Abstract
Heparan sulfate (HS) is a sulfated glycosaminoglycan located on the surface and extracellular matrix of mammalian cells. HS is constituted of highly N-sulfated domains (NS domains) interrupted by lower sulfation domains. The arrangement of these domains dictates the function of HS which is mainly involved in binding proteins and regulating their biological activities. Heparin, a heparan sulfate analogue present in mast cells, resembles the NS domains of HS but lacks the alternating high and low sulfation architecture. Because the NS domains that range up to hexadecasaccharide in size are the main protein binders, heparin has been used as a model for HS in protein binding studies. Heparan sulfate, however, is the more physiologically relevant modulator of growth factor-receptor interactions. In this work, liquid chromatography-mass spectrometry was used to compare the compositions of affinity purified heparin and HS octasaccharides with anticoagulant activities versus library octasaccharides. The fine structures of the biologically active HS compositions were then compared against those of library octasaccharides using low energy collision induced dissociation tandem mass spectrometry. This approach confirmed isomeric enrichment of these compositions and, most importantly, produces ions diagnostic of their biological activity.
Heparin and heparan sulfate (HS) are sulfated glycosaminoglycans that are produced by the same biosynthetic steps. The extent of modifications differs markedly between the two polymers leading to their structural and functional differences. The biosynthetic modifications go nearly to completion for heparin that is expressed primarily in mast cell granules. Heparan sulfate consists primarily of highly N-sulfated heparin-like domains (NS domains) interrupted by N-acetylated low sulfation domains (NA) and mixed (NA/NS) domains. Hence, HS tends to be more chemically heterogeneous than heparin (1). The highly polydisperse nature of these compounds can be explained by the facts that only a subset of sites are modified, the inefficiency of the enzymatic reactions, the lack of proofreading activity of the modifying enzymes, the presence of tissue dependant-isozymes with different specificities and finally by the presence of post-biosynthetic chain modifying enzymes that include the 6-endosulfatases (2) and mammalian heparanase (3). In addition, other physiological conditions such as the availability of the sulfate donor substrate 3′-phosphoadenosine 5′-phosphosulfate (4) and oxidative reactions (5, 6) play an important role in defining the final modification state of HS.
Heparin and HS are involved in a multitude of physiological processes spanning embryonic development, adult physiological processes, cancer progression and pathogen invasion (7, 8). At the molecular level, they exert most of their biological functions through interaction with proteins at the cell surface and in the extracellular matrix including enzymes, enzyme inhibitors, growth factors/morphogens and cognate receptors (9). While the involvement of HS in numerous biological functions is well known, there are only few examples of sequence specific interactions where binding requirements have been partially elucidated. Previously, a pentasaccharide with a distinguishing central 3-O sulfation was defined as the minimal sequence required for high affinity binding to Antithrombin III (ATIII) (10–14). The well defined ATIII-heparin binding requirement have been more recently challenged by alternative views whereby the mere charge density plays a significant role in dictating the affinity of ATIII to heparin (15). Studies on growth factors binding to HS highlighted the importance of specific sulfation positions such as N- and 2-O sulfates for FGF2 (16–18) and 6-O-sulfates for FGF1 (19) but in contrast others have shown that interaction affinities correlate with the overall degree of sulfation (20). More recently, the requirement of the nonsulfated domains for protein binding is gaining great importance. The emerging binding paradigm entails that not only the high sulfation character of NS domains is required for binding but also the arrangement and spacing pattern dictated by intervening NA and hybrid NA/NS domains (21). This model is being confirmed by new examples of heparin-binding proteins such as the C terminal endostatin fragment of collagen XVIII that has been suggested to bind NS domains separated by one GlcNAc residue (22) and other multimeric cytokines, namely platelet growth factor-4 (23), interleukin-8 (24) and interferon-γ (25).
The physiological importance of heparin and HS (collectively called heparinoids) results in an imminent need to understand the structures of their protein binding domains. Mass spectrometric analysis of heparinoids capable of binding proteins has been the subject of great interest. Approaches to study heparinoid protein binders by mass spectrometry contain a first step of purifying binders prior to mass spectrometric analysis. Several methods to purify protein binders have been used including immobilized proteins on affinity columns, hydrophobic trapping (26), gel filtration (27) and filter trapping (28, 29). Alternative methods consist of spraying the protein-oligosaccharide complexes (30–32). As detection and characterization of heparinoid binding proteins has undergone considerable progress in the MS dimension, structural determination of these oligosaccharides by tandem mass spectrometry has lagged behind. To date, tandem mass spectrometry (MS2) of heparinoids has been limited to small oligosaccharides up to hexasaccharides (33–37). For larger oligosaccharides, there is a need to reduce the size before MS2 can be applied. For example, MS has been used to investigate binding of heparin and HS octasaccharides to chemokines (28, 30). However, the structural data of the heparin octasaccharide binder was obtained by the traditional approach of digestion into disaccharides followed by collision induced dissociation tandem mass spectrometry (CID MS2) of the disaccharide products (38, 39). The difficulties of subjecting large heparinoid oligosaccharides to direct tandem analysis can be explained by their heterogeneity and polydisperse nature in addition to their adduction propensity and lability of sulfate groups during tandem analysis. Nonetheless, as the involvement of NS domains that can range up to hexadecamer in size in protein binding is becoming clearer (40), there seems to be a necessity to apply MS2 on larger oligosaccharides.
In previous work, we optimized a combined size exclusion-hydrophobic trapping affinity purification method followed by liquid chromatography/mass spectrometry (LC/MS) for determining the compositions of heparin hexasaccharides that bind ATIII (41). Furthermore, we took advantage of the ability of the LC to separate the differentially sulfated heparin hexasaccharides to correct for the sulfate loss that occurs in source and hence an accurate quantification for each hexasaccharide composition was generated. In the present study, higher complexity oligosaccharides namely octasaccharides were investigated for their ability to bind and modulate ATIII using high resolution high mass accuracy MS circumventing the sulfate loss problem. The scope of heparinoids analyzed was extended to include HS in addition to heparin because the former is the main physiological protein regulator in vivo. The choice of octasaccharides reflects a balance between length appropriate for protein binding studies and degree of structural complexity that is amenable to analysis. In addition, tandem mass spectrometry was used to compare the fine structures of the biologically active HS octasaccharides that bind ATIII. The conclusion is that tandem mass spectrometric analysis of these octasaccharides produces fragments diagnostic of biological activity and hence permits the differentiation of two samples that differ in their ability to modulate ATIII. The data demonstrate a more general approach for comparative analysis of extended NS domains that uses patterns of tandem mass spectrometric product ions that constitute a fingerprint for their protein binding activity.
Materials and method
Materials
Porcine intestinal mucosa heparin (sodium salt, 182USP/mg) was obtained from Sigma-Aldrich (St. Louis, MO). Porcine intestinal mucosa heparan sulfate was purchased from Celsus laboratories, Inc (Cincinnati, OH). Heparin lyase I and III purified from Flavobacterium heparinum were from IBEX (Montreal, QC). Antithrombin III was a gift from GTC Biotherapeutics (Framingham, MA). Actichrome heparin anti-FXa was from American Diagnostica Inc. (Stamford, CT). Amide 80 packing material was purchased from TOSOH Bioscience LLC (Montgomeryville, PA). Dialysis cellulose acetate membranes were purchased from the Nest group (Southborough, MA).
