Abstract
Accumulation of sterols in membranes of the endoplasmic reticulum (ER) leads to the accelerated ubiquitination and proteasomal degradation of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a rate-limiting enzyme in synthesis of cholesterol and nonsterol isoprenoids. This degradation results from sterol-induced binding of reductase to the Insig-1 or Insig-2 proteins of ER membranes. We previously reported that in immortalized human fibroblasts (SV-589 cells) Insig-1, but not Insig-2, recruits gp78, a membrane-bound RING-finger ubiquitin ligase. We now report that both Insig-1 and Insig-2 bind another membrane-bound RING-finger ubiquitin ligase called Trc8. Knockdown of either gp78 or Trc8 in SV-589 cells through RNA interference (RNAi) inhibited sterol-induced ubiquitination of reductase and inhibited sterol-induced degradation by 50–60%. The combined knockdown of gp78 and Trc8 produced a more complete inhibition of degradation (> 90%). Knockdown of gp78 led to a three to fourfold increase in levels of Trc8 and Insig-1 proteins, which opposed the inhibitory action of gp78. In contrast, knockdown of Trc8 had no effect on gp78 or Insig-1. The current results suggest that sterol-induced ubiquitination and proteasomal degradation of reductase is dictated by the complex interplay of at least four proteins: Insig-1, Insig-2, gp78, and Trc8. Variations in the concentrations of any one of these proteins may account for differences in cell- and/or tissue-specific regulation of reductase degradation.
Keywords: cholesterol metabolism, ER-associated degradation
Sterol-induced ubiquitination and proteasomal degradation of the endoplasmic reticulum (ER)-localized enzyme 3-hydroxy-3-methylglutaryl coenzyme A (HMG CoA) reductase is one of several mechanisms through which mammalian cells modulate synthesis of cholesterol and nonsterol isoprenoids (1, 2). Ubiquitination results from sterol-induced binding of reductase to one of two ER membrane proteins called Insig-1 and Insig-2 (3, 4). Insig binding is mediated entirely by the NH2-terminal membrane domain of reductase, a region that consists of eight membrane-spanning segments and precedes a large, COOH-terminal catalytic domain that projects into the cytosol (5, 6). Insig-1 binds to a membrane-bound, RING (really interesting new gene)-finger ubiquitin ligase called gp78 (7, 8), whereas Insig-2 does not detectably bind to gp78 (9). In the presence of sterols, gp78 associates with reductase through a mechanism that requires the presence of Insig-1 (10). These observations indicate that when cellular sterol levels rise, Insig-1 brings gp78 to reductase for subsequent ubiquitination and extraction from membranes for proteasomal degradation.
RNA interference (RNAi)-mediated knockdown of gp78 in SV-589 cells [a line of immortalized human fibroblasts (11)] markedly inhibited ubiquitination of reductase, but decreased degradation of the enzyme by only 50–60% (10). This partial effect, coupled with the inefficiency of gp78-Insig-2 binding, led us to consider the possible existence of another ubiquitin ligase that contributes to reductase degradation. The membrane-bound, RING-finger ubiquitin ligase Trc8 (12, 13) binds to both Insigs when the proteins are overexpressed together in cells (14). In the current studies, we confirm these findings and examine a role for Trc8 in reductase ubiquitination and degradation. Knockdown of Trc8 through RNAi blocks sterol-induced ubiquitination of reductase and partially inhibits its sterol-induced degradation. When gp78 and Trc8 are both knocked down by RNAi, sterol-mediated reductase degradation is inhibited by 90%. Knockdown of gp78 inhibits the degradation of Trc8 as well as that of Insig-1, allowing these proteins to accumulate. This finding may explain the partial effect of gp78 knockdown on reductase degradation. Our current data suggest that gp78 and Trc8 combine to mediate sterol-accelerated degradation of reductase. The complex interplay of these two ubiquitin ligases, together with the two Insigs have important implications for variations in cell and/or tissue-specific regulation of reductase activity.
Results
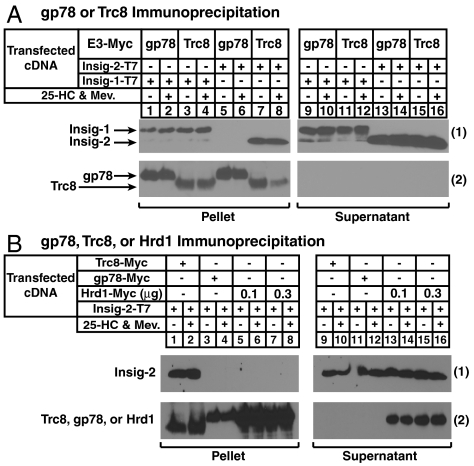
The previous finding that Trc8 binds to both Insig-1 and Insig-2 (14) prompted us to design an experiment that compares the association of Insigs with two membrane-bound ubiquitin ligases, gp78 and Trc8. CHO-7 cells were transfected with Insig-1 or Insig-2 containing COOH-terminal T7 epitopes together with Trc8 or gp78 COOH-terminally tagged with Myc epitopes. The cells were depleted of sterols and subsequently treated in the absence or presence of the oxysterol 25-hydroxycholesterol (25-HC) plus mevalonate to provide nonsterol isoprenoids that augment reductase degradation (4). Detergent lysates were immunoprecipitated with anti-Myc-coupled agarose beads to pull-down transfected gp78 or Trc8 and coprecipitated Insig-1 or Insig-2 was analyzed by immunoblot. Insig-1, but not Insig-2, coimmunoprecipitated with gp78 (Fig. 1 A, 1, lanes 1, 2, 5, and 6), a result consistent with previous findings (9). In contrast, Trc8 coprecipitated Insig-1 and Insig-2 with roughly equal efficiency (Fig. 1 A, 1, lanes 3, 4, 7, and 8). Comparative analysis of wild type and various truncation or deletion mutants of Trc8 revealed that the membrane domain of the enzyme mediates its association with both Insigs (Fig. S1).
