal” is the Rosetta energy for the homodimer, “ΔGbind” is the difference in energy between the complex and two monomers of a model (ΔGbind = EAB - EA - EB), “No. buried-unsatisfied” is the number of polar atoms that are not solvent accessible and do not form hydrogen bonds to another atom in the protein, and “Polar interface area, %” represents the amount of solvent-accessible surface area of polar atoms hidden at the interface (SASApolar/SASAtotal).
Fig. 2.
Computational designs used in experiments. (A) Overall topology of computational designs. The γ-adaptin appendage domain (Protein Data Bank ID 2A7B) is used as the scaffold for the designed interface. Coloring (purple and green) highlights the symmetric chains in the model. The solvent-excluded side of the interface is shown in detail for βdimer1 (B), βdimer2 (C), βdimer3 (D), and βdimer4 (E). Selected side chains are shown in sticks. Black dashed lines represent hydrogen bonds at the interface; the six main-chain hydrogen bonds are not shown.
Determining Oligomeric Status.
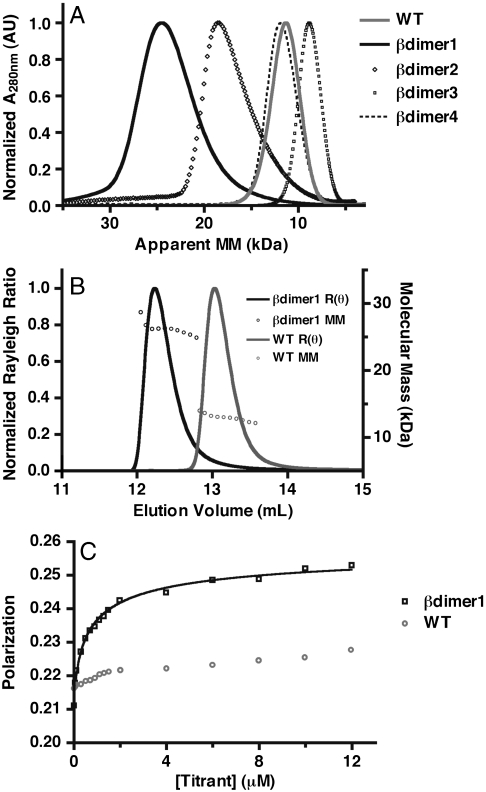
We first assessed the oligomeric state of the four designs and the wild-type protein by size-exclusion chromatography. The molecular mass of a monomeric protein based on sequence is 13.5 kDa. The wild type, βdimer3, and βdimer4 eluted near the expected molecular mass for a monomeric protein, whereas βdimer1 and βdimer2 eluted close to the size expected for a dimer (Fig. 3A and Table 2). We were unable to perform additional experiments with βdimer2 and βdimer4 because they did not express at sufficient levels.
Fig. 3.
Experimental determination of molecular mass in solution. (A) Size-exclusion chromatography (Superdex 75) of the designs and wild-type protein. Absorbance has been normalized based on maximum value, the apparent molecular mass (MM) is based on a standard curve obtained from globular proteins. (B) Size-exclusion chromatography (Superdex 75) followed by multiangle light scattering of wild type (gray) and βdimer1 (black). Rayleigh ratio [R(θ)] (solid lines) has been normalized based on maximum value; MM (open circles) is calculated from light scattering and refractive index. The average molecular mass is 26 kDa for βdimer1, 14 kDa for the wild type. (C) Measurement of dimer dissociation constant of βdimer1 using a fluorescence polarization assay. Bodipy-labeled βdimer1 was titrated with unlabeled βdimer1 (black) and wild-type protein (gray), and the change in polarization was fit to a homodimerization model (see SI Materials and Methods). The calculated homodimer dissociation constant for βdimer1 is 1.0 ± 0.1 μM (SEM).
Table 2.
molecular mass of designed homodimers in solution
| Protein |
Molecular mass, kDa | |||
| Monomer (calculated) | SEC | AUC | SEC/MALS | |
| Wild type | 13.6 | 11 | 12 | 13 |
| βdimer1 | 13.6 | 26 | 26 | 26 |
| βdimer2 | 13.7 | 21 | — | — |
| βdimer3 | 13.7 | 10 | 16 | 13 |
| βdimer4 | 13.6 | 12 | — | — |
The molecular mass in solution measured for the wild-type protein and homodimer designs measured with using SEC, AUC, and MALS. “Monomer (calculated)” is the molecular mass expected based on sequence. The βdimer2 and βdimer4 proteins did not express in significant quantities for additional molecular mass determination.