Porcine intestinal mucosa heparin depolymerization
A quantity of 100 mg of heparin was digested with heparin lyase I in 1ml 100mM Ammonium acetate PH 7.4 containing 0.1mg/ml BSA at 37°C. The reaction was stopped at 30% completion as judged by UV absorbance at 232 nm. The digestion mixture was fractionated on a preparative size exclusion chromatography (SEC) column (170cm × 1.5 cm; Bio-Rad, Hercules, CA) with 100mM ammonium bicarbonate buffer flowing at 40μl/min (42). The octasaccharide (dp8) fraction was collected and desalted by dialysis with a 500Da cutoff membrane.
Porcine intestinal mucosa heparan sulfate depolymerization
A quantity of 100 mg of HS was digested exhaustively with heparin lyase III in 1 ml sodium acetate buffer (pH 7) supplemented with calcium acetate to a concentration of 5 mM. The digestion mixture was size fractionated and the octasaccharide fraction collected and desalted as described above.
Binding assay
ATIII (1 nmol) was mixed with heparin (10 nmol) or HS (15 nmol) octasaccharide and the binders recovered as described in (41). In brief, ATIII was incubated with the octasaccharide library fraction and the protein-octasaccharide complex separated by size exclusion chromatography. Subsequently, the complex was applied to a reverse phase cartridge and washed with medium stringency salt solution (200mM ammonium acetate). Finally, the bound octasaccharides were eluted with a high stringency salt solution (2M ammonium acetate). The experiments were performed in triplicate and the results reported as averages with standard deviation.
Liquid chromatography-mass spectrometry (LC-MS)
Heparin and HS oligosaccharides were separated by amide hydrophilic interaction chromatography (HILIC) with on-line mass spectrometric detection. The column used was packed in house with 3μm beads amide-80 packing material (TOSOH Biosciences) into a 250 μm × 15cm capillary. The oligosaccharides (40 pmol of library fraction) were trapped on a 250 μm × 5cm column for 5 minutes at 15 μL /min flow rate of 30% A. The trapping event allowed direct injection of the bound fractions highly concentrated in salt without prior desalting. The oligosaccharides were eluted from the column with gradient starting at 30 % and ending at 65 % solvent A over a period of 30 minutes at a flow of 1.5 μL/min delivered by a Waters NanoAquity (Milford, MA). Solvent A consists of 50 mM ammonium formate, pH 4.4. Solvent B is 95% acetonitrile and 5% A.
Mass spectrometric analysis was accomplished using a Thermo-Fisher Scientific-LTQ Orbitrap operating in the negative high resolution mode. The instrument ionization source was interfaced to an automated Triversa Nanomate robot (Advion Biosystems, Inc., Ithaca, NY) operating with a spray voltage of −1.4 kV. The instrument was tuned using Arixtra (Organon Sanofi-Synthelabo LLC, NJ), an octasulfated synthetic pentasaccharide standard. The instrument voltages were optimized to abolish sulfate loss during ion transfer between the trap and the Orbitrap. Data acquisition was done with a resolution of 30000 and mass accuracy was better than 5ppm.
Tandem Mass Spectrometry
The heparinoid library fractions as well as the ATIII bound oligosaccharides were cleaned from residual salt by extensive dialysis prior to tandem mass spectrometric analysis. The samples were nano-sprayed in 50:50 methanol: water using the Triversa Nanomate from Advion (Ithaca, NY) operating at −1.3 kV. The tandem analysis was performed at 30000 resolution in the negative ion mode using a Thermo-Fisher Scientific LTQ Orbitrap by collision induced dissociation (CID) with collision energy of 20. When necessary, successive isolation windows were applied in order to optimize the isolation interval to obtain clean isolations of the compounds of interest.
Biological activity
The octasaccharide recovered from the binding protocol were desalted by dialysis and tested for the ability to regulate ATIII activity as described (41).
Results
LC MS for quantification of octasaccharides that bind ATIII
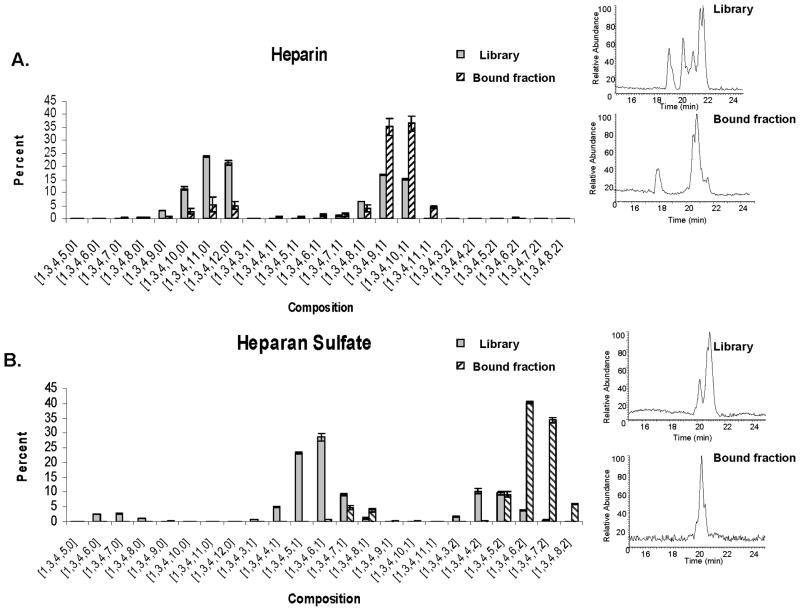
The first aim of this work was to identify the compositions of degree of polymerization (dp) 8 from heparin and HS libraries that bind ATIII. To prepare the octasaccharide libraries, full length heparin and HS were digested respectively with heparin lyase I and III. The digestion products were separated by preparative size exclusion chromatography and the octasaccharide fraction was collected and desalted (supplementary Figure 1). In order to purify binders from the octasaccharide libraries, ATIII was incubated with the SEC purified dp8 fraction and the protein-oligosaccharide complex separated using a high performance SEC followed by a hydrophobic trapping assay (28). The ATIII-bound octasaccharides were then eluted using a high salt buffer. The libraries consisted of distributions of differentially sulfated and acetylated octasaccharides. These compositions were quantified using HILIC LC/MS (41) (Figure 1). The octasaccharides eluted from the column in order of increasing polarity directly proportional to the number of sulfates on the molecule (41). This on-line separation minimized ion suppression effects. As a result, the LC/MS offered an accurate and reproducible comparative platform that was used to show enrichment of ATIII binding compositions by comparing their relative abundances to those present in the library octasaccharide fraction.
Figure 1. Octasaccharide compositional distribution in library versus ATIII bound fractions.
A. Heparin.
B. Heparan Sulfate
Each composition is given under the following format [ΔHexA, HexA, GlcN, SO3, Ac]. The insets show the base peak chromatograms of the LC/MS.
The change in the distribution of compositions that occurs after the affinity enrichment protocol indicates that in the heparin library most compositions contain high affinity binders whereas in HS some compositions are better binders than other. The compositions of heparin versus HS dp8 that bind ATIII differ dramatically; the sets of binding compositions in heparin versus HS are mutually exclusive.
Figure 1A shows the profiles of the heparin octasaccharide library compared with the heparin ATIII bound fraction. The dp8 compositions are abbreviated using a numerical code where numbers correspond respectively to the chemical groups in the following chemical formula [ΔHexA, HexA, GlcN, SO3, Ac]. The heparin octasaccharides consist primarily of N-sulfated or singly N-acetylated compositions. The two most abundant compositions in the library fraction are N-sulfated with eleven and twelve sulfate groups, with respective abundances of 23.8 % and 21.4%. In contrast, the most abundant compositions in the bound fraction are singly N-acetylated with nine and ten sulfate groups, accounting together for more than 70% of the bound fraction relative to only 32% of the library.
A similar but more pronounced enrichment is observed in the HS bound fraction compared to the HS octasaccharide library. The HS library consists primarily of singly and doubly N-acetylated compositions (Figure 1B). The most abundant binders are doubly N-acetylated with 6 and 7 sulfate groups. The respective abundances of these compositions are 40.3% and 34.3%. While such compositions in the library collectively account for 4.4% of the total, they represent nearly 75% of the total in the ATIII-bound fraction. The specificity of binding was tested by negative control experiments where the same binding protocol was performed with heat inactivated ATIII (data not shown). No oligosaccharides were recovered after the combined SEC-hydrophobic trapping workup, showing that unspecific binders can not survive the steps of the used protocol.
The change that occurs to the profile of compositions in the library fraction subsequent to the binding protocol shows that the distribution of high affinity binders is different between the library and the bound fractions. Additionally, the protocol is able to enrich with binders that are minor or fall under the detection limit in the library such as heparin [1,3,4,11,1] and HS [1,3,4,8,2].
Biological activity of ATIII-binding oligosaccharides
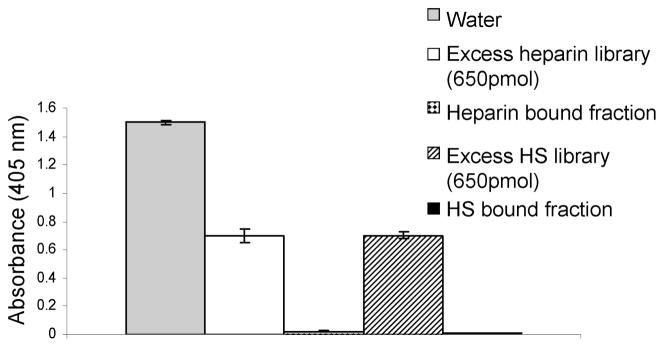
The biological activity of the affinity purified octasaccharides, was assayed using a Factor Xa (FXa) in vitro inhibition assay. Heparin binds ATIII, inducing a change in its conformation and allowing it to act as a potent inhibitor for serine proteases involved in the blood coagulation cascade such as FXa. The end result is the inhibition of the reactions involved in the blood coagulation cascade (43, 44). In this in vitro assay, FXa is incubated with and allowed to hydrolyze a synthetic substrate yielding a colored product detected at 405 nm. When ATIII and oligosaccharides with an ATIII binding site are added to the incubation reaction, ATIII is able to inhibit the enzymatic activity of FXa and hence color formation is suppressed. The ability of the bound fraction to modulate the ATIII inhibitory activity on FXa is shown in Figure 2. The presence of ATIII alone in the incubation reaction yields a basal level of FXa activity evaluated at 1.4 AU. An excess amount of both the heparin and HS libraries exerts an activating effect on ATIII allowing it to inhibit FXa and decrease color formation. The heparinoid bound fractions have the ability to potentiate the ATIII mediated inhibition of FXa to a much higher degree compared to both controls.
Figure 2. Biological activity of heparin and HS bound fractions.
The ATIII mediated inhibitory activity of heparin and HS octasaccharides on FXa was studied using an in vitro colorimetric assay. FXa is able to hydrolyze a synthetic substrate yielding a colored product detected at 405nm. When FXa is incubated with its substrate in the presence of ATIII and biologically active heparinoids, its hydrolytic activity is inhibited. The 405nm absorbance is inversely related to the anti-FXa activity of heparinoid octasaccharides used in the experiments. Experiments were done in triplicate.
In general, using the same digestion conditions, a porcine intestinal mucosa heparin dp8 library is expected to be richer in ATIII binding sites compared to the same size and same origin HS library. When added in the same quantities, the former will induce a higher FXa inhibition compared to the latter. Our data show that both heparin and HS libraries have comparable in-vitro biological activities. This can be explained by the differential digestion conditions used to generate the heparin and HS octasaccharide libraries from full length heparinoids preparations. In fact, heparin octasaccharides were generated by partial digestion of full length heparin using heparin lyase I cleavage. Heparin lyase I is known to cleave within the ATIII binding site and inactivate it (45, 46). Hence after this procedure, the binding sites remaining in the library are much less than the theoretical number of binding sites expected if the enzyme did not cleave within the ATIII binding site. In contrast, HS octasaccharides are generated by cleavage of parental HS chains using Heparin lyase III. This enzyme is known to preserve the ATIII binding sites (45, 46). Hence, it is not surprising that the heparin octasaccharide library did not inhibit Fxa more than the HS library.
The former results confirm that the binding property of the affinity purified octasaccharides is paralleled by an ability to activate ATIII in vitro.
Tandem MS for comparison of fine structure of ATIII binding and library octasaccharides
The complexity of heparin and HS octasaccharide fractions is not only the result of the multiple compositions that arise from different degree of sulfation and acetylation in a size purified library fraction, but also the distribution of isomeric structures. This means that a single composition with defined number of sulfates and acetates is a collection of positional isomers. Consequently, the fragments yielded during tandem analysis of a precursor ion originate from an isomeric mixture. To date, no separation method can resolve such heparinoid isomers to purity. Additionally, sulfate groups tend to dissociate during CID analysis limiting the number of meaningful fragment ions and hence sequence coverage. Nonetheless, this method produces valuable information comparing the structures of the protein-binding versus library octasaccharides. This technique proved useful to show that the distributions of isomers in the two samples are different and, most importantly, could generate product ion patterns that were diagnostic of biological activity.
The degree to which sulfate groups undergo tandem mass spectrometric dissociation to produce losses of SO3 (80 u) is inversely related to the charge state of the oligosaccharide during ionization (35). This is because, as the charge state increases, the repulsion between the different sulfate groups increases making glycosidic bonds more susceptible to dissociation. Static nanospray has the advantage of yielding higher charge states for the same oligosaccharide composition compared to that observed using on-line LC/MS, where the charge of the analytes is neutralized by ammonium adducts contained in the mobile phase. This approach was therefore used to compare heparinoid octasaccharide isomers in protein binding fractions versus library.
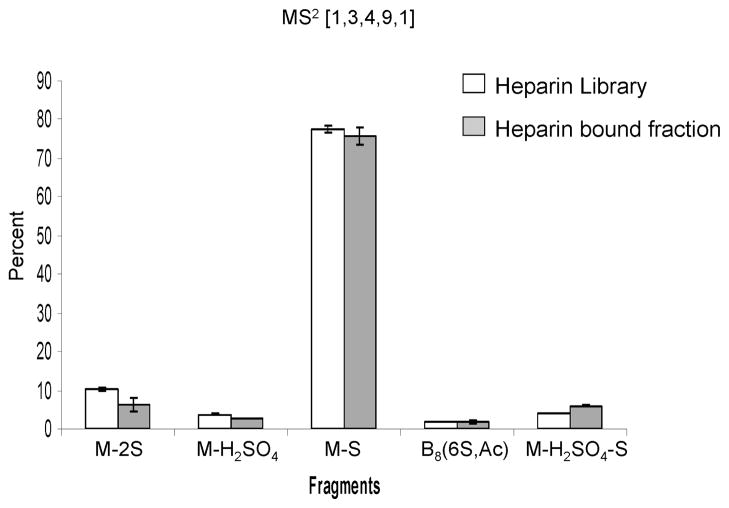
The compositions subjected to tandem MS were selected based on the ability to obtain a clean isolation of the precursor ion. Heparins are very highly sulfated and the compositions that could be isolated and fragmented are the [1,3,4,10,0], [1,3,4,11,0], [1,3;4,12,0], [1,3,4,9,1], [1,3,4,10,1]. The most abundant charge state observed was [M-6H]6−. Given the high sulfate content of heparin octasaccharides, at this charge state losses of SO3 from the precursor ion were the most abundant product ions and those corresponding to glycosidic bond and cross ring cleavages were of very low abundance. This is illustrated by the fragments yielded by tandem analysis of the least sulfated heparin composition that was isolated [1,3,4,9,1] (Figure 3). In addition to the challenges facing fragmentation of heparins, the expression of this latter is limited to mast cells, and HS is the more relevant regulator of protein activity in vivo. For these reasons, effort was focused on tandem mass spectrometric analysis of HS octasaccharides. In HS, the average number of sulfate groups per disaccharide across the chain is close to 1 (47) compared to 2.7 for heparin (48). Although, the average sulfation within the NS domains is greater than the average sulfation across the entire chains, it still does not exceed that of heparin. The composition distribution data in Figure 1 provides an estimation of the average sulfation of a heparin and HS octasaccharide. An octasaccharide from heparin has an average of 2.6 sulfates per disaccharide which is consistent with the literature values. In HS, an NS domain of dp 8 has an average sulfation of 1.4 per disaccharide. As a result, the abundances of ions produced by losses of SO3 from the precursor are expected to be significantly less abundant for the charge states observed using negative nanospray ionization.
Figure 3. MS2 of heparin [1,3,4,9,1].
The fragments yielded upon fragmentation of this lowly sulfated heparin composition with a 6− charge state are majorly loss of sulfate.
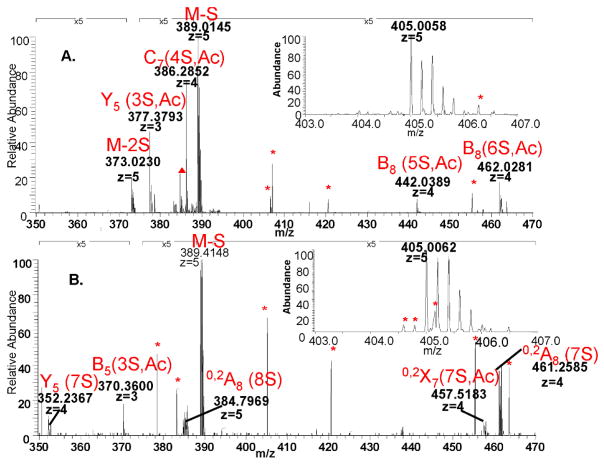
Low energy CID fragmentation was performed on three ATIII-binding compositions, namely [1,3,4,7,1], [1,3,4,8,1] and [1,3,4,6,2] as [M-5H]5− ions. The respective m/z of the isolated precursors with their accuracies is shown in supplementary Table 1. All the mass accuracies are lower than +/− 5ppm which is in the accepted range of the instrument. At that charge state losses of SO3 are observed but glycosidic bond and cross-ring cleavages also produce abundant ions. The tandem mass spectra show that the isomeric compositions corresponding to ATIII-bound and library octasaccharides produce ion patterns that differ significantly in abundance values (Figures 4, 5 and 6). The differences in product ion abundances reflect the difference isomeric glycoforms present in the ATIII-bound versus library fractions.
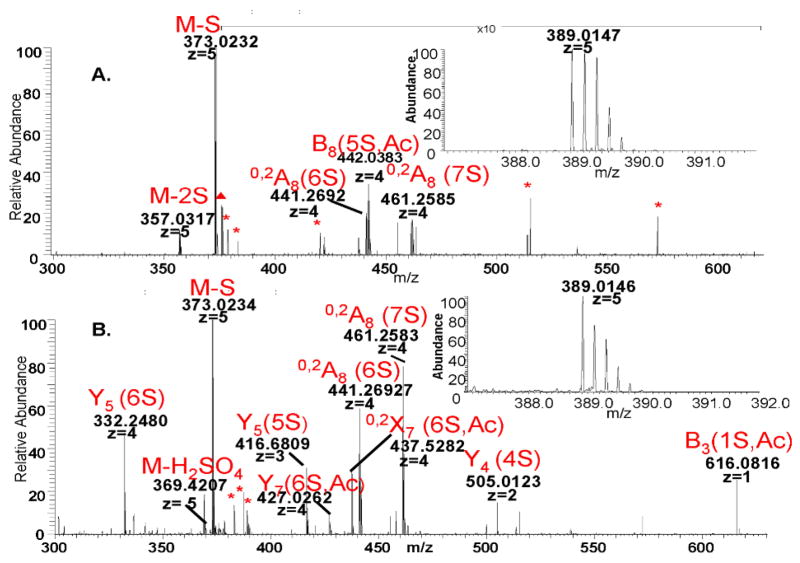
Figure 4. CID MS2 spectrum of [1,3,4,7,1] composition in the library versus the bound fraction.
The insets show the isolation of the precursor ion. The peaks are labeled with their corresponding structure, (
 ) if they could not be assigned to a structure and (*) if they are background noise.
) if they could not be assigned to a structure and (*) if they are background noise.
A. Tandem mass spectrum of [1,3,4,7,1] in the library. One interval of the spectrum has been magnified as indicated in the figure for better visibility of the fragments.
B. Tandem mass spectrum of [1,3,4,7,1] in the bound fraction. The isotopic pattern of the precursor (inset) is skewed compared to that of the precursor in the library due to an artifact introduced upon application of the isolation window.
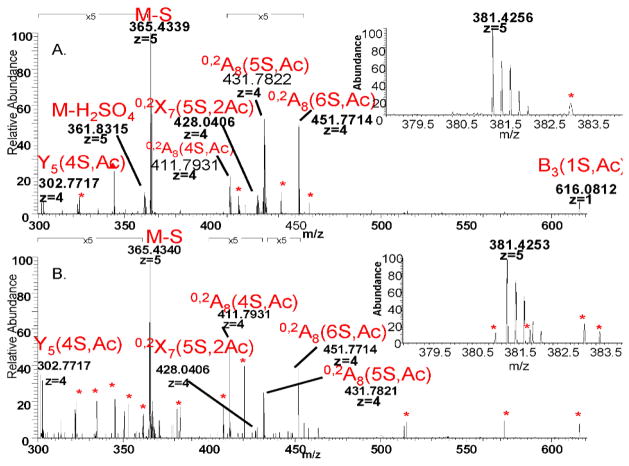
Figure 5. CID MS2 spectrum of HS [1,3,4,8,1] composition. The legend is similar to that in Figure 4.
A. library
B. ATIII-bound fraction. The ions co-isolated with the precursor (inset) do not fall into a pattern that may be assigned to a charge state and do not correspond to expected values for heparin/HS oligosaccharides. Hence they are labeled as co-isolated noise.
Figure 6. CID MS2 spectrum of HS [1,3,4,6,2] composition in the library versus the bound fraction. The legend is similar to that in Figure 4.
A. Library
B. ATIII-bound fraction.
In the insets of figures 4 and 5, the isolated precursors show discrepancies in their isotopic distribution between the library and the bound fraction. The isotopic pattern of an ion in isolation mode within the ion trap tends to be skewed depending on the isolation parameters applied. The isolation window used to isolate the precursor from the library in the ion trap of the mass spectrometer is different from the one used to isolate the same precursor from the bound fraction. The center of the window of isolation and its width are dictated by the peaks that surround the precursor in the MS dimension. Since the library and the bound fraction have different MS profiles, the former parameters are adjusted accordingly to obtain clean isolations of the precursors. Supplementary Figure 2 shows that the precursors have identical isotopic distributions in the MS mode but this distribution tends to be altered upon application of an isolation window.
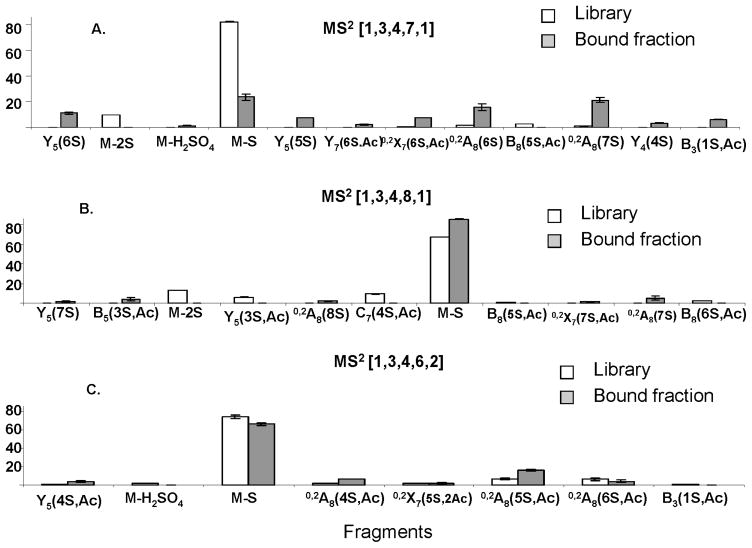
Figure 7 shows comparisons of the product ions abundances for ATIII-bound and library octasaccharide compositions. Note that the standard Domon-Costello nomenclature is used (49) with the number of sulfate (S) and acetate (Ac) groups specified in parentheses. Supplementary figure 3 shows the Domon-Costello product ions possible for the octasaccharide [1,3,4,7,2] carrying the ATIII binding site. Several product ions differentiate the ATIII-bound versus library [1,3,4,7,1] compositions (Fig. 7A). These include [M-2(SO3)-5H]5− and B8(5S,Ac). Similarly, the fragmentation of the affinity purified [1,3,4,7,1] yields specific fragments Y5(6S), M-H2SO4, Y5(5S), Y7(6S,Ac), Y4(4S) and B3(1S,Ac). Other product ions that increased in abundance in the ATIII-bound [1,3,4,7,1] composition include 0,2X7(6S,Ac), 0,2A8(6S) and 0,2A8(7S). Dissociation of the [1,3,4,8,1] composition in the library yields product ions of negligible abundance values in the corresponding ATIII-bound fraction (Fig 7B). These ions are [M-2SO3]5−, Y5(3S,Ac), C7(4S,Ac), B8(5S,Ac) and B8(6S,Ac). The product ions specific to ATIII-bound [1,3,4,8,1] are Y5(7S), B5(3S,Ac), 0,2A8(8S), 0,2X7(7S,Ac) and 0,2A8(7S). A similar observation applies to the fragmentation of the doubly acetylated composition [1,3,4,6,2] (Fig 7C). Library-specific fragments are M-H2SO4, and B3(1S,Ac). The bound fraction is greatly enriched Y5(4S,Ac) and 0,2A8(4S,Ac) and 0,2A8(5S,Ac).
Figure 7. Enrichment of specific isomers in the bound fractions.
Comparison of CID MS2 profiles of three HS compositions between the library and the bound fractions. The y axis represents the percent abundance of an ion relative to the total abundance of all ions combined.
A. [1,3,4,7,1]
B. [1,3,4,8,1]
C. [1,3,4,6,2]
Tandem mass spectrometric dissociation of a given HS octasaccharide composition in a library versus an ATIII-bound fraction provides proof for isomer enrichment. For [1,3,4,7,1], the library fragments show only fragments consistent with an acetate group on the reducing end of the octasaccharide, namely 0,2A8(6/7S). In contrast, a considerable proportion of the bound [1,3,4,7,1] product ions indicate that the acetate group is at the non-reducing end which is in agreement with the non-reducing end acetate group found in the ATIII binding site. These fragments are Y5(4/5/6S) and B3(1S,Ac).
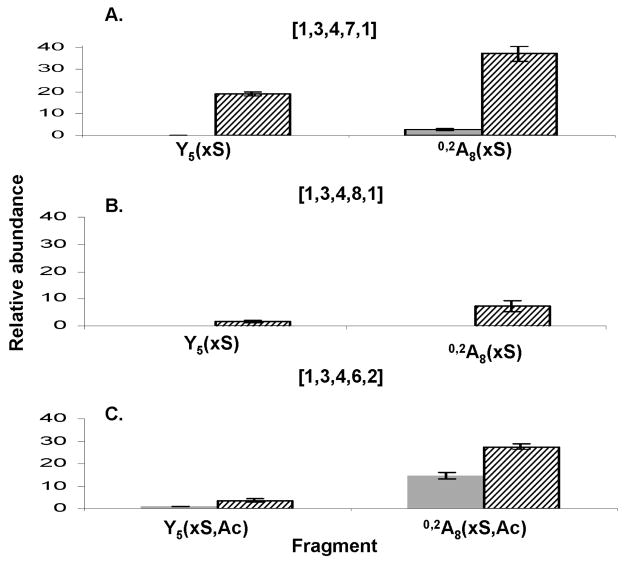
These examples demonstrate the presence of product ions the abundances of which are diagnostic for ATIII-binding octasaccharide compositions. This is shown by the enrichment of unacetylated Y5 (Y5 xS) and 0,2A8 (0,2A8 xS) where x indicates variable number of sulfate groups, in the ATIII-binding compositions [1,3,4,7,1] and [1,3,4,8,1]. Similarly, the singly acetylated Y5 (Y5 xS,Ac) and 0,2A8 (0,2A8 xS,Ac) are enriched in the doubly acetylated binding composition [1,3,4,6,2]. This is illustrated by the presence of Y5(5/6S), 0,2A8(6/7S), enriched in the bound [1,3,4,7,1] (Fig 8A), Y5(7S) and 0,2A8(7/8)S uniquely present in the bound [1,3,4,8,1] (Fig. 8B) and Y5(4S,Ac) and 0,2A8 (4/5S,Ac) greatly enriched in the bound [1,3,4,6,2] 405 The data demonstrate that the summed abundances for all sulfation and acetylation forms of Y5 and 0,2A8 product ions correlate to the propensity for ATIII binding. Thus, the octasaccharide structures that bind ATIII give rise to a characteristic glycosidic bond dissociation pattern. This effect is likely to arise from the both the specificity of ATIII and the reduction of the polydispersity of the ATIII-bound octasaccharides relative to the library fractions.
Figure 8. Enrichment of diagnostic ions in the bound fraction.
The relative abundances of all the ions of the general formula Y5(xS) on the one hand and 0,2A8(xS) on the other have been summed together to produce the respective summed relative abundances. X indicates the number of sulfates. The figure shows the enrichment of these ions for the singly acetylated compositions [1,3,4,7,1] (A) and [1,3,4,8,1] (B). A new standard deviation has been calculated from the individual standard deviations of the relative abundances of each individual ion.
In (C), the same procedure was applied to Y5(xS,Ac) and 0,2A8(xS,Ac) in the doubly acetylated composition [1,3,4,6,2].
Discussion
These results show that heparin compositions that bind ATIII are considerably disparate compared to the HS binders in terms of sulfation and acetylation content. The most potent binding heparin compositions yielded by the assay are [1,3,4,9,1] and [1,3,4,10,1]. Original studies on heparin oligosaccharide capable of binding ATIII used partially depolymerized heparin by nitrous acid cleavage. The most potent ATIII binding octasaccharide sequence was IdoA-GlcNAc6S-GlcA-GlcNS3S6S-IdoA2S-GlcNS6S-IdoA2S-aMan6S-ol. A lower affinity sequence was IdoA2S-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS-3S,6S-IdoA2S-aMan6S-ol (12, 13). This latter octasaccharide lacks the essential reducing end N-sulfate of the ATIII binding site which was lost due to anhydromannose formation at the oligosaccharide reducing end subsequent to nitrous acid cleavage and hence its lower affinity. These two structures are both singly acetylated with 9 sulfate groups. In both these octasaccharides, variants where the GlcNS3S6S residue is replaced by GlcNS3S occur resulting in singly acetylated binding octasaccharides with a 8 sulfate groups (13). Hence, nitrous acid depolymerization of heparin generates singly acetylated binders with 8 and 9 sulfates. Given that the heparin octasaccharide binders in this study are generated by partial heparin lyase I digestion, they contain N-sulfated GlcN at the reducing end (50). Because this residue is lost after nitrous acid depolymerization, it is not surprising that the most potent binders have 9 and 10 sulfate groups. Although acetylation is not required for binding, its preponderance at the reducing end in ATIII binding octasaccharides is explained by requirement of the downstream GlcA for binding. In fact, when GlcNAc lies upstream of GlcA, the epimerase cannot act on GlcA to convert it into IdoA (10). In summary, singly acetylated heparin octasaccharides with 9 and 10 sulfates are in agreement with previous findings.
The role of HS as an antithrombotic agent has gained great importance in the past decade. Anticoagulant HS, like heparin, possesses a pentasaccharide sequence that is able to bind and activate ATIII in vitro. HS at the surface of endothelial cells is thought to provide the vascular wall with antithrombotic properties. However, the in vivo anticoagulant role has been a subject of controversy. Knock-out mice deficient in the 3-O sulfotransferase-1 (3-OST-1) do not show pro-coagulant phenotype (51). However, the conclusions of these studies do not abolish the role of HS as the physiological anticoagulant because there is residual activity that might be due to compensation mechanisms by other isoforms of 3-OST-1. The structure of hexasaccharides generated by partial digestion of HS from F9 embryonic carcinoma cells with lyases II and III is summarized by ΔUA-GlcNAc6S-GlcA-GlcNS 3S +/−6S-IdoA 2S-GlcNS 6S (52). Octasaccharides of porcine intestinal mucosa HS with anticoagulant activity have not been studied previously. The results show that the most abundant ATIII-binding HS octasaccharides have respectively 6 and 7 sulfate groups, in agreement with hexasaccharide findings. However, in the present study, the HS octasaccharide library fraction was generated using heparin lyase III digestion. The primary site of cleavage of lyase III is GlcNAc/NS-GlcA (50, 53). Because unsulfated GlcA occurs preferentially adjacent to a GlcNAc, a GlcNAc-GlcA is expected to be a more abundant disaccharide unit in HS than GlcNS-GlcA. Therefore, the oligosaccharides generated by lyase III digestions are expected to have a reducing end GlcNAc residue. Hence, for the ATIII-binding HS octasaccharides bearing two acetate groups, a non-reducing end GlcNAc-GlcA disaccharide is likely, in addition to reducing end GlcNAc due to enzyme bias. However, this does not exclude the possibility that the acetyl group can be internal. This explains the observation of abundant ATIII-binding HS octasaccharides bearing two acetate groups.
These results also demonstrate that product ion patterns constitute fingerprints that differentiate HS oligosaccharide compositions among different sources. In the present case, the product ion profiles of [1,3,4,7,1], [1,3,4,8,1] and [1,3,4,6,2] precursors in the ATIII bound fraction contain diagnostic ions of their protein binding biological activity. In order to be useful, diagnostic ions should have the property of being more abundant in the protein bound fraction and indicate the presence of structural elements common to the three biologically active compositions. It is apparent from the tandem mass spectrometric profiles of the HS octasaccharides that the protein binding structures have characteristic patterns of glycosidic bond dissociation relative to the library fractions.
The significance of the combined LC/MS compositional analysis with the tandem mass spectrometric dimension is in the resulting ability to identify compositions that bind and modulate the activity of a protein of interest and product ions diagnostic for oligosaccharide structures responsible for this activity. It is anticipated that this approach will be applicable to the determination of protein binding properties of HS oligosaccharides with other protein of interest. Here, ATIII was used to purify binders that exist in very low abundance in heparinoid fractions due to their presence in only a small percentage of heparinoid chains (1–10% of HS isolated from tissues and 30% of pharmaceutical heparin bind ATIII) in addition to their restrictive binding mechanism (54). If this system is used with proteins that show more relaxed binding criteria such as growth factors, it is likely that a greater number of binding compositions will be observed. However, the applications of the HS diagnostic ion approach are not limited to the differentiation of protein binding oligosaccharides. Previously, tandem mass spectrometric product ions diagnostic of tissue type and disease state have been observed for chondroitin/dermatan sulfate oligosaccharides (55). In the same fashion, HS is known to exhibit structural modifications with regard to disease stage (56, 57), cell type (58, 59), their tissue of origin (60) and developmental stage (61). These modifications are the result of changes that affect the biosynthetic machinery combined with alterations in the activity of the HS modifying enzymes in the extracellular compartment. It is anticipated that diagnostic product ions will be useful in drawing a fingerprint of NS domains from different sources and hence constitute markers of physiological conditions that alter HS structure.
Supplementary Material
Supplementary table with m/z of precursor ions isolated and their mass accuracies.
Supplementary Figure 1 showing the size exclusion chromatograms for heparin and HS digestions.
Supplementary Figure 2 comparing the MS and MS2 isotopic distributions of isolated precursor ions.
Acknowledgments
Funding from NIH grants P41RR10888, S10RR020946 are gratefully acknowledged.
We thank GTC Biotherapeutics (Framingham, MA) for providing ATIII.
Abbreviations
- HS
Heparan sulfate
- NS domain
N-sulfated domains
- NA domain
N-acetylated domains
- AT III
Antithrombin III
- MS
Mass spectrometry
- MS2
Tandem mass spectrometry
- CID
Collision induced dissociation
- LC/MS
Liquid chromatography/Mass spectrometry
- SEC
Size exclusion chromatography
- Dp
Degree of polymerization
- HILIC
Hydrophilic interaction liquid chromatography
- ΔHexA
Unsaturated hexuronic acid
- HexA
Hexuronic acid
- GlcN
Glucosamine
- Ac
Acetate
- FXa
Factor Xa
Footnotes
Supporting information available:
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Varki A, Cummings R, Esko J, Freeze H, Hart G, Marth G. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. [PubMed] [Google Scholar]
- 2.Ai X, Do AT, Lozynska O, Kusche-Gullberg M, Lindahl U, Emerson CP., Jr QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. The Journal of cell biology. 2003;162:341–351. doi: 10.1083/jcb.200212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vlodavsky I, Goldshmidt O, Zcharia E, Atzmon R, Rangini-Guatta Z, Elkin M, Peretz T, Friedmann Y. Mammalian heparanase: involvement in cancer metastasis, angiogenesis and normal development. Seminars in cancer biology. 2002;12:121–129. doi: 10.1006/scbi.2001.0420. [DOI] [PubMed] [Google Scholar]
- 4.Carlsson P, Presto J, Spillmann D, Lindahl U, Kjellen L. Heparin/heparan sulfate biosynthesis: processive formation of N-sulfated domains. The Journal of biological chemistry. 2008;283:20008–20014. doi: 10.1074/jbc.M801652200. [DOI] [PubMed] [Google Scholar]
- 5.Fransson LA, Belting M, Cheng F, Jonsson M, Mani K, Sandgren S. Novel aspects of glypican glycobiology. Cell Mol Life Sci. 2004;61:1016–1024. doi: 10.1007/s00018-004-3445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belting M, Mani K, Jonsson M, Cheng F, Sandgren S, Jonsson S, Ding K, Delcros JG, Fransson LA. Glypican-1 is a vehicle for polyamine uptake in mammalian cells: a pivital role for nitrosothiol-derived nitric oxide. The Journal of biological chemistry. 2003;278:47181–47189. doi: 10.1074/jbc.M308325200. [DOI] [PubMed] [Google Scholar]
- 7.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annual review of biochemistry. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 8.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 9.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annual review of biochemistry. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl U, Thunberg L, Backstrom G, Riesenfeld J, Nordling K, Bjork I. Extension and structural variability of the antithrombin-binding sequence in heparin. The Journal of biological chemistry. 1984;259:12368–12376. [PubMed] [Google Scholar]
- 11.Casu B, Oreste P, Torri G, Zoppetti G, Choay J, Lormeau JC, Petitou M, Sinay P. The structure of heparin oligosaccharide fragments with high anti-(factor Xa) activity containing the minimal antithrombin III-binding sequence. Chemical and 13C nuclear-magnetic-resonance studies. The Biochemical journal. 1981;197:599–609. doi: 10.1042/bj1970599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thunberg L, Backstrom G, Lindahl U. Further characterization of the antithrombin-binding sequence in heparin. Carbohydrate research. 1982;100:393–410. doi: 10.1016/s0008-6215(00)81050-2. [DOI] [PubMed] [Google Scholar]
- 13.Atha DH, Stephens AW, Rimon A, Rosenberg RD. Sequence variation in heparin octasaccharides with high affinity for antithrombin III. Biochemistry. 1984;23:5801–5812. doi: 10.1021/bi00319a020. [DOI] [PubMed] [Google Scholar]
- 14.Atha DH, Lormeau JC, Petitou M, Rosenberg RD, Choay J. Contribution of monosaccharide residues in heparin binding to antithrombin III. Biochemistry. 1985;24:6723–6729. doi: 10.1021/bi00344a063. [DOI] [PubMed] [Google Scholar]
- 15.Seyrek E, Dubin PL, Henriksen J. Nonspecific electrostatic binding characteristics of the heparin-antithrombin interaction. Biopolymers. 2007;86:249–259. doi: 10.1002/bip.20731. [DOI] [PubMed] [Google Scholar]
- 16.Maccarana M, Casu B, Lindahl U. Minimal sequence in heparin/heparan sulfate required for binding of basic fibroblast growth factor. The Journal of biological chemistry. 1993;268:23898–23905. [PubMed] [Google Scholar]
- 17.Habuchi H, Suzuki S, Saito T, Tamura T, Harada T, Yoshida K, Kimata K. Structure of a heparan sulphate oligosaccharide that binds to basic fibroblast growth factor. The Biochemical journal. 1992;285(Pt 3):805–813. doi: 10.1042/bj2850805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turnbull JE, Fernig DG, Ke Y, Wilkinson MC, Gallagher JT. Identification of the basic fibroblast growth factor binding sequence in fibroblast heparan sulfate. The Journal of biological chemistry. 1992;267:10337–10341. [PubMed] [Google Scholar]
- 19.Kreuger J, Prydz K, Pettersson RF, Lindahl U, Salmivirta M. Characterization of fibroblast growth factor 1 binding heparan sulfate domain. Glycobiology. 1999;9:723–729. doi: 10.1093/glycob/9.7.723. [DOI] [PubMed] [Google Scholar]
- 20.Jastrebova N, Vanwildemeersch M, Rapraeger AC, Gimenez-Gallego G, Lindahl U, Spillmann D. Heparan sulfate-related oligosaccharides in ternary complex formation with fibroblast growth factors 1 and 2 and their receptors. The Journal of biological chemistry. 2006;281:26884–26892. doi: 10.1074/jbc.M600806200. [DOI] [PubMed] [Google Scholar]
- 21.Lindahl U. Heparan sulfate-protein interactions--a concept for drug design? Thrombosis and haemostasis. 2007;98:109–115. [PubMed] [Google Scholar]
- 22.Kreuger J, Matsumoto T, Vanwildemeersch M, Sasaki T, Timpl R, Claesson-Welsh L, Spillmann D, Lindahl U. Role of heparan sulfate domain organization in endostatin inhibition of endothelial cell function. The EMBO journal. 2002;21:6303–6311. doi: 10.1093/emboj/cdf638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stringer SE, Gallagher JT. Specific binding of the chemokine platelet factor 4 to heparan sulfate. The Journal of biological chemistry. 1997;272:20508–20514. doi: 10.1074/jbc.272.33.20508. [DOI] [PubMed] [Google Scholar]
- 24.Spillmann D, Witt D, Lindahl U. Defining the interleukin-8-binding domain of heparan sulfate. The Journal of biological chemistry. 1998;273:15487–15493. doi: 10.1074/jbc.273.25.15487. [DOI] [PubMed] [Google Scholar]
- 25.Lortat-Jacob H, Turnbull JE, Grimaud JA. Molecular organization of the interferon gamma-binding domain in heparan sulphate. The Biochemical journal. 1995;310(Pt 2):497–505. doi: 10.1042/bj3100497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Y, Sweeney MD, Saad OM, Leary JA. Potential inhibitors of chemokine function: analysis of noncovalent complexes of CC chemokine and small polyanionic molecules by ESI FT-ICR mass spectrometry. Journal of the American Society for Mass Spectrometry. 2006;17:524–535. doi: 10.1016/j.jasms.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Harmer NJ, Robinson CJ, Adam LE, Ilag LL, Robinson CV, Gallagher JT, Blundell TL. Multimers of the fibroblast growth factor (FGF)-FGF receptor-saccharide complex are formed on long oligomers of heparin. The Biochemical journal. 2006;393:741–748. doi: 10.1042/BJ20050985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu Y, Sweeney MD, Saad OM, Crown SE, Hsu AR, Handel TM, Leary JA. Chemokine-glycosaminoglycan binding: specificity for CCR2 ligand binding to highly sulfated oligosaccharides using FTICR mass spectrometry. The Journal of biological chemistry. 2005;280:32200–32208. doi: 10.1074/jbc.M505738200. [DOI] [PubMed] [Google Scholar]
- 29.Maccarana M, Lindahl U. Mode of interaction between platelet factor 4 and heparin. Glycobiology. 1993;3:271–277. doi: 10.1093/glycob/3.3.271. [DOI] [PubMed] [Google Scholar]
- 30.Schenauer MR, Yu Y, Sweeney MD, Leary JA. CCR2 chemokines bind selectively to acetylated heparan sulfate octasaccharides. The Journal of biological chemistry. 2007 doi: 10.1074/jbc.M703387200. [DOI] [PubMed] [Google Scholar]
- 31.Abzalimov RR, Dubin PL, Kaltashov IA. Glycosaminoglycans as naturally occurring combinatorial libraries: developing a mass spectrometry-based strategy for characterization of anti-thrombin interaction with low molecular weight heparin and heparin oligomers. Analytical chemistry. 2007;79:6055–6063. doi: 10.1021/ac0710432. [DOI] [PubMed] [Google Scholar]
- 32.Fermas S, Gonnet F, Varenne A, Gareil P, Daniel R. Frontal analysis capillary electrophoresis hyphenated to electrospray ionization mass spectrometry for the characterization of the antithrombin/heparin pentasaccharide complex. Analytical chemistry. 2007;79:4987–4993. doi: 10.1021/ac070146h. [DOI] [PubMed] [Google Scholar]
- 33.Ruiz-Calero V, Moyano E, Puignou L, Galceran MT. Pressure-assisted capillary electrophoresis-electrospray ion trap mass spectrometry for the analysis of heparin depolymerised disaccharides. Journal of chromatography. 2001;914:277–291. doi: 10.1016/s0021-9673(00)01181-x. [DOI] [PubMed] [Google Scholar]
- 34.Pope RM, Raska CS, Thorp SC, Liu J. Analysis of heparan sulfate oligosaccharides by nano-electrospray ionization mass spectrometry. Glycobiology. 2001;11:505–513. doi: 10.1093/glycob/11.6.505. [DOI] [PubMed] [Google Scholar]
- 35.Zaia J, Costello CE. Tandem mass spectrometry of sulfated heparin-like glycosaminoglycan oligosaccharides. Analytical chemistry. 2003;75:2445–2455. doi: 10.1021/ac0263418. [DOI] [PubMed] [Google Scholar]
- 36.de Agostini AI, Dong JC, de Vantery Arrighi C, Ramus MA, Dentand-Quadri I, Thalmann S, Ventura P, Ibecheole V, Monge F, Fischer AM, HajMohammadi S, Shworak NW, Zhang L, Zhang Z, Linhardt RJ. Human follicular fluid heparan sulfate contains abundant 3-O-sulfated chains with anticoagulant activity. The Journal of biological chemistry. 2008;283:28115–28124. doi: 10.1074/jbc.M805338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chai W, Leteux C, Westling C, Lindahl U, Feizi T. Relative susceptibilities of the glucosamine-glucuronic acid and N-acetylglucosamine-glucuronic acid linkages to heparin lyase III. Biochemistry. 2004;43:8590–8599. doi: 10.1021/bi036250k. [DOI] [PubMed] [Google Scholar]
- 38.Sweeney MD, Yu Y, Leary JA. Effects of sulfate position on heparin octasaccharide binding to CCL2 examined by tandem mass spectrometry. Journal of the American Society for Mass Spectrometry. 2006;17:1114–1119. doi: 10.1016/j.jasms.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 39.Meissen JK, Sweeney MD, Girardi M, Lawrence R, Esko JD, Leary JA. Differentiation of 3-O-sulfated heparin disaccharide isomers: identification of structural aspects of the heparin CCL2 binding motif. Journal of the American Society for Mass Spectrometry. 2009;20:652–657. doi: 10.1016/j.jasms.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turnbull J, Powell A, Guimond S. Heparan sulfate: decoding a dynamic multifunctional cell regulator. Trends in cell biology. 2001;11:75–82. doi: 10.1016/s0962-8924(00)01897-3. [DOI] [PubMed] [Google Scholar]
- 41.Naimy H, Leymarie N, Bowman MJ, Zaia J. Characterization of heparin oligosaccharides binding specifically to antithrombin III using mass spectrometry. Biochemistry. 2008;47:3155–3161. doi: 10.1021/bi702043e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ziegler A, Zaia J. Size-exclusion chromatography of heparin oligosaccharides at high and low pressure. Journal of chromatography. 2006;837:76–86. doi: 10.1016/j.jchromb.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 43.Damus PS, Hicks M, Rosenberg RD. Anticoagulant action of heparin. Nature. 1973;246:355–357. doi: 10.1038/246355a0. [DOI] [PubMed] [Google Scholar]
- 44.Bjork I, Ylinenjarvi K, Olson ST, Bock PE. Conversion of antithrombin from an inhibitor of thrombin to a substrate with reduced heparin affinity and enhanced conformational stability by binding of a tetradecapeptide corresponding to the P1 to P14 region of the putative reactive bond loop of the inhibitor. The Journal of biological chemistry. 1992;267:1976–1982. [PubMed] [Google Scholar]
- 45.Conrad HE. Heparin Binding Proteins. Academic Press; New York: 1998. [Google Scholar]
- 46.Sasisekharan R, Raman R, Prabhakar V. Glycomics approach to structure-function relationships of glycosaminoglycans. Annual review of biomedical engineering. 2006;8:181–231. doi: 10.1146/annurev.bioeng.8.061505.095745. [DOI] [PubMed] [Google Scholar]
- 47.Gandhi NS, Mancera RL. The structure of glycosaminoglycans and their interactions with proteins. Chemical biology & drug design. 2008;72:455–482. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 48.Capila I, Linhardt RJ. Heparin-protein interactions. Angewandte Chemie (International ed. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 49.Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate Journal. 1988;5:397–409. [Google Scholar]
- 50.Desai UR, Wang HM, Linhardt RJ. Specificity studies on the heparin lyases from Flavobacterium heparinum. Biochemistry. 1993;32:8140–8145. doi: 10.1021/bi00083a012. [DOI] [PubMed] [Google Scholar]
- 51.HajMohammadi S, Enjyoji K, Princivalle M, Christi P, Lech M, Beeler D, Rayburn H, Schwartz JJ, Barzegar S, de Agostini AI, Post MJ, Rosenberg RD, Shworak NW. Normal levels of anticoagulant heparan sulfate are not essential for normal hemostasis. The Journal of clinical investigation. 2003;111:989–999. doi: 10.1172/JCI15809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L, Yoshida K, Liu J, Rosenberg RD. Anticoagulant heparan sulfate precursor structures in F9 embryonal carcinoma cells. The Journal of biological chemistry. 1999;274:5681–5691. doi: 10.1074/jbc.274.9.5681. [DOI] [PubMed] [Google Scholar]
- 53.Linhardt RJ, Turnbull JE, Wang HM, Loganathan D, Gallagher JT. Examination of the substrate specificity of heparin and heparan sulfate lyases. Biochemistry. 1990;29:2611–2617. doi: 10.1021/bi00462a026. [DOI] [PubMed] [Google Scholar]
- 54.Liu J, Pedersen LC. Anticoagulant heparan sulfate: structural specificity and biosynthesis. Applied microbiology and biotechnology. 2007;74:263–272. doi: 10.1007/s00253-006-0722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hitchcock AM, Yates KE, Costello CE, Zaia J. Comparative glycomics of connective tissue glycosaminoglycans. Proteomics. 2008;8:1384–1397. doi: 10.1002/pmic.200700787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nadanaka S, Kitagawa H. Heparan sulphate biosynthesis and disease. Journal of biochemistry. 2008;144:7–14. doi: 10.1093/jb/mvn040. [DOI] [PubMed] [Google Scholar]
- 57.Sanderson RD, Yang Y, Kelