“Two independent groups of investigators have found evidence of an enzyme… which synthesizes DNA from an RNA template. This discovery, if upheld, will have important implications… information transfer from DNA to RNA can be inverted.” 1970, preamble to refs. 1 and 2.
These words accompanied two articles describing the discovery of RNA-dependent DNA polymerase, now known as reverse transcriptase (RT), in the virions of RNA tumor viruses. RT activity was recognized independently by David Baltimore (1) and Howard Temin and Satoshi Mizutani (2). In five printed pages these authors collectively challenged the unidirectionality of macromolecular synthesis, from DNA to RNA to protein, the central dogma of molecular biology. Indeed by demonstrating that DNA synthesis can be templated by RNA, they overturned the central dogma. However, there was no way of predicting how in the ensuing 4 decades scientists would uncover the essential role of RT in shaping genomes and making possible the diversity of life on earth.
This was the topic of the colloquium “Telomerase and Retrotransposons: Reverse Transcriptases that Shaped Genomes,” held September 29–30, 2010, which forms the basis of this special feature. We now know that approximately half of the human genome and a significant portion of every other eukaryotic genome is generated by RTs, which are present in two major forms: telomerases, which synthesize the ends of linear eukaryotic chromosomes, and retrotransposon RTs, which deposit copies of themselves throughout the genetic landscape and generate pseudogenes with spectacularly diverse roles in gene regulation. There was no inkling 40 years ago that misregulation of RTs can cause chromosomal catastrophes that are characteristic of cancer, neurological disorders, and aging, as occurs when telomerase and retrotransposon RTs escape normal controls. A central question in genome biology then is how the activity of RTs has been harnessed, regulated, and modified throughout evolution to generate diversity while maintaining cellular balance and genome integrity.
The colloquium addressed the evolution and interrelationships of RTs of viruses, transposons, telomeres, and cellular genes, while exploring RT structure, function, and its bearing on development, aging, and disease. It remains debatable whether telomerases represent domesticated retrotransposons or whether these chromosome-capping agents radiated into mobile genetic elements. Nevertheless, their phylogenetic and functional relatedness is clear (Fig. 1), and indeed, retrotransposons can provide alternative means of maintaining chromosome ends, as will become evident from the following.
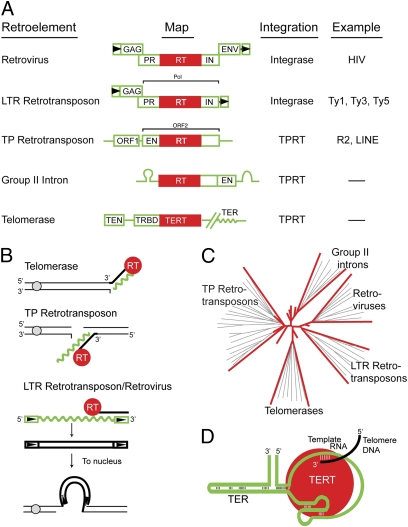
Fig. 1.
Featured retroelements. (A) Architecture of retroelements. RNA maps (5′–3′, green) show RT (red). The schematic is not to scale. Stem-loops flanking the group II intron RT represent catalytic RNA, and the wavy line represents the RNA component of telomerase (TER). The hatches indicate that TERT and TER are encoded by separated genes. Boxed triangles designate LTRs. Coding sequences are GAG, Gag protein; PR, protease; IN, integrase; ENV, envelop protein; EN, endonuclease; TEN, telomerase essential N terminus; and TRBD, telomerase RNA-binding domain. (B) Initiation of cDNA synthesis by RT. RTs are shown at telomere and within chromosome. TERT acts on 3′-OH at a telomere, whereas the RT of EN-proficient TP retrotransposons acts on the 3′-OH at a double-strand break. The green wavy line indicates RNA. (C) Phylogram of RT classes. Phylogram shows clustering of RT members, as described in detail in the article by Gladyshev and Arkhipova (12). (D) Schematic of telomerase. Telomerase RNA (TER, green) with its pseudoknot structure has the template region base-paired to telomere DNA (black) in the active site of telomerase RT (TERT, red). The diagram is not to scale.
The colloquium and this compendium of articles reflect the lifestyle of retroviruses, retrotransposons, and telomeres. The retrotransposons we consider are the retroviral-like elements that contain long terminal repeats [LTR retrotransposons (e.g., Ty elements)] and those that do not contain LTRs [e.g., long interspersed nuclear elements (LINE) elements and group II introns]. These non-LTR retrotransposons are also referred to as target-primed (TP) elements, for their mechanism of chromosome integration (3). The architecture of the different retroelements under consideration is schematized in Fig. 1A, indicating that they are all molecular mosaics, with different functional modules associated with RT. RT is fused to protease and integrase domains in both retroviruses and LTR retrotransposons, whereas RT is juxtaposed to endonuclease domains in TP retrotransposons and some group II introns. Retrotransposition of a TP retrotransposon or group II intron is usually initiated by endonucleolytic cleavage, exposing a 3′-OH that serves as the primer for reverse transcription, with the RNA of the element being the template (Fig. 1B). This process of target-primed reverse transcription (TPRT) (4) is also used for cDNA synthesis at telomeres, where the telomerase RT (TERT) uses a 3′-OH that occurs naturally at chromosome ends to prime TPRT. In contrast, LTR retrotransposons and retroviral RNA templates are first reverse transcribed, and then the cDNA is inserted into the genome via integrase (5).
Evolutionary Relationships
It has been appreciated for some time that telomerase and retrotransposon RTs exhibit a number of structural and mechanistic similarities and are likely to share a common origin (6–11). Phylogenetic analyses reinforce a deep-rooted connection between these two families of polymerases (Fig. 1C). Irina Arkhipova's lecture, as well as the accompanying article on a distinct group of RTs (12), explored evolutionary space between the large class of RTs that are associated with diverse selfish mobile retroelements and telomerase RT. She described an “evolutionary intermediate,” a unique family of single-copy RT genes that is not only distinct from the telomerase RT gene but also shows none of the hallmarks of mobile element association. This RT gene family seems to have ancient origins and is found sporadically throughout all eukaryotic kingdoms. Thus, telomerase is not the only single-copy gene that evolved from an ancestral RT gene. This finding lends credence to the idea that different classes of retroelements evolved independently from divergent single-copy RT genes.
Another connection between telomeres and retrotransposons was provided by Mary-Lou Pardue, who described Drosophila telomeres, which are maintained not by telomerase but rather by repeated retrotransposition of a small set of retroelements HeT-A, TART, and TAHRE (13). In a “natural” experiment in the genus Drosophila, and perhaps other insects, the loss of telomerase apparently led to the utilization of other RTs that have coevolved with their host cells to provide a robust mechanism for maintaining chromosome ends. Although HeT-A, TART, and TAHRE resemble typical non-LTR retrotransposons, they have several distinctive features contributing to their effectiveness at the telomere. Further evidence of collaboration between the host cell and these elements is seen in the sequence of a fragment of a telomere array, moved into a centromere region by an ancient transposition and later remodeled to resemble the chromatin of other centromere regions—chromatin that is very different from that of the analogous sequence remaining at telomeres.
However, this rather astonishing feat of substitution does not seem to occur frequently, because loss of telomerase in most organisms results in the activation of recombination pathways at telomeres. These pathways have been extensively characterized in yeast but not in mammalian cells (14). Tammy Morrish (Greider laboratory) described her ongoing study of the genetic requirement for telomere recombination using tumor and immortalized cells derived from telomerase knockout mice. Her preliminary data point to the involvement of the recombination function of RAD50. An intriguing and unresolved question is whether in this setting retrotransposons could also play a role in capping the chromosome ends.
Beyond the recurring questions concerning the telomerase–retrotransposon relationship, unusual aspects of telomerase and telomere evolution are emerging. Dorothy Shippen presented a case of telomerase RNA (TER) duplication and functional specialization. In the plant Arabidopsis thaliana, one of the two TERs acts as a repressor rather than a stimulator of telomere elongation, possibly by forming an inactive complex with the catalytic TERT (15). This finding illustrates the adaptability of telomerase components.
The versatility of the telomerase ribonucleoprotein (RNP) is further reflected in the diversity of DNA sequences that it synthesizes. Neal Lue highlighted the extraordinary variability of the telomere repeat units in a group of budding yeasts that include both Saccharomyces and Candida species. Whereas the repeat units are 5–8 bp and G-rich in most phyla, the ones in budding yeast can be as long as 25 bp and exhibit little nucleotide bias on either strand (16). The selection pressures that resulted in telomere sequence divergence in these and other organisms are not understood and are worthy of further analyses.
Another example of the plasticity of retroelements is the group II intron, which is thought to be of bacterial origin and is widely hypothesized to be ancestral to the prevalent eukaryotic spliceosomal introns. Group II intron RNAs are unusual retroelements in that they are ribozymes that encode RT. After self-splicing, the intron can reverse-splice into a DNA target and be reverse transcribed by TPRT, similar to TP-retrotransposition and telomere synthesis (3, 17). Marlene Belfort's lecture addressed the paradox that arises from the compelling hypothesis that group II introns, found exclusively in prokaryotes and eukaryotic organelles, are the progenitors of spliceosomal introns, which are unique to and abundant in eukaryotic nuclei. To understand why a eukaryotic nucleus is inhospitable to group II introns even though their apparent descendants are ubiquitous, the Belfort group introduced a group II intron into nuclear genes (18). The group II intron was spliced; however, splicing was primarily cytoplasmic, and for reasons that remain enigmatic, the spliced transcript was not translated. Perhaps gene compartmentalization in nuclei, exclusion of group II introns, and biogenesis of spliceosomal introns were all driven by the need to protect genes from the mutagenic effects of group II retroelements.
Telomerases and Retrotransposons Function Within the Context of Highly Structured RNPs
A detailed understanding of telomerase mechanisms will ultimately require high-resolution structural information. Impressive progress toward achieving this goal was made by the determination of the atomic resolution structure of TERT from Tribolium castaneum (19). At the colloquium and in an accompanying perspective, Juli Feigon summarized advances on the structure of human telomerase RNA (TER) and the RNP (20, 21). The noncoding TER not only supplies the template for reverse transcription but also stimulates the polymerization by TERT through mechanisms that remain poorly understood (Fig. 1D). Two conserved TER structural elements seem to be particularly important for the stimulatory function: a template/pseudoknot domain (the core domain) and a stem-terminus element (STE) (22). Structural models of subdomains of these elements have been determined in recent years, primarily by NMR. Feigon presented a structure model of the entire core domain based on structures and dynamics of all of the helical subdomains, which revealed an intriguing V-shaped conformation. Additionally, on the basis of the structure of the Tribolium castaneum TERT, she proposed two plausible views of the relative disposition of the core domain and TERT. A parallel advance on the STE is reported by Julian Chen and colleagues in an accompanying article (23). Using a unique photoaffinity cross-linking approach and mass spectrometry, they identified three nucleotide–amino acid contacts between a vertebrate STE (known as CR4/5) and the corresponding TERT, thus bringing into sharper focus the architecture of the telomerase RNP.
Another powerful approach for elucidating the assembly and mechanisms of RNPs is single-molecule FRET, as illustrated in an accompanying article by Xiaowei Zhuang (24). By labeling specific residues within the Tetrahymena telomerase RNA with fluorescent donors and acceptors, Zhaung and coworkers were able to monitor the conformation of a pseudoknot in the enzyme complex. They showed that the TERT protein prevents the misfolding of RNA and that only RNPs with properly formed pseudoknots are catalytically active. These observations provide compelling support for the importance of a correctly folded pseudoknot in telomerase activity, a recurrent idea that has lacked decisive confirmation.
Although knowledge of the structure of retrotransposon RNPs is less advanced than that of telomerase, the group II intron RNP is beginning to yield to biophysical analysis. By purifying the intron RNA in complex with the intron-encoded protein from its native Lactococcal host, the Belfort group in collaboration with the Joachim Frank (Columbia University) and Greg Van Duyne (University of Pennsylvania) laboratories has obtained a glimpse of an active RNP. Cryo-EM, size-exclusion chromatography, and sedimentation analyses revealed the intron RNP precursor as a large, loosely packed structure, in contrast to the compact spliced intron RNP (25). These results suggest that a major conformational change and RNP compaction is required to achieve the catalytically active state.
Tricks to Accomplish the Complex Task of Reverse Transcription
A unique property of telomerase as an RT is its ability to make consecutive copies of telomeric repeats using a short template (Fig. 1D), referred to as repeat addition processivity. To accomplish this task, telomerase must undergo a translocation reaction, as follows: after each round of reverse transcription, the RNA–DNA hybrid dissociates and TERT realigns the 3′ end of the DNA to the RNA template, via a putative anchor site in TERT, to enable the next round of polymerization. Insights into these molecular gymnastics were provided by two presentations. Kathy Collins described her investigation of the human telomerase core complex through coexpression of truncated TERT protein and TER RNA domains, as well as analysis of the resulting RNP. Particularly noteworthy was her finding that the telomerase essential N-terminal (TEN) domain of TERT is dispensable for nucleotide addition but can complement the rest of the complex in trans to reconstitute a highly processive telomerase (26). This finding reinforces the long-standing hypothesis that the TEN domain, which is known to have a weak single-stranded DNA-binding activity, may serve as the anchor site of telomerase that enables the RNP to trap telomeric DNA during repeated cycles of extension and RNA–DNA dissociation. Julian Chen addressed the fate of the RNA–DNA hybrid using a clever “template-free” assay, in which the template region of the RNA was selectively removed from the enzyme preparation and supplied in trans as a part of an RNA–DNA hybrid substrate. By analyzing the binding properties and activities of this reconfigured telomerase and a series of mutants, he concluded that a key determinant of translocation efficiency is the ability of the telomerase active site to reengage the RNA–DNA hybrid (27). This proposition in turn suggests that the active site is temporarily disengaged from the RNA template region during translocation and that the telomerase RNP is even more dynamic than previously envisioned.
A key difference between retrotransposons and telomerase is that retrotransposon RTs are encoded by the RNA template used for reverse transcription, whereas telomerase RT and RNA are encoded by separate genes (Fig. 1A). Thus, retrotransposons face the problem of partitioning their RNA genomes between translation and reverse transcription. The RNA of LTR retrotransposons is used as a template for synthesis of not only RT but also of the structural protein Gag, which forms a virus-like particle (VLP) wherein the RNA is reverse transcribed. In her presentation, Suzanne Sandmeyer proposed that partitioning of the Ty3 LTR retrotransposon occurs when translating Ty3 RNA interacts with mRNA translation suppressors and decay proteins that concentrate in stress granules and mRNA processing bodies. Indeed, shortly after induction of Ty3 expression, Ty3 Gag and RNA colocalize in microscopically distinct cytoplasmic foci that resemble stress granules and processing bodies, and contain many of the same sequestration and decay factors, but are formed under separate circumstances. Sandmeyer has named these foci “retrosomes,” reflecting the observation that components of other retrotransposons, including the human L1 element and eukaryotic retroviruses, localize to similar cytoplasmic bodies. It is likely that sequestration of Ty3 RNA with stress granule/processing body proteins is mediated by the VLP structural protein, Gag, because neither Ty3 RNA nor Gag proteins localize to retrosomes if Gag binding to Ty3 RNA is blocked (28). Retrosomes may function to concentrate Ty3 VLP components to ensure efficient VLP assembly.
The RNA genomes of LTR retrotransposons and retroviruses are reverse transcribed within RNP complexes in the cytoplasm, and then the cDNA is integrated into the host genome. Host proteins transcribe the retrotransposon or integrated provirus. This generates another round of template RNAs for protein synthesis. The RNAs can also be used to generate additional genomes for VLPs (LTR retrotransposons) or viral particles (retroviruses). This highly complex mode of viral particle replication requires that reverse transcription is coordinated with other obligatory steps during cellular replication. Insights into this regulation was provided by Xiaowei Zhuang, who described elegant single-molecule experiments in which FRET was applied to monitor the dynamics of individual HIV-1 RT initiation complexes (29). Her studies reveal a dynamic interaction between HIV-1 RT and viral RNA:tRNA template-primer during initiation of reverse transcription. While paused in the initiation phase, RT flips between two opposite orientations: one is competent for extension of the primer, whereas the other is not. Once a stem-loop structure located upstream of the primer-binding site in the HIV-1 RNA is traversed, only the orientation that places the active site of RT at the site of primer extension is favored, and elongation is rapid. Zhuang speculated that polymerase pausing at the stem-loop may serve to stall reverse transcription until the virion has adopted its mature form and enters a new cell, whereupon the reverse transcript can be introduced into a naïve genome.
Tom Eickbush spoke about the evolution, expression, and mechanism of reverse transcription of the R2 element, a site-specific TP retrotransposon that inserts into a conserved region of 28S ribosomal RNA of the insect Bombyx mori. R2 elements are present in most animal lineages and are transmitted vertically. They are expressed as part of the 28S rRNA transcript. The 28S-R2 cotranscript is processed at the 5′ end by self-cleavage by a double pseudoknot-bearing ribozyme in the R2 RNA (30). The single R2 ORF encodes a protein with RT, endonuclease, and DNA binding domains. By following the TPRT activity of the purified B. mori R2 ORF, it was shown that the R2 ORF binds both the 5′ and 3′ end of R2 RNA, and it was proposed that RNA binding at these sites coordinates first- and second-strand cDNA synthesis (31). Eickbush discussed his recent experiments identifying RNA binding motifs in the R2 ORF immediately N-terminal of the conserved RT motifs. Mutations in this region eliminated TPRT activity, providing further support for a critical role for the RNA template in the TPRT reaction. Interestingly, the position of the RNA binding domains relative to the RT motifs in the R2 ORF are the same as the domains of telomerase, named CP, QFP, and T, that are necessary for RNP formation (32).
Since Eickbush's discovery that the RT of the R2 TP retrotransposon uses the 3′-OH of nicked target DNA as its primer (4), scientists have noted the mechanistic similarities between retrotransposition and telomere addition. Indeed, a human L1 TP retrotransposon lacking a functional endonuclease (EN) domain can retrotranspose to dysfunctional telomeres (10). In his lecture and the accompanying article (33), John Moran described studies on the factors involved in initiating RT activity of the human L1 TP retrotransposon. Using partially purified RT–RNA complexes from cells expressing L1 (34), Kulpa and Moran showed that L1 RT prebound to its RNA template uses DNA substrates that mimic telomeric ends as primers. Moreover, the L1 RNP, like telomerase, is associated with a nuclease activity that can process the end of the target DNA before reverse transcription is initiated. The ability of EN-defective L1 RNPs to prime reverse transcription from processed telomeric sequences in vitro and the retrotransposition of EN-defective L1 to telomeric ends in vivo imply that EN-independent L1 RT activity resembles that of ancestral TP retrotransposons and telomerase. Thus, under defined circumstances, the L1 RNP, like telomerase, can carry out RNA-mediated DNA repair to provide an interesting case of functional mimicry.
Targeting Specificity
In contrast to telomeres that are typically confined to chromosome ends, the retrotranscripts of viruses and transposons can target a plethora of sites. The host–element interactions that govern the integration of reverse transcripts into host genomes by a retrovirus and a retrovirus-like transposon are varied and fascinating. Dan Voytas spoke about integration specificity of the yeast retrovirus-like transposons, Ty5 and Ty1. Ty5 preferentially targets heterochromatic regions of the yeast genome via an interaction between Ty5 integrase and the silencing protein Sir4. As reported in the accompanying article (35), high-throughput DNA sequencing of a library of Ty5 integration sites in the Saccharomyces cerevisiae genome yielded a surprising finding: nearly one-quarter of Ty5 retrotransposition events are euchromatic. Further analysis of both heterochromatic and euchromatic insertions uncovered a secondary Ty5 target site, in DNase I-sensitive, nucleosome-free regions. In the accompanying article (35), Voytas speculates that Sir4 brings the cDNA–integrase complex to the vicinity of heterochromatin, but specific sites of insertion are governed by the accessibility of the DNA. DNA is most accessible in the nucleosome-free regions flanking ORFs; hence both the primary and this newly revealed secondary target site bias give rise to Ty5 retrotransposition events that rarely disrupt essential coding sequences. This interpretation is bolstered by the finding that ploidy has no effect on the chromosomal distribution of Ty5, indicating that coding sequences are not necessarily cold spots because of selection against deleterious events. In contrast to Ty5, Ty1 was shown to preferentially integrate into nucleosomal DNA upstream of pol III-transcribed genes, a pol II gene-poor region.
The human protein LEDGF, a bipartite protein consisting of integrase and chromatin-binding domains, binds HIV-1 integrase and tethers the preintegration complex containing the HIV-1 reverse transcript and integrase to genomic sites. The consequence of LEDGF tethering is that HIV-1 reverse transcripts typically integrate within active transcription units (36). Steve Hughes and collaborators Alan Engelman (Dana-Farber Institute) and David Allis (Rockefeller University) have fused the integrase-binding domain of LEDGF to chromatin-binding domains of other proteins (37) to test the idea that the HIV-1 target site bias is determined primarily by the specificity of the chromatin-binding domain of the host protein with which HIV-1 integrase interacts. In his talk, Hughes discussed swapping experiments involving the plant homeodomain PHD finger motif of the histone demethylase JARID1A, the PHD finger and atypical bromodomain region of the histone methyltransferase MLL, and the chromodomain from polycomb group protein Cbx7. Fusions to the integrase-binding domain of LEDGF were expressed in mouse cells that lack LEDGF. The results of these experiments demonstrate that chromatin-binding domain swapping in LEDGF redirects HIV-1 integration to novel regions of the genome. The successful redirection of HIV-1 integration events raises the possibility that lentiviral gene therapy vectors could be directed to nondeleterious regions of the human genome.
Regulation by Telomere Proteins and Accessory Factors
The regulation of telomerase by auxiliary components of the complex and telomere-bound proteins is an especially active area of investigation. Many such factors have been described in diverse systems. Kathy Collins summarized her studies in elucidating the protein–protein and protein–nucleic interactions within the Tetrahymena telomerase RNP and between the RNP and telomeric DNA. Recent highlights include the identification and characterization of a telomere adaptor subcomplex and a replication protein A (RPA)-related subunit (Teb1) that interact with the core complex to form a holoenzyme with a high degree of repeat addition processivity (38, 39). RPA is a non–sequence-specific single-stranded DNA-binding protein complex that participates in numerous DNA transactions, such as replication, repair, and recombination. The RPA-like subunit in telomerase, however, exhibits a strong preference for single-stranded telomeric DNA and apparently exploits this high-affinity binding to facilitate telomerase–telomere interactions. In an accompanying article (40), Collins and collaborator Ming Lei (University of Michigan) report the crystal structures of multiple oligonucleotide/oligosaccharide binding (OB) fold domains in Teb1 and examined their roles in DNA binding and processivity enhancement. The results offer a strikingly detailed illustration of how the non–sequence-specific RPA (presumably the ancestor of Teb1) may be adapted to serve telomere-specific functions through the acquisition of altered DNA-binding properties.
The budding yeast S. cerevisiae, with its constellation of genetic tools, has also offered a productive system for tackling telomerase regulation. Three proteins of importance are the telomere end-binding protein Cdc13 and the telomerase regulatory proteins Est1 and Est3. All three proteins are essential for telomere extension in vivo, but their precise mechanisms of action were debated (41). In a noteworthy development, an interaction between Cdc13 and Est1, which has long been postulated to serve the recruitment of telomerase to telomeres (42), has at last been recapitulated in vitro. Ginger Zakian reported at the colloquium and in an accompanying article (43) on a yeast overexpression system that enabled purification of both Cdc13 and Est1, which indeed interact specifically with each other with a Kd of ≈250 nM. Intriguingly, mutations in CDC13 and EST1 that abolished telomere maintenance in vivo did not have an obvious effect on this physical interaction, suggesting that a step other than recruitment may be affected. Besides interacting with Cdc13, Est1 has also been implicated in promoting the assembly of Est3 into the telomerase complex (44, 45). Using a similar approach, Zakian and colleagues also demonstrated a direct and specific physical interaction between Est1 and Est3, thus providing a plausible mechanism for how this assembly function is achieved (46).
In contrast to Est1, the action of Est3 is perhaps less controversial but more obscure. Intriguing similarities between this protein and a domain of the mammalian telomere binding protein TPP1 had been noted earlier, although the functional and evolutionary implications are unclear (47, 48). Although early experiments on S. cerevisiae Est3 failed to disclose a contribution of this protein to telomerase primer extension activity in vitro, both the Lundblad and Lue laboratories subsequently reported stimulatory effects of Est3 from other budding yeasts, Saccharomyces castellii and Candida albicans (44, 49). At the colloquium and in an accompanying article (50), Neal Lue presented studies of two unusual Est3 homologs in Candida parapsilosis and Lodderomyces elongisporus, which bear unique N- and C-terminal extensions not found in other Est3 homologs. He reported robust interactions between these Est3s and the corresponding TEN domains, the putative “anchor site” of TERTs. Moreover, although Est3 alone does not bind DNA, it can be cross-linked to DNA when associated with the TEN domain. Thus, Est3 may potentially extend the contact surface between the telomerase holoenzyme and telomeric DNA. Although the precise mechanisms of Est3 and other regulatory factors remain to be worked out, it now seems that the regulation of telomerase is achieved by domains and factors that modulate or augment the RNP–DNA interactions.
The mechanisms of Est3 were also discussed by Vicki Lundblad, whose extensive mutagenesis analysis of the Saccharomyces protein uncovered distinct surfaces of the protein that mediate separate functions (47). In addition, Lundblad described her study of an RPA-like trimeric complex named CST (Cdc13-Stn1-Ten1) that serves critical telomere-specific functions (51). The complex binds to the terminal single-stranded, G-rich overhangs and regulates numerous activities that impact telomere length and structure (e.g., telomerase, recombination, and nucleases). The repeated discovery of RPA-like proteins at telomeres suggests a critical need to protect and/or manipulate single-stranded DNAs at chromosome ends.
Beyond the Genome: Retroelements During Development, Aging, and Disease
There is a growing awareness of retroelements as determinants of cell fate. The Ty1 LTR retrotransposon in S. cerevisiae is known to be a powerful player in genome plasticity. Ty1 elements are associated with DNA fragile sites, and reverse transcripts of Ty1 and cellular mRNAs are frequently found at the junctions of genome rearrangements. In her talk and the accompanying article (52), Joan Curcio reported that she and colleagues Patrick Maxwell (Rensselaer Polytechnic Institute) and William Burhans (Roswell Park Cancer Institute) have found that Ty1 retromobility increases during chronological aging. This increase is associated with higher levels of chromosomal rearrangements (detected via a loss-of-heterozygosity assay) and chromosome loss in old cells. When retrotransposition is blocked by a variety of mutations or an RT inhibitor, aging-associated genome instability is diminished, and in some cases lifespan is significantly extended. It is possible that unrepaired retrotransposition events lead to chromosomal rearrangements; alternatively, retrotranscripts synthesized by Ty1 may heal spontaneous chromosome breaks, leading to DNA deletions, break-induced recombination, or chromosomal rearrangements. Although lifespan is a complex phenotype influenced by a variety of genetic and environmental factors, Curcio's work raises the possibility that genome instability associated with retrotransposition and RNA-mediated DNA repair mechanisms plays a role in limiting lifespan.
Human L1 elements, which constitute ≈17% of the human genome, will, predictably, be inherited by future generations when they occur in germ cells. However, the discovery by Rusty Gage and colleagues that L1 is mobile in human neural progenitor cells in culture and that adult brain tissue, unlike other somatic tissues, sustains massive L1 retrotransposition, was unexpected and remarkable (53). L1 retrotransposition in neural progenitor cells generates genetic mosaicism that likely underlies the extraordinary diversity of neurons. Gage described how the down-regulation of the transcription factor Sox2 during the transition from neural progenitor cells to differentiated neurons opens a brief window for L1 mobilization to occur. Given the ability of L1 transposition to seed major changes in the genome, it is likely that retrotransposition in neural stem cells is highly regulated. Indeed, Gage and colleagues have evidence for an unusually high level of L1 mobility in brain tissue of patients with two distinct neurological disorders. The postmortem tissue from patients with Rett syndrome, a neurodevelopmental disorder arising from mutation of the MeCP2 gene, as well as induced pluripotent cells derived from Rett syndrome fibroblasts, have approximately twice the normal copy number of L1 retrotransposons (54). In the accompanying article (55), Gage and colleagues extend this finding to brain tissue of patients with ataxia telangiectasia (AT), a disorder that results in mutations in the DNA damage-signaling gene, ATM. They show that the retrotransposition efficiency of a human L1 element endogenously expressed in ATM-deficient human cell lines or in ATM knockout transgenic mice is increased, suggesting that the higher L1 copy number in the brains of AT patients could be due to elevated retrotransposition. The findings indicate an association between overactive L1 retrotransposition and neurological disease, although in both cases it remains to be determined whether unusually high retrotransposon copy number is the cause of the neurological symptoms or a consequence of the underlying mutations.
Disruption of telomerase function is also responsible for a collection of human diseases characterized by defective tissue renewal, including aplastic anemia, pulmonary fibrosis, and liver cirrhosis. Perhaps the prototype of such diseases is a heritable disorder known as dyskeratosis congenita (DC). DC patients suffer from bone marrow failures and typically have a characteristic triad of abnormal skin pigmentation, nail dystrophy, and oral leukoplakia (56). The realization that DC is caused by telomere loss came initially from the identification of disease-linked TERC (telomerase RNA) and DKC1 (dyskerin, an RNA-binding telomerase subunit) mutations in some patients (57). This conclusion was subsequently bolstered by the discovery of mutations in other telomerase subunit genes in distinct cohorts of patients. At the colloquium, Steve Artandi reported missense mutations in yet another telomerase subunit named TCAB1, which he had recently found to be required for the trafficking of the telomerase RNP to the Cajal body and also for telomere maintenance (58, 59). It seems all but certain that continued studies of telomerase mechanisms and assembly will have profound impacts on understanding the molecular basis and pathogenesis of telomere-related diseases.
Beyond specific diseases, telomerase has also garnered attention as a major player in cellular pathways that impact aging and cancer. These connections have received compelling support from numerous cell-based and whole-animal studies (60). Although many of the effects of telomerase on cancer and aging, and hence on genome stability, are clearly mediated through its activities at telomeres, a number of intriguing studies have pointed to extratelomeric functions of telomerase subunits, especially the TERT protein. In this vein, Bill Hahn explored the activity of an alternative complex formed by TERT, BRG1 (a chromatin remodeling ATPase implicated in the transcriptional regulation of many target genes), and NS/GNL3L (two related proteins that are highly expressed in proliferative, multipotential cells). He found that overexpression of NS or GNL3L increased the fraction of TICs (tumor initiating cells or tumor stem cells) in a cell population (61). Consistent with working through a non–telomere-related pathway, the effect was not accompanied by telomere length alterations and was abolished by knockdown of BRG1 and TERT but not TERC (the TER gene). It should be noted that the nontelomere functions of telomerase subunits have remained controversial and are seemingly at odds with the results of a number of transgenic mouse studies (62). Nevertheless, the potentially dramatic implications of such functions in human diseases guarantee continued experimentation and eventual resolution of these issues and arguments.
Conclusions
A significant portion of most eukaryotic genomes is derived from RNA. Formation of this “retrogenome” involves the synthesis of reverse transcripts and the subsequent or accompanying incorporation of cDNA into the host genome. From the foregoing it is clear that retroviruses, retrotransposons, and telomerase all played a role in this genomic sculpting. Telomerase and TP retrotransposons can use their RNA templates to synthesize novel DNA at the ends of chromosomal breaks and thus are capable of maintaining and molding the genome. The phylogenetic and functional relatedness of these retroelements contrasts with adaptations of structure and mechanism that are tailored to specific needs. Details of evolutionary relatedness of these diverse retroelements, contrasted with their distinctiveness, and their role in determining cell fate are described above and in the following articles.
In the journal Nature's News and Views article written 4 decades ago, “Central Dogma Reversed” (63), the prescient editor predicted that the discoveries of Baltimore and Temin “are likely to generate one of the largest bandwagons molecular biology has seen.” The editor went on to ask, “Do uninfected eukaryotic cells or bacteria contain similar RNA dependent DNA polymerases?” to those in virions. The meeting, celebrating the 40th anniversary of the discovery of RT, fittingly keynoted by Dr. David Baltimore, answered this question. Similarly, this compendium of articles describes the magnitude, complexity, and beauty of the bandwagon.
Acknowledgments
We thank Maryellen Carl and Rebecca McCarthy for their help with the manuscript, and Matt Stanger for rendering the figure. Work in the authors’ laboratories was supported by National Institutes of Health Grants GM39422 and GM4484 (to M.B.), GM52072 (to M.J.C.), and GM62631 (to N.F.L.).
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Telomerase and Retrotransposons: Reverse Transcriptases that Shaped Genomes” held September 29–30, 2010, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at www.nasonline.org/telomerase_and_retrotransposons.
References
- 1.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 2.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 3.Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu Rev Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 5.Bushman F. Gene Transfer by Retroviruses. Lateral DNA Transfer: Mechanisms and Consequences. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2002. pp. 169–212. [Google Scholar]
- 6.Pardue ML, Danilevskaya ON, Traverse KL, Lowenhaupt K. Evolutionary links between telomeres and transposable elements. Genetica. 1997;100:73–84. [PubMed] [Google Scholar]
- 7.Eickbush TH. Telomerase and retrotransposons: Which came first? Science. 1997;277:911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura TM, Cech TR. Reversing time: Origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- 9.Gladyshev EA, Arkhipova IR. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci USA. 2007;104:9352–9357. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrish TA, et al. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 11.Curcio MJ, Belfort M. The beginning of the end: Links between ancient retroelements and modern telomerases. Proc Natl Acad Sci USA. 2007;104:9107–9108. doi: 10.1073/pnas.0703224104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gladyshev EA, Arkhipova IR. A widespread class of reverse transcriptase-related cellular genes. Proc Natl Acad Sci USA. 2011;108:20311–20316. doi: 10.1073/pnas.1100266108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pardue M-L, DeBaryshe PG. Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci USA. 2011;108:20317–20324. doi: 10.1073/pnas.1100278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 15.Cifuentes-Rojas C, Kannan K, Tseng L, Shippen DE. Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proc Natl Acad Sci USA. 2011;108:73–78. doi: 10.1073/pnas.1013021107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Lue NF. Plasticity of telomere maintenance mechanisms in yeast. Trends Biochem Sci. 2010;35:8–17. doi: 10.1016/j.tibs.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambowitz AM, Zimmerly S. Group II introns: Mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol. 2011;3:a003616. doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chalamcharla VR, Curcio MJ, Belfort M. Nuclear expression of a group II intron is consistent with spliceosomal intron ancestry. Genes Dev. 2010;24:827–836. doi: 10.1101/gad.1905010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q, Kim N-K, Peterson RD, Wang Z, Feigon J. Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Proc Natl Acad Sci USA. 2010;107:18761–18768. doi: 10.1073/pnas.1013269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q, Kim N-K, Feigon J. Architecture of human telomerase RNA. Proc Natl Acad Sci USA. 2011;108:20325–20332. doi: 10.1073/pnas.1100279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackburn EH, Collins K. Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol. 2011;3:a003558. doi: 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bley CJ, et al. RNA–protein binding interface in the telomerase ribonucleoprotein. Proc Natl Acad Sci USA. 2011;108:20333–20338. doi: 10.1073/pnas.1100270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mihalusova M, Wu JY, Zhuang X. Functional importance of telomerase pseudoknot revealed by single-molecule analysis. Proc Natl Acad Sci USA. 2011;108:20339–20344. doi: 10.1073/pnas.1017686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang T, et al. The group II intron ribonucleoprotein precursor is a large, loosely packed structure. Nucleic Acids Res. 2011;39:2845–2854. doi: 10.1093/nar/gkq1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robart AR, Collins K. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol Cell. 2011;42:308–318. doi: 10.1016/j.molcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi X, et al. RNA/DNA hybrid binding affinity determines telomerase template-translocation efficiency. EMBO J, 2011. [DOI] [PMC free article] [PubMed]
- 28.Larsen LS, et al. Ty3 nucleocapsid controls localization of particle assembly. J Virol. 2008;82:2501–2514. doi: 10.1128/JVI.01814-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S, Harada BT, Miller JT, Le Grice SF, Zhuang X. Initiation complex dynamics direct the transitions between distinct phases of early HIV reverse transcription. Nat Struct Mol Biol. 2010;17:1453–1460. doi: 10.1038/nsmb.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eickbush DG, Eickbush TH. R2 retrotransposons encode a self-cleaving ribozyme for processing from an rRNA cotranscript. Mol Cell Biol. 2010;30:3142–3150. doi: 10.1128/MCB.00300-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christensen SM, Ye J, Eickbush TH. RNA from the 5′ end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc Natl Acad Sci USA. 2006;103:17602–17607. doi: 10.1073/pnas.0605476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosoy D, Peng Y, Mian IS, Lue NF. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J Biol Chem. 2003;278:3882–3890. doi: 10.1074/jbc.M210645200. [DOI] [PubMed] [Google Scholar]
- 33.Kopera HC, Moldovan JB, Morrish TA, Garcia-Perez JL, Moran JV. Similarities between long interspersed element-1 (LINE-1) reverse transcriptase and telomerase. Proc Natl Acad Sci USA. 2011;108:20345–20350. doi: 10.1073/pnas.1100275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 35.Baller JA, Gao J, Voytas DF. Access to DNA establishes a secondary target site bias for the yeast retrotransposon Ty5. Proc Natl Acad Sci USA. 2011;108:20351–20356. doi: 10.1073/pnas.1103665108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciuffi A, et al. Methods for integration site distribution analyses in animal cell genomes. Methods. 2009;47:261–268. doi: 10.1016/j.ymeth.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferris AL, et al. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc Natl Acad Sci USA. 2010;107:3135–3140. doi: 10.1073/pnas.0914142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min B, Collins K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell. 2009;36:609–619. doi: 10.1016/j.molcel.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min B, Collins K. Multiple mechanisms for elongation processivity within the reconstituted tetrahymena telomerase holoenzyme. J Biol Chem. 2010;285:16434–16443. doi: 10.1074/jbc.M110.119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng Z, et al. Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc Natl Acad Sci USA. 2011;108:20357–20361. doi: 10.1073/pnas.1113624108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bianchi A, Shore D. How telomerase reaches its end: Mechanism of telomerase regulation by the telomeric complex. Mol Cell. 2008;31:153–165. doi: 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 42.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 43.Wu Y, Zakian VA. The telomeric Cdc13 protein interacts directly with the telomerase subunit Est1 to bring it to telomeric DNA ends in vitro. Proc Natl Acad Sci USA. 2011;108:20362–20369. doi: 10.1073/pnas.1100281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu M, Yu EY, Singh SM, Lue NF. Mutual dependence of Candida albicans Est1p and Est3p in telomerase assembly and activation. Eukaryot Cell. 2007;6:1330–1338. doi: 10.1128/EC.00069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osterhage JL, Talley JM, Friedman KL. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat Struct Mol Biol. 2006;13:720–728. doi: 10.1038/nsmb1125. [DOI] [PubMed] [Google Scholar]
- 46.Tuzon CT, Wu Y, Chan A, Zakian VA. The Saccharomyces cerevisiae telomerase subunit Est3 binds telomeres in a cell cycle- and Est1-dependent manner and interacts directly with Est1 in vitro. PLoS Genet. 2011;7:e1002060. doi: 10.1371/journal.pgen.1002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J, Mandell EK, Tucey TM, Morris DK, Lundblad V. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat Struct Mol Biol. 2008;15:990–997. doi: 10.1038/nsmb.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu EY, Wang F, Lei M, Lue NF. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol. 2008;15:985–989. doi: 10.1038/nsmb.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J, Mandell EK, Rao T, Wuttke DS, Lundblad V. Investigating the role of the Est3 protein in yeast telomere replication. Nucleic Acids Res. 2010;38:2279–2290. doi: 10.1093/nar/gkp1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yen W-F, Chico L, Lei M, Lue NF. Telomerase regulatory subunit Est3 in two Candida species physically interacts with the TEN domain of TERT and telomeric DNA. Proc Natl Acad Sci USA. 2011;108:20370–20375. doi: 10.1073/pnas.1017855108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 52.Maxwell PH, Burhans WC, Curcio MJ. Retrotransposition is associated with genome instability during chronological aging. Proc Natl Acad Sci USA. 2011;108:20376–20381. doi: 10.1073/pnas.1100271108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 54.Muotri AR, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coufal NG, et al. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc Natl Acad Sci USA. 2011;108:20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walne AJ, Dokal I. Dyskeratosis Congenita: A historical perspective. Mech Ageing Dev. 2008;129:48–59. doi: 10.1016/j.mad.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 57.Armanios M. Syndromes of telomere shortening. Annu Rev Genomics Hum Genet. 2009;10:45–61. doi: 10.1146/annurev-genom-082908-150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Venteicher AS, et al. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhong F, et al. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev. 2011;25:11–16. doi: 10.1101/gad.2006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blasco MA. Telomeres and human disease: Ageing, cancer and beyond. Nat Rev Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 61.Okamoto N, et al. Maintenance of tumor initiating cells of defined genetic composition by nucleostemin. Proc Natl Acad Sci USA. 2011;108:20388–20393. doi: 10.1073/pnas.1015171108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Friedman KL. Telomerase reverse transcriptase and Wnt signaling. Mol Cell Biol. 2011;31:2366–2368. doi: 10.1128/MCB.05462-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.News and Views. Central dogma reversed. Nature. 1970;226:1198–1199. doi: 10.1038/2261198a0. [DOI] [PubMed] [Google Scholar]