To confirm the results from size-exclusion, we performed sedimentation equilibrium experiments using a Beckman XL-I analytical ultracentrifuge (AUC-SE). The wild-type protein, βdimer1, and βdimer3 were spun at 46,400 × g until equilibrium was reached. Three concentrations of protein (20, 40, and 60 μM) were used for each sample. Equilibrium absorbance profiles at 280 nm were used to determine molecular mass. The profiles for all three proteins were well fit by a single species model (Fig. S1). The molecular mass determined from the equilibrium profile of the wild-type protein and βdimer3 were 12 and 16 kDa, respectively, close to that expected for a monomer. The molecular mass of βdimer1 was found by the same method to be 26 kDa, near that expected for a homodimer (Table 2).
We further tested the solution molecular mass of βdimer1, βdimer3, and the wild-type protein by size-exclusion chromatography (SEC) followed by multiangle light scattering (MALS). Each protein came off the size-exclusion column as a single peak. Light scattering and refractive index were used to determine the molecular mass of the peak (Fig. 3B). The results were similar to the SEC experiment described above. βdimer3 and the wild-type protein were determined to have a molecular mass of 13 kDa, whereas βdimer1 had a molecular mass of 26 kDa (Table 2). These results further confirmed that βdimer1 forms a homodimer, but the wild type and βdimer3 do not.
Homodimer Binding Affinity.
We used a fluorescence polarization assay to measure the dimer dissociation constant of βdimer1. Briefly, we expressed βdimer1 with the mutation S62C and labeled it with thiol reactive Bodipy. The monomer-dimer equilibrium was monitored by titrating excess unlabeled protein into dilute, Bodipy-labeled, βdimer1 protein and observing the increase in polarization from the formation of a slowly rotating dimeric species. βdimer1 was titrated with wild-type protein as a control. The change in polarization upon binding was fit to a homodimerization model (SI Materials and Methods). βdimer1 had a dimer dissociation constant of 1.0 μM, whereas the wild-type protein showed little to no interaction with βdimer1 (Fig. 3C). To confirm this result, we performed AUC-SE with βdimer1 at concentrations of 0.8, 1.5, and 2.0 μM. The data were fit to a monomer-dimer self-association model that produced a dimer dissociation constant of 0.96 μM (Fig. S2), which closely matches the dissociation constant determined by fluorescence polarization.
Crystal Structure of the homodimer.
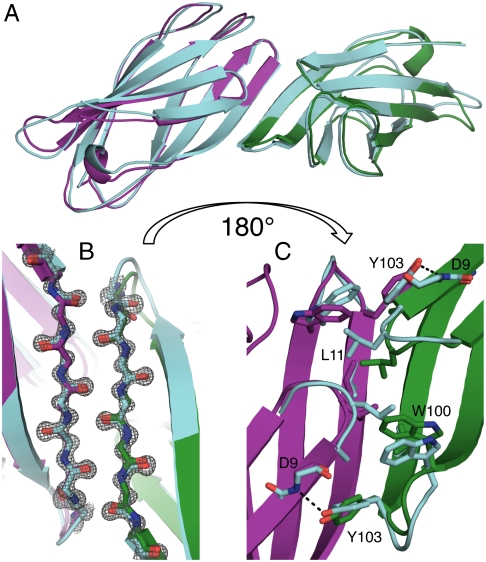
We determined the crystal structure of βdimer1 using molecular replacement and diffraction data to a resolution of 1.09 Å (Table S1). The coordinates of the dimer design were used as the search model for molecular replacement. The asymmetric unit contained two molecules of βdimer1 protein, henceforth called chain A and chain B. These two chains in the crystal structure interact in a manner that is remarkably similar to the model of βdimer1 (Fig. 4A) with an rmsd between the crystal structure and model of 1.0 Å for all backbone atoms. The intermolecular β-strand pairing found at the interface between the two chains in the crystal structure matches that of the designed model (Fig. 4B). The conformation of the interacting strands in the crystal structure of βdimer1 show only minor differences when compared to the wild-type protein (Fig. S3), indicating that substantial backbone rearrangement is not required for the formation of the homodimer.
Fig. 4.
Comparison of βdimer1 computational model to crystal structure. (A) Overlay of the βdimer1 computational model (green and purple) and crystal structure (cyan). The backbone atom rmsd for the entire structure is 1.0 Å. (B) Backbone–backbone interactions between the interface-forming β-strands viewed from the solvent-accessible side of the intermolecular β-sheet. The 2Fo - Fc electron density (gray) is contoured to 2σ. (C) Detailed view of designed side chains forming interactions on the solvent-excluded side of interacting β-strands. A black dashed line represents the interface-spanning hydrogen-bond between D9 and Y103.
The conformations of the interface residues in the crystal structure were well predicted by the computational model (Fig. 4C). The conformations of the designed hydrophobic side chains in the crystal structure closely match those of the designed structure including L11 from one chain packing between L11 and W100 from the other chain. The computational model also accurately predicted an interface-spanning hydrogen bond between the backbone nitrogen of D9 and the hydroxyl oxygen on Y103 (Fig. 4C). The solvent-exposed side of the paired strands presents an interesting divergence from the model. The side chains of residues E108 and Q106 appear to form hydrogen bonds from the side-chain nitrogen on Q106 to a carboxyl oxygen on E108, and from the other carboxyl oxygen on E108 to the side-chain oxygen on Q106 (Fig. S4), suggesting the carboxyl group of E108 is protonated.
One possible pitfall of using β-strands to mediate an interaction is the possibility for register shifts between the paired strands. It is interesting to consider what structural elements in βdimer1 set the register between the two proteins. Docking the chains with constraints to force a register shift in either direction yielded no models with backbone–backbone hydrogen bonds at the interface. A shift in one direction introduces a clash between the side chains and backbone atoms of L11 in both chains, whereas a shift in the opposite direction creates a clash between residues Y8 and L11 on one chain with Y103 on the interacting chain (Fig. S5).
Discussion
Our results demonstrate that protein–protein interactions can be engineered by selecting protein scaffolds with complementary, surface-exposed β-strands, then computationally designing the interface residues to form favorable interactions. One of the four homodimers we designed formed a stable dimer in solution. The crystal structure of this protein closely matches the design model.
We intentionally selected designs where the intermolecular interactions, other than the paired β-strands, were either predominantly polar or predominantly nonpolar. Although the energy and metric scores for these interfaces were similar (Table 1), only the designs with predominantly hydrophobic interfaces formed dimers in solution (Table 2). These results are unsurprising given previous observations that homodimeric interfaces are more hydrophobic than heterodimeric interfaces (36) and that new hydrogen-bond networks are difficult to design (37). A closer inspection of the computational model of βdimer3 revealed that four interface-spanning hydrogen bonds, between designed side chains and backbone atoms, were suboptimal. The N–H bond vector from the hydrogen-bond donors was more than 60° out of plane with the lone-pair electrons on the acceptor carbonyl oxygens. The N–H bond vector is typically in plane with the accepting electrons in crystal structures of natural proteins (38). This deviation is not penalized in the current implementation of the hydrogen-bond energy evaluation in Rosetta.
Some previous attempts at computational protein interface design have been plagued by problems controlling binding orientation, including a complete 180° rotation from the design model (39) or existence of multiple low-energy binding conformations (9). The β-strand pairing addresses these issues by constraining the possible geometry of the interface, as illustrated by the high similarity between the computational model and experimentally determined structure of βdimer1. One challenge of the β-strand pairing approach is that the paired strands must have complementary curvatures in order to form low-energy hydrogen bonds across the interface. This requirement limits the number of naturally occurring proteins that can be redesigned to form new homo- or heterodimers. One potential way to escape this limitation is by designing de novo scaffolds that have edge strands with the appropriate curvature for a target interaction.
Homodimerization illustrates an important step in protein evolution. Many protein–protein interfaces are built on the progression from a monomer to symmetric homodimer to asymmetric homodimer to heterodimer (24, 40). In fact, the majority of protein interfaces common across the three kingdoms of life are symmetric homodimers (23). Our results demonstrate that it is possible to make the first step in this process without disturbing the backbone conformation of the monomer. A logical next step in this path is to redesign the interface of the constructed homodimer to form a heterodimer.
The method of finding complementary, surface-exposed, β-strands presented here could be extended to other aspects of protein design. This protocol could be used to design a protein to bind a natural protein with an exposed β-strand, or build higher order oligomeric structures.
Materials and Methods
Search Method for Homodimer Scaffolds.
To find possible starting structures for homodimer design, we computationally scanned through a set of 5,500 high-resolution crystal structures. All computational steps were performed in the Rosetta3 suite of macromolecular protein modeling software (41). We defined a β-strand as surface-exposed if it met three criteria: (i) five sequential residues had β-strand secondary structure as judged by the Database of Secondary Structure of Proteins algorithm (42); (ii) there were no backbone–backbone hydrogen bonds formed by every other residue in the strand; and (iii) every other residue had fewer than 16 neighboring residues, or had 16–30 neighbors and a SASA per atom greater than 2.0 Å2. Residues are defined as neighbors if their Cβ to Cβ distance is < 10 Å. An example command line used to find the exposed strands follows:
./exposed_strand_finder.<exe> -database <rosetta_database> -l <list_of_inputs> -ignore_unrecognized_res true -packing::pack_missing_sidechains -out::nooutput > exposed_list
We then created a potential homodimer. The axis of an exposed β-strand was defined as a vector from the Cα atom on the first residue of the strand to the Cα atom on the final residue of the strand. Another vector is defined at the center residue of the strand from the carbonyl carbon to carbonyl oxygen. A final vector is drawn perpendicular to the two vectors described through the Cα atom of the residue at center of the strand. The antiparallel homodimer is constructed by copying the protein and rotating the copy 180° about this axis. The copied chain is then translated away from the original by 6.0 Å to create a starting point for evaluation. The copied chain was then translated along the axis of the exposed strand in steps of 7 Å to identify alternate conformations that have no clashing backbone atoms. As a final filter, we check to make sure there are no backbone–backbone clashes between the two chains. The identification of exposed β-strands generated from the above protocol was used as input for the next step. The command line used to make potential homodimers with interacting strands of five residues was
./homodimer_maker.<exe> -database <rosetta_database> –s <pdb_file> -run::chain <chain_char> -sheet_start <start_residue_#> -sheet_stop <last_residue_#> -window_size 5
To narrow down the list of alignments, we ran short symmetric design simulations followed by side-chain and backbone minimization. These alignments were then filtered for designs that possessed an interface area of at least 850 Å2, two or fewer polar atoms not forming hydrogen bonds at the interface, and a calculated ΔGbinding of less than -15.0 Rosetta energy units. The protocol used for this step is similar to the one for full design below.
Homodimer Design and Selection.
The computational homodimer interface design strategy is similar to the Dock Design Minimize Interface protocol used previously for heterodimer design (9). Each step employed Rosetta’s symmetry protocols, which can perform symmetric protein–protein docking, symmetric design, and side-chain/backbone minimization (31, 32). The homodimer model generated above was used to generate the symmetry definition and starting structure for interface design. First, the protein is symmetrically docked against itself to sample rigid-body degrees of freedom. After the docking step, all residues within 8 Å of the other chain were symmetrically designed. Finally, the backbone and side chains of all interface residues were minimized.
An example command line for this protocol is
./homodimer_design.<exe> -database <rosetta_database> -s <pdb_file> -symmetry:symmetry_definition <symmdef> -nstruct 5000 –pack_min_runs 4 -make_ala_interface false –find_bb_hbond_E true –no_his_his_pairE true -disallow_res CGP –use_input_sc –ex1 –ex2 –docking:docking_local_refine true –docking:sc_min true –docking:dock_ppk false –symmetry:perturb_rigid_body_dofs 3 5 –out:file:fullatom
Evaluation of Designs.
We selected which computational designs to express based on several metrics. As a first criterion, we selected designs that were in the top 10% in backbone–backbone hydrogen-bond energy across the interface, total Rosetta energy, and calculated ΔGbind. We then calculated additional metrics including interface energy density (ΔGbind/SASA), RosettaHoles score (33), and number of buried-unsatisfied at the interface. To pick out the final designs to test experimentally, we visually inspected the designs that scored better than native interfaces in all of these metrics.
DNA Construct and Protein Production.
Genes encoding the wild-type protein and the four designs were synthesized by GenScript USA and subcloned into the pQE-80L vector as 6x-His-maltose-binding protein fusions as described previously (9). All proteins were expressed in BL21(DE3) cells. Expression and purification methods are outlined in SI Materials and Methods.
Multiangle Light Scattering.
Samples of βdimer1, βdimer3, and the wild-type protein were concentrated to approximately 300 µM (4 mg/mL) and injected onto a size-exclusion column connected to a MALS instrument and a refractometer. Instrument and data fitting methods are described in SI Materials and Methods.
Analytical Ultracentrifugation Sedimentation Equilibrium.
The molecular mass of wild-type protein, βdimer1, and βdimer3 were found by spinning these proteins at concentrations of 20, 40, and 60 μM and monitoring absorbance at 280 nm. The homodimer dissociation constant was found by spinning βdimer1 at concentrations of 0.8, 1.5, and 2.0 μM and monitoring absorbance at 215 nm. The entire AUC protocol and data fitting methods can be found in SI Materials and Methods.
Fluorescence Polarization Assay.
A variant of βdimer1 with the mutation S62C was produced for labeling with thiol reactive Bodipy(507/545)-iodoacetamide (Molecular Probes). The change in polarization was monitored as unlabeled βdimer1 and wild-type protein were then titrated into Bodipy-labeled βdimer1. Full experimental details and the model fitting method are described in SI Materials and Methods.
Crystallization of βdimer1.
The hanging-drop vapor diffusion method was used to crystallize βdimer1 at 20 °C. The final model contains two molecules in the asymmetric unit with all residues defined in the electron density, except for residues 23–26 in both molecules. Crystallization conditions and structure refinement techniques are given in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We would like to thank I. André for developing and providing support for symmetry-related protocols in Rosetta. This work was supported by US National Institutes of Health Grant R01-GM073960.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: X-ray coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3ZY7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115124108/-/DCSupplemental.
References
- 1.Karanicolas J, Kuhlman B. Computational design of affinity and specificity at protein–protein interfaces. Curr Opin Struct Biol. 2009;19:458–463. doi: 10.1016/j.sbi.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandell DJ, Kortemme T. Computer-aided design of functional protein interactions. Nat Chem Biol. 2009;5:797–807. doi: 10.1038/nchembio.251. [DOI] [PubMed] [Google Scholar]
- 3.Havranek JJ, Harbury PB. Automated design of specificity in molecular recognition. Nat Struct Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- 4.Grigoryan G, et al. Computational design of virus-like protein assemblies on carbon nanotube surfaces. Science. 2011;332:1071–1076. doi: 10.1126/science.1198841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS. High-resolution protein design with backbone freedom. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 7.Fleishman SJ, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jha RK, et al. Computational design of a PAK1 binding protein. J Mol Biol. 2010;400:257–270. doi: 10.1016/j.jmb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sammond DW, et al. Computational design of the sequence and structure of a protein-binding peptide. J Am Chem Soc. 2011;133:4190–4192. doi: 10.1021/ja110296z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds KA, et al. Computational redesign of the SHV-1 β-Lactamase/β-Lactamase inhibitor protein interface. J Mol Biol. 2008;382:1265–1275. doi: 10.1016/j.jmb.2008.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S, et al. Nonnatural protein–protein interaction-pair design by key residues grafting. Proc Natl Acad Sci USA. 2007;104:5330–5335. doi: 10.1073/pnas.0606198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Der BS, Kuhlman B. From computational design to a protein that binds. Science. 2011;332:801–802. doi: 10.1126/science.1207082. [DOI] [PubMed] [Google Scholar]
- 14.Guharoy M, Chakrabarti P. Secondary structure based analysis and classification of biological interfaces: Identification of binding motifs in protein–protein interactions. Bioinformatics. 2007;23:1909–1918. doi: 10.1093/bioinformatics/btm274. [DOI] [PubMed] [Google Scholar]
- 15.Ni YG, et al. A PCSK9-binding antibody that structurally mimics the EGF (A) domain of LDL-receptor reduces LDL cholesterol in vivo. J Lipid Res. 2011;52:78–86. doi: 10.1194/jlr.M011445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbreth RN, et al. Isoform-specific monobody inhibitors of small ubiquitin-related modifiers engineered using structure-guided library design. Proc Natl Acad Sci USA. 2011;108:7751–7756. doi: 10.1073/pnas.1102294108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richardson JS, Richardson DC. Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tereshko V, et al. Toward chaperone-assisted crystallography: Protein engineering enhancement of crystal packing and X-ray phasing capabilities of a camelid single-domain antibody (VHH) scaffold. Protein Sci. 2008;17:1175–1187. doi: 10.1110/ps.034892.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koide S. Engineering of recombinant crystallization chaperones. Curr Opin Struct Biol. 2009;19:449–457. doi: 10.1016/j.sbi.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhlman B, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 21.Nauli S, et al. Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2. Protein Sci. 2002;11:2924–2931. doi: 10.1110/ps.0216902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nesloney CL, Kelly JW. Progress towards understanding β-sheet structure. Bioorg Med Chem. 1996;4:739–766. doi: 10.1016/0968-0896(96)00051-x. [DOI] [PubMed] [Google Scholar]
- 23.Kim WK, Henschel A, Winter C, Schroeder M. The many faces of protein–protein interactions: A compendium of interface geometry. PLoS Comput Biol. 2006;2:e124. doi: 10.1371/journal.pcbi.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 25.Hoskins J, Lovell S, Blundell TL. An algorithm for predicting protein–protein interaction sites: Abnormally exposed amino acid residues and secondary structure elements. Protein Sci. 2006;15:1017–1029. doi: 10.1110/ps.051589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loughlin WA, Tyndall JDA, Glenn MP, Fairlie DP. β-strand mimetics. Chem Rev. 2004;104:6085–6118. doi: 10.1021/cr040648k. [DOI] [PubMed] [Google Scholar]
- 27.Khakshoor O, Nowick JS. Artificial β-sheets: Chemical models of β-sheets. Curr Opin Chem Biol. 2008;12:722–729. doi: 10.1016/j.cbpa.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sievers SA, et al. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature. 2011;475:96–101. doi: 10.1038/nature10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khakshoor O, Demeler B, Nowick JS. Macrocyclic β-sheet peptides that mimic protein quaternary structure through intermolecular β-sheet interactions. J Am Chem Soc. 2007;129:5558–5569. doi: 10.1021/ja068511u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khakshoor O, et al. X-ray crystallographic structure of an artificial β-sheet dimer. J Am Chem Soc. 2010;132:11622–11628. doi: 10.1021/ja103438w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiMaio F, Leaver-Fay A, Bradley P, Baker D, Andre I. Modeling symmetric macromolecular structures in Rosetta3. PLoS One. 2011;6:e20450. doi: 10.1371/journal.pone.0020450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das R, et al. Simultaneous prediction of protein folding and docking at high resolution. Proc Natl Acad Sci USA. 2009;106:18978–18983. doi: 10.1073/pnas.0904407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheffler W, Baker D. RosettaHoles: Rapid assessment of protein core packing for structure prediction, refinement, design and validation. Protein Sci. 2009;18:229–239. doi: 10.1002/pro.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueller-Dieckmann C, Panjikar S, Tucker PA, Weiss MS. On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength. Acta Crystallogr D Biol Crystallogr. 2005;61:1263–1272. doi: 10.1107/S0907444905021475. [DOI] [PubMed] [Google Scholar]
- 35.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 36.Bahadur RP, Chakrabarti P, Rodier F, Janin J. A dissection of specific and non-specific protein–protein interfaces. J Mol Biol. 2004;336:943–955. doi: 10.1016/j.jmb.2003.12.073. [DOI] [PubMed] [Google Scholar]
- 37.Joachimiak LA, Kortemme T, Stoddard BL, Baker D. Computational design of a new hydrogen bond network and at least a 300-fold specificity switch at a protein–protein interface. J Mol Biol. 2006;361:195–208. doi: 10.1016/j.jmb.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 38.Kortemme T, Morozov AV, Baker D. An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein–protein complexes. J Mol Biol. 2003;326:1239–1259. doi: 10.1016/s0022-2836(03)00021-4. [DOI] [PubMed] [Google Scholar]
- 39.Karanicolas J, et al. A de novo protein binding pair by computational design and directed evolution. Mol Cell. 2011;42:250–260. doi: 10.1016/j.molcel.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ben-Shem A, Frolow F, Nelson N. Evolution of photosystem I—from symmetry through pseudosymmetry to asymmetry. FEBS Lett. 2004;564:274–280. doi: 10.1016/S0014-5793(04)00360-6. [DOI] [PubMed] [Google Scholar]
- 41.Leaver-Fay A, et al. Rosetta3: An object oriented suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.