Fig. 1.
Association of Insig-1 and Insig-2 with membrane-bound ubiquitin ligases. (A) CHO-7 cells were set up on day 0 at 5 × 105 cells per 60-mm dish in medium A containing 5% LPDS. On day 1, cells were transfected in 5% LPDS with 50 ng/dish of pCMV-Insig-1-T7, 500 ng/dish of pCMV-Insig-2-T7, 3 ng/dish of pCMV-gp78-Myc, and 30 ng/dish of pCMV-Trc8-Myc as indicated and described in Materials and Methods. The total amount of DNA/dish was adjusted to 3 μg by the addition of empty pcDNA3 vector. Following incubation for 6 h at 37 °C, cells were depleted of sterols by direct addition of medium A containing 5% LPDS, 10 μM sodium compactin, and 50 μM sodium mevalonate (final concentration). After 16 h at 37 °C, cells were refed identical medium containing 10 μM MG-132 in the absence or presence of 1 μg/mL 25-HC plus 10 mM mevalonate and incubated for an additional 5 h. Cells were subsequently harvested for preparation of detergent lysates that were immunoprecipitated with anti-Myc-coupled agarose beads. Immunoprecipitation pellet and supernatant fractions were subjected to SDS-PAGE and immunoblot analysis with anti-T7 IgG (against Insig-1 and Insig-2) or IgG-9E10 (against gp78 and Trc8). (B) CHO-7 cells were set up on day 0, transfected with pCMV-Insig-2-T7 (150 ng), pCMV-Trc8-Myc (100 ng), pCMV-gp78-Myc (10 ng), and pCMV-Hrd1-Myc (0.3 and 1 μg) as indicated, depleted of sterols on day 1, and treated with 25-HC plus mevalonate on day 2 as described in (A). Following treatments, cells were harvested for immunoprecipitation with anti-Myc-coupled agarose beads. Resulting pellet and supernatant fractions were immunoblotted with anti-T7 IgG (against Insig-2) or IgG-9E10 (against Trc8, gp78, and Hrd1).
The amino acid sequence of the membrane domain of human gp78, which mediates its binding to Insig-1 (10), bears significant homology to that of another membrane-bound ubiquitin ligase called Hrd1. Despite this homology, Insig-1 does not bind to Hrd1 (10). Fig. 1B shows that Insig-2 associated with Trc8 (1, lanes 1 and 2), but it failed to associate detectably with either gp78 (lanes 3 and 4) or with Hrd1 (lanes 5–8).
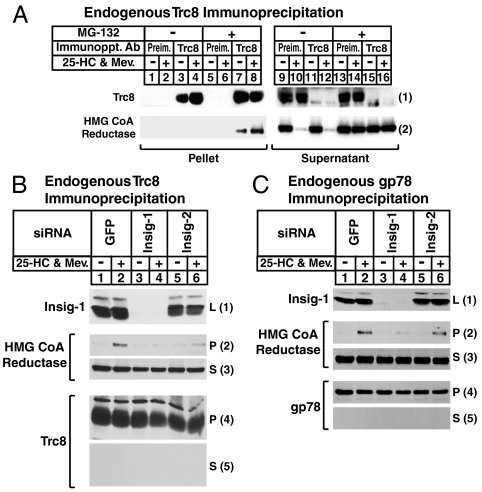
We next examined the association of endogenous Trc8 with reductase. Sterol-depleted SV-589 cells were treated in the absence or presence of 25-HC plus mevalonate and the proteasome inhibitor MG-132 prior to lysis and immunoprecipitation with polyclonal anti-Trc8 or control, preimmune IgG. Immunoblot analysis revealed specific pull-down of endogenous Trc8 (Fig. 2 A, 1, compare lanes 1, 2, 5, and 6 with lanes 3, 4, 7, and 8). In the absence of MG-132, 25-HC plus mevalonate caused the disappearance of reductase as expected (Fig. 2 A, 2, lanes 10 and 12), and this was blocked by MG-132 (lanes 14 and 16). In lysates from MG-132-treated cells, reductase appeared in the pellet fraction of the anti-Trc8 immunoprecipitation (Fig. 2 A, 2, lane 7), which was enhanced by 25-HC plus mevalonate (lane 8).
Fig. 2.
Sterol-induced, Insig-mediated binding of Trc8 to HMG CoA reductase. (A) SV-589 cells were set up on day 0 at 2 × 105 cells per 100-mm dish in medium B supplemented with 10% FCS. On day 3, cells were depleted of sterols through incubation for 16 h at 37 °C in medium B containing 10% LPDS, 50 μM compactin, and 50 μM mevalonate. The cells were then switched to identical medium containing 10 μM MG-132 for 2 h, after which they received 1 μg/mL 25-HC plus 10 mM mevalonate as indicated and incubated for an additional 2 h at 37 °C. Cells were then harvested and immunoprecipitated with IgG-556 (against Trc8); resulting pellet and supernatant fractions were subjected to immunoblot analysis with IgG-3D10 (against Trc8) and IgG-A9 (against reductase). (B and C) SV-589 cells were set up on day 0 as described in (A). On days 1 and 2, the cells were transfected in medium B containing 10% FCS with the indicated siRNA as described in Materials and Methods. After the second transfection on day 2, cells were depleted of sterols as described in (A) and then switched to identical medium containing 10 μM MG-132 in the absence or presence of 1 μg/mL 25-HC plus 10 mM mevalonate. Following incubation for 2 h at 37 °C, cells were harvested and lysates subjected to immunoprecipitation with either IgG-556 [against Trc8, (B)] or IgG-740F [against gp78, (C)]. Resulting pellet (P) and supernatant (S) fractions as well as total lysates (L) were subjected to immunoblot analysis with IgG-3D10 (against Trc8), IgG-A9 (against reductase), and IgG-740F, and IgG-17H1 (against Insig-1). The two bands for Insig-1 result from alternative use of initiating methionines (21). The larger band results from translation initiation at residue 1, whereas the smaller band results from initiation at residue 37.
To determine the requirement of each Insig for sterol-induced binding of Trc8 to reductase, we conducted the RNAi experiment shown in Fig. 2B. SV-589 cells were transfected with duplexes of small interfering RNAs (siRNAs) targeting mRNAs encoding either green fluorescent protein (GFP), which is not expressed in the cells, Insig-1, or Insig-2. The cells were then treated with MG-132 in the absence or presence of 25-HC plus mevalonate, after which they were lysed and immunoprecipitated with anti-Trc8. The results show that Insig-1 protein levels were appropriately reduced by transfection of its corresponding siRNA (Fig. 2 B, 1, lanes 3 and 4); we could not measure Insig-2 protein levels inasmuch as antibodies capable of detecting the endogenous protein are not currently available. However, a 50–70% reduction in the amount of Insig-2 mRNA is routinely achieved through RNAi as determined by quantitative real-time PCR (4). In cells transfected with GFP siRNA, reductase coprecipitated with Trc8 in a 25-HC plus mevalonate-dependent fashion (Fig. 2 B, 2, lane 2). This coprecipitation was markedly reduced in cells that received siRNAs against Insig-1 (Fig. 2 B, 2, lane 4) and to a lesser extent in cells transfected with the Insig-2 siRNA (lane 6). In the experiment of Fig. 2C, gp78 immunoprecipitations were conducted in control, Insig-1, or Insig-2 knockdown cells. Sterol-induced binding of reductase to gp78 was abrogated by Insig-1 knockdown (Fig. 2 C, 2, compare lanes 2 and 4), but not by Insig-2 knockdown (lane 6).
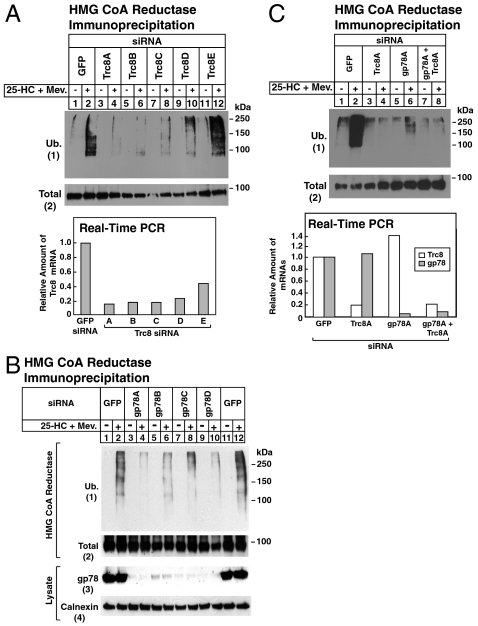
The contributions of Trc8 and gp78 to the sterol-induced ubiquitination of endogenous reductase were next examined in Fig. 3. In Fig. 3A, SV-589 cells were transfected with the GFP siRNA or one of five siRNAs targeting different regions of the Trc8 mRNA. Following sterol depletion, cells were treated with MG-132 in the absence or presence of 25-HC plus mevalonate. Detergent lysates were then subjected to antireductase immunoprecipitation, followed by immunoblot analysis with antiubiquitin. In cells transfected with control GFP siRNA, 25-HC plus mevalonate stimulated reductase ubiquitination (Fig. 3 A, 1, lane 2). Sterol-induced ubiquitination of reductase was significantly reduced in cells transfected with four of the five siRNAs targeting Trc8 (Fig. 3 A, 1, lanes 4, 6, 8, and 10). The magnitude of this inhibition correlated with the efficiency of Trc8 knockdown, which was determined by quantitative real-time PCR of Trc8 mRNA. Similarly, knockdown of gp78 using four different siRNAs inhibited ubiquitination of reductase (Fig. 3 B, 1 compare lanes 2 and 12 with lanes 4, 6, 8, and 10). Fig. 3C shows that individual knockdown of Trc8 or gp78 inhibited ubiquitination of reductase to a similar extent as the simultaneous knockdown of both ligases (1, lanes 2, 4, 6, and 8).
Fig. 3.
Inhibition of sterol-induced ubiquitination of HMG CoA reductase by RNA interference-mediated knockdown of Trc8 and gp78. SV-589 cells were set up on day 0, transfected with the indicated siRNAs on days 1 and 2, and depleted of sterols as described in Fig. 2A. Sterol-depleted cells were subsequently incubated for 30 min in medium B containing 10% LPDS, 50 μM compactin, and 10 μM MG-132 in the absence or presence of 1 μg/mL 25-HC plus 10 mM mevalonate as indicated. Cells were harvested and immunoprecipitated with polyclonal anti-reductase, followed by immunoblot analysis with IgG-A9 (against reductase) and IgG-P4D1 (against ubiquitin). Total RNA from 25-HC plus mevalonate-treated cells in (A) and (C) was subjected to first-strand cDNA synthesis and real-time PCR. Each value for cells transfected with the indicated siRNA represents the amount of the indicated mRNA relative to that in control cells transfected with the GFP siRNA.
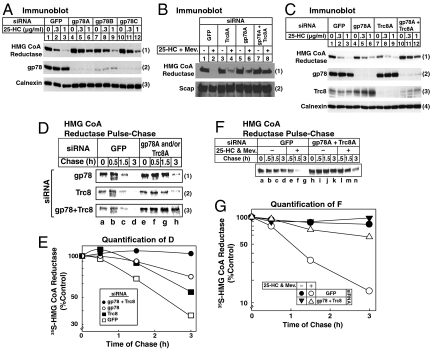
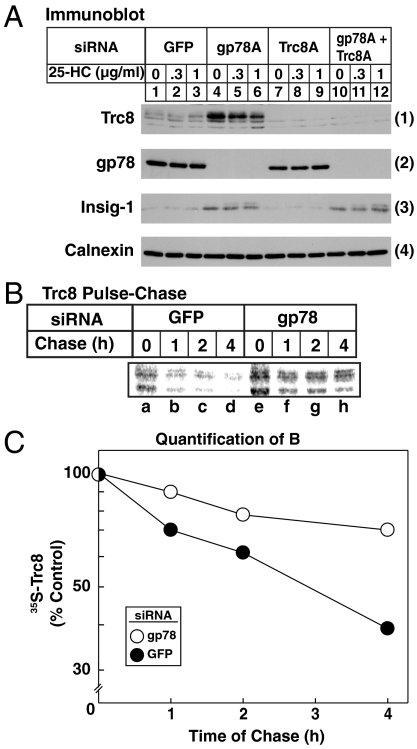
RNAi experiments were next conducted in the absence of MG-132 to explore the roles of gp78 and Trc8 in sterol-accelerated degradation of reductase (Fig. 4). Consistent with our previous observations (10), knockdown of gp78 with three different siRNAs partially inhibited, but did not abolish, reductase degradation (Fig. 4 A, 1, compare lanes 1–3 with lanes 4–12). Knockdown of Trc8 also partially inhibited reductase degradation (Fig. 4 B, 1, compare lanes 4 and 2). In striking contrast, the combined knockdown of gp78 and Trc8 abolished reductase degradation almost completely (Fig. 4 B, 1, lane 8). In the experiment of Fig. 4C, the inhibitory effect of individual gp78 and Trc8 knockdowns was even less pronounced (1, lanes 4–9), yet the combined knockdown of both ligases abolished reductase degradation (lanes 10–12). Immunoblotting revealed the appropriate reduction in gp78 and Trc8 when cells were transfected with the corresponding siRNA (Fig. 4 C, 2 and 3, lanes 4–12), whereas levels of the loading controls Scap and calnexin remained unchanged (Fig. 4 B and C, 2 and 4, respectively).
Fig. 4.
Trc8 and gp78 are required for sterol-accelerated degradation of HMG CoA reductase as revealed by RNA interference. SV-589 cells were set up for experiments on day 0, transfected with the indicated siRNAs on days 1 and 2 (A, C, D, and F) or on days 1 and 3 (B), and depleted of sterols as described in Fig. 2A. (A–C) Sterol-depleted cells were incubated for 2 h in medium B supplemented with 10% LPDS and 50 μM compactin in the absence or presence of the indicated concentration of 25-HC in (A and C) or 0.5 μg/mL 25-HC plus 10 mM mevalonate in (B). Following incubation for 2–3 h at 37 °C, cells were harvested for subcellular fractionation. Aliquots of the membrane fractions (normalized for equal protein loaded/lane) were subjected to SDS-PAGE and immunoblot analysis with IgG-A9 (against reductase), IgG-R139 (against Scap), IgG-740F (against gp78), IgG-556 (against Trc8), and anti-calnexin IgG. (D and F) Sterol-depleted cells were preincubated with methionine/cysteine-free medium B containing 10% LPDS and 50 μM compactin for 1 h at 37 °C. After pulse-labeling for 30 min at 37 °C in identical medium containing 130 μCi/mL of [35S] methionine, cells were chased in medium B supplemented with 10% LPDS, 50 μM compactin, 0.5 mM unlabeled methionine, and 1 mM cysteine in the absence or presence of 1 μg/mL 25-HC plus 10 mM mevalonate as indicated. Following the indicated time of chase, cells were harvested and subjected to anti-reductase immunoprecipitation, followed by SDS-PAGE and transfer of proteins to a nitrocellulose filter. The filter was exposed to an imaging plate at room temperature, scanned in a Storm 820 PhosphorImager, and the image was photographed. (E and G) [35S] Radioactivity in the scanned gels from Fig. 4 D and F corresponding to reductase was quantified by densitometry. The intensity of reductase during the pulse [lanes a and e in (D); lanes a and h in (F)] was arbitrarily set at 100%.
We next conducted a pulse-chase experiment to directly demonstrate the effect of gp78 and Trc8 knockdown on sterol-accelerated reductase degradation. Sterol-depleted cells transfected with siRNAs were pulse-labeled for 30 min with 35S-labeled methionine plus cysteine, after which they were washed and switched to medium containing 25-HC plus mevalonate and an excess of unlabeled methionine and cysteine. Radiolabeled reductase was monitored by antireductase immunoprecipitation, followed by SDS-PAGE and visualization in a phosphorimager. In control cells transfected with GFP siRNA, 25-HC plus mevalonate caused the rapid disappearance of 35S-labeled reductase that was almost complete after 3 h (Fig. 4 D, 1–3, lanes a–d). This disappearance was significantly blunted in cells transfected with either gp78 or Trc8 siRNA (Fig. 4 D, 1 and 2, respectively, lanes e–h). In cells transfected with both gp78 and Trc8 siRNA, degradation was markedly slowed and labeled reductase persisted throughout the chase (Fig. 4 D, 3, lanes e–h). These results were quantified by scanning of images in a phosphorimager and the data are plotted in Fig. 4E. Fig. 4 F and G show that the rapid degradation of reductase was dependent on the presence of 25-HC plus mevalonate and again, it was abolished by the simultaneous knockdown of gp78 and Trc8.
A notable observation in gp78 knockdown cells was the significant increase in the amount of Trc8 (Fig. 4 C, 3, lanes 4–6 and 10–12), suggesting that degradation of Trc8 might be mediated by gp78. In the experiment of Fig. 5A, cells were subjected to individual or combined knockdown of gp78 and Trc8 and subsequently analyzed by immunoblot following treatment with cycloheximide in the absence or presence of 25-HC plus mevaloante. In control-transfected cells, a low level of Trc8 was observed (Fig. 5 A, 1, lanes 1–3), and knockdown of gp78 led to a significant increase in the protein (lanes 4–6). Insig-1 was also increased after knockdown of gp78, but not Trc8 (Fig. 5 A, 3, compare lanes 1–3 with lanes 4–6 and 7–9), a result consistent with the previously established role of gp78 in Insig-1 degradation (9). To confirm a role for gp78 in degradation of Trc8, a pulse-chase experiment was conducted. The results show that Trc8 was rapidly degraded in control cells transfected with GFP siRNA (Fig. 5 B, lanes a–d; C). Degradation of Trc8 was markedly slowed by RNAi-mediated knockdown of gp78 (Fig. 5B, lanes e–h; C)
Fig. 5.
Stabilization of Trc8 by RNA interference-mediated knockdown of gp78. SV-589 cells were set up for experiments on day 0, transfected with the indicated siRNAs on days 1 and 2 as described in Fig. 3. (A) Following sterol depletion, cells were incubated in medium B supplemented with 10% LPDS, 50 μM compactin, and 50 μM cycloheximide. After 1 h at 37 °C, cells received the indicated concentration of 25-HC and incubated an additional 2 h. Cells were subsequently harvested for subcellular fractionation; resulting membrane fractions were subjected to immunoblot analysis with IgG-556 (against Trc8), IgG-740F (against gp78), IgG-17H1 (against Insig-1), and anti-calnexin IgG. Results are representative of at least two independent experiments. (B) Cells were pulse-labeled for 30 min in medium B containing 10% FCS and 130 μCi/mL [35S] methionine and subsequently chased in medium B supplemented with 10% FCS, 0.5 mM unlabeled methionine, and 1 mM cysteine. Following the indicated time of chase, cells were harvested, subjected to anti-Trc8 immunoprecipitation, and analyzed as described in Fig. 4. (C) [35S] Radioactivity in the scanned gel from Fig. 5B corresponding to the two bands of Trc8 was quantified by densitometry. The nature of these two bands is currently unknown. The intensity of reductase during the pulse (Fig. 5B, lanes a and e) was arbitrarily set at 100%.
Discussion
Our previous studies established that sterol-dependent binding of Insigs to HMG CoA reductase results in recruitment of a membrane-associated ubiquitin ligase that initiates ubiquitination of reductase, marking it for proteasomal degradation (3, 15). We subsequently discovered that gp78, a RING-finger ubiquitin ligase with multiple membrane-spanning segments (7, 8), efficiently binds to Insig-1, but not to Insig-2 (3, 9). Further examination provided several lines of evidence indicating a role for gp78 in sterol-induced ubiquitination and degradation of reductase: (i) overexpression of a dominant-negative version of gp78 consisting of the truncated membrane domain, which binds to Insig-1 but cannot ubiquitinate, blocked sterol-accelerated degradation of reductase; (ii) gp78 formed a complex with reductase in an Insig-1-dependent, sterol-regulated fashion; and (iii) RNAi-mediated knockdown of gp78 prevented reductase ubiquitination and inhibited the enzyme’s degradation by 50–60% (10). It is important to note that gp78 also mediates the ubiquitination and degradation of Insig-1 and accordingly, knockdown of gp78 leads to a marked increase in Insig-1 protein (9). Reductase degradation is known to be strictly dependent on Insig/reductase ratios (16); increased levels of Insig-1 render reductase more sensitive to sterol-accelerated degradation in normal cells. Thus, the increased Insig-1 resulting from gp78 knockdown could account for the partial effect of the treatment on reductase degradation.
In addition to increased levels of Insig-1, compensation by another ubiquitin ligase could contribute to continued degradation of reductase in gp78 knockdown cells. This scenario is especially intriguing considering that while Insig-2 is absolutely required for reductase degradation (17), the protein does not efficiently bind gp78 (9). Such a putative ubiquitin ligase would be predicted to efficiently bind to Insig-2 and work together with gp78 to mediate sterol-accelerated reductase degradation. In the current studies, we identify membrane-associated Trc8 as a candidate for this ubiquitin ligase. Coimmunoprecipitation experiments show that unlike gp78, Trc8 binds to both Insig-1 and Insig-2 with similar efficiency when the proteins are overexpressed together in cells (Fig. 1A). Immunoprecipitation of endogenous Trc8 (Fig. 2 A and B) or gp78 (Fig. 2C) brought down endogenous reductase in a sterol-regulated fashion. Consistent with overexpression studies, RNAi experiments reveal that reductase-Trc8 binding is mediated by both Insig-1 and Insig-2 (Fig. 2B), whereas reductase-gp78 binding is mediated primarily by Insig-1 (Fig. 2C).
The current studies revealed an apparent inconsistency between the results of experiments in which reductase ubiquitination was measured (Fig. 3) and experiments in which reductase degradation was measured (Fig. 4). Knockdown of either gp78 or Trc8 appeared to block ubiquitination nearly completely, yet the individual knockdowns had only partial effects on reductase degradation. Indeed, complete inhibition of reductase degradation required simultaneous knockdown of both ligases. We attribute this apparent discrepancy to the inefficiency of the assay for ubiquitinated reductase. Even with the addition of MG-132, only a very small amount of reductase is capable of being trapped in a ubiquitinated form at any one time. This inefficiency is revealed by immunoblots for reductase, which do not show the high molecular weight smear characteristic of ubiquitinated proteins (see total reductase blots in Fig. 3). Ubiquitination can only be detected when reductase is immunoprecipitated, subjected to SDS-PAGE, and blotted with antiubiquitin (as in Fig. 3). Thus, we suggest that knockdown of gp78 or Trc8 individually left a significant fraction of ubiquitinated reductase that was below the limit of detection by our assay.
An intriguing finding of our studies is the stabilization of Trc8 in gp78 knockdown cells (Figs. 4C and 5A), which was confirmed by pulse-chase analysis (Fig. 5B). The gp78-mediated degradation of Trc8 may provide a mechanism to allow Trc8 to rise when gp78 levels fall, thereby maintaining reductase degradation. Future studies examining the response to gp78 knockdown are required, especially considering that Insig-1 is also significantly stabilized by knockdown of gp78 (Fig. 5) and could work together with Trc8 to ubiquitinate reductase when gp78 is absent. Moreover, we cannot rule out the possibility that levels of Insig-2 protein are modulated by gp78 knockdown, inasmuch as antibodies capable of detecting endogenous Insig-2 are not currently available.
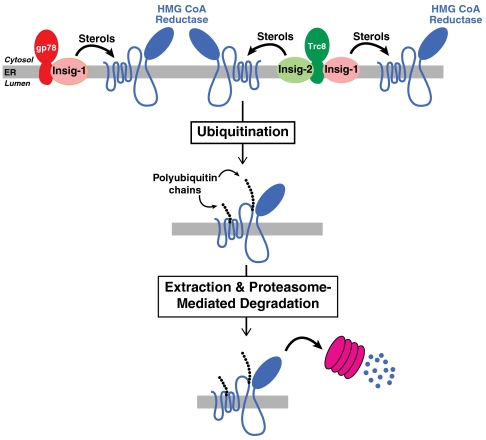
Based on the current results, we propose a working model for reductase degradation (Fig. 6). In this model, two membrane-bound RING-finger ubiquitin ligases, gp78 and Trc8, can mediate ubiquitination and degradation of reductase. The gp78-dependent ubiquitination of reductase is mediated primarily by Insig-1, whereas Trc8-dependent ubiquitination is mediated by both Insig-1 and Insig-2. Consequently, the ratios of Trc8/gp78 and of Insig-1/Insig-2 are predicted to be key determinants of reductase degradation, thereby creating a mechanism for cell type and/or tissue-specific regulation of the reaction.
Fig. 6.
Model for sterol-accelerated ubiquitination and degradation of HMG CoA reductase mediated by Trc8 and gp78. In sterol-treated cells, Insig-1 bridges gp78 to reductase and either Insig-1 or Insig-2 bridges Trc8 to reductase for ubiquitination. The resultant gp78- and Trc8-mediated ubiquitination marks reductase for extraction from ER membranes and subsequent delivery to proteasomes for degradation.
In SV-589 cells used in this study, quantitative real-time PCR revealed that gp78 expression (Ct = 23.2) exceeds that of Trc8 (Ct = 24.7) by ∼3-fold and Insig-1 expression (Ct = 19.6) exceeds that of Insig-2 (Ct = 25.3) by ∼50-fold. These observations provide a plausible explanation as to why knockdown of gp78 more potently inhibits reductase degradation than Trc8 knockdown in SV-589 cells (Fig. 4 B–D). An open question for investigation in future studies is whether Trc8 mediates ubiquitination and degradation of Insig-2. Insulin is known to strongly repress Insig-2 expression in the liver of animals, which can be partly attributed to down-regulation of the liver-specific Insig-2a transcript (18, 19). However, if Trc8 targets Insig-2 for ubiquitination, one cannot rule out the possibility that the reaction is enhanced by insulin and contributes to the reduction in Insig-2 that occurs in the presence of the hormone. Future studies examining the coordinated control of reductase ubiquitination by gp78 and Trc8, gp78-mediated control of Trc8 turnover, and the potential role for Trc8 in Insig-2 ubiquitination and degradation will provide key insights into molecular mechanisms governing the maintenance of cellular cholesterol homeostasis.
Materials and Methods
Cell Culture.
Chinese hamster ovary (CHO)-7 cells, CHO-K1 cells adapted for growth in lipoprotein-deficient serum (LPDS), were maintained in monolayer in medium A (1∶1 mixture of Ham’s F-12 medium and Dulbecco’s modified Eagle’s medium containing 100 units/mL penicillin and 100 mg/mL streptomycin sulfate) supplemented with 5% (vol/vol) LPDS at 37 °C, 8–9% CO2. SV-589 cells, immortalized human fibroblasts expressing the SV40 large T antigen (11), were grown in monolayer at 37 °C, 5% CO2 in medium B (Dulbecco’s modified Eagle’s medium containing 1000 mg glucose/L, 100 units/mL penicillin, and 100 mg/mL streptomycin sulfate) containing 10% fetal calf serum (FCS).
Transfection, Immunoprecipitation, Cell Fractionation, and Immunoblot Analysis.
Transfection of CHO-7 cells with FuGENE6 reagent (Roche Applied Science) was performed as described (3). Conditions of subsequent incubations are described in figure legends. Following incubations, triplicate dishes of cells for each variable were harvested and pooled for analysis. Immunoprecipitation with anti-T7-coupled agarose beads (Novagen), anti-Myc-coupled agarose beads (Sigma), or anti-Trc8 IgG plus Protein A/G agarose (Santa Cruz Biotechnology) and subcellular fractionation by differential centrifugation was performed as described previously (20). Aliquots of supernatant and pellet fractions from immunoprecipitations and 100,000 × g membrane pellets from subcellular fractionations were subjected to SDS-PAGE and immunoblot analysis. Primary antibodies used for immunoblot analysis are described in SI Materials and Methods.
Ubiquitination and pulse-chase analysis.
Conditions of incubations are described in figure legends. To examine reductase ubiquitination, triplicate dishes of cells were harvested following incubations, lysed, and subjected to immunoprecipitation with polyclonal antireductase and immunoblot analysis as described (4). To examine degradation of reductase or Trc8 by pulse-chase analysis, cells were pulse-labeled in methionine/cysteine-free medium B as described in the figure legends; pulse-chase analysis and subsequent immunoprecipitation with antireductase or anti-Trc8 was carried out as previously described (4). Immunoprecipitates were subjected to SDS-PAGE and transferred to nitrocellulose filters, which were exposed to an imaging plate at room temperature and scanned in a Storm 820 PhosphorImager (Amersham Biosciences).
Other methods.
Additional materials and methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Drs. Michael S. Brown and Joseph L. Goldstein for their encouragement and insightful advice. We also thank Tammy Dinh and Kristina Brasher for excellent technical assistance; and Lisa Beatty, Muleya Kapaale, Shomanike Head, and Ijeoma Onwuneme for help with tissue culture. This work is supported by a grant from the National Institutes of Health (Grant HL20948). R.A.D.-B. is a W.M. Keck Foundation Distinguished Young Investigator in Medical Research and an Early Career Scientist of the Howard Hughes Medical Institute. P.V.S. is supported by the Summer Undergraduate Research Fellowship program at the University of Texas Southwestern Medical Center.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112831108/-/DCSupplemental.
References
- 1.DeBose-Boyd RA. Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 2008;18:609–621. doi: 10.1038/cr.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown MS, Goldstein JL. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980;21:505–517. [PubMed] [Google Scholar]
- 3.Sever N, Yang T, Brown MS, Goldstein JL, DeBose-Boyd RA. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol Cell. 2003;11:25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 4.Sever N, et al. Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. J Biol Chem. 2003;278:52479–52490. doi: 10.1074/jbc.M310053200. [DOI] [PubMed] [Google Scholar]
- 5.Liscum L, et al. Domain structure of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a glycoprotein of the endoplasmic reticulum. J Biol Chem. 1985;260:522–530. [PubMed] [Google Scholar]
- 6.Roitelman J, Olender EH, Bar-Nun S, Dunn WA, Jr, Simoni RD. Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum. J Cell Biol. 1992;117:959–973. doi: 10.1083/jcb.117.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimizu K, et al. The autocrine motility factor receptor gene encodes a novel type of seven transmembrane protein. FEBS Lett. 1999;456:295–300. doi: 10.1016/s0014-5793(99)00966-7. [DOI] [PubMed] [Google Scholar]
- 8.Fang S, et al. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JN, Song B, DeBose-Boyd RA, Ye J. Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase, gp78. J Biol Chem. 2006;281:39308–39315. doi: 10.1074/jbc.M608999200. [DOI] [PubMed] [Google Scholar]
- 10.Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005;19:829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto T, et al. The human LDL receptor: a cysteine-rich protein with multiple Alu sequences in its mRNA. Cell. 1984;39:27–38. doi: 10.1016/0092-8674(84)90188-0. [DOI] [PubMed] [Google Scholar]
- 12.Gemmill RM, et al. The hereditary renal cell carcinoma 3;8 translocation fuses FHIT to a patched-related gene, TRC8. Proc Natl Acad Sci USA. 1998;95:9572–9577. doi: 10.1073/pnas.95.16.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorick KL, et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96(20):11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JP, et al. The TRC8 ubiquitin ligase is sterol regulated and interacts with lipid and protein biosynthetic pathways. Mol Cancer Res. 2010;8:93–106. doi: 10.1158/1541-7786.MCR-08-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song BL, DeBose-Boyd RA. Ubiquitination of 3-Hydroxy-3-methylglutaryl-CoA Reductase in Permeabilized Cells Mediated by Cytosolic E1 and a Putative Membrane-bound Ubiquitin Ligase. J Biol Chem. 2004;279:28798–28806. doi: 10.1074/jbc.M402442200. [DOI] [PubMed] [Google Scholar]
- 16.Lee PCW, Liu P, Li WP, DeBose-Boyd RA. Amplification of the gene for SCAP, coupled with Insig-1 deficiency, confers sterol resistance in mutant Chinese hamster ovary cells. J Lipid Res. 2007;48:1944–1954. doi: 10.1194/jlr.M700225-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Lee PC, Sever N, DeBose-Boyd RA. Isolation of sterol-resistant Chinese hamster ovary cells with genetic deficiencies in both Insig-1 and Insig-2. J Biol Chem. 2005;280:25242–25249. doi: 10.1074/jbc.M502989200. [DOI] [PubMed] [Google Scholar]
- 18.Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc Natl Acad Sci USA. 2003;100:3155–3160. doi: 10.1073/pnas.0130116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelking LJ, et al. Schoenheimer effect explained—feedback regulation of cholesterol synthesis in mice mediated by Insig proteins. J Clin Invest. 2005;115:2489–2498. doi: 10.1172/JCI25614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jo Y, Sguigna PV, Debose-Boyd RA. Membrane-associated ubiquitin ligase complex containing gp78 mediates sterol-accelerated degradation of 3-Hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 2011;286:15022–15031. doi: 10.1074/jbc.M110.211326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang T, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2003;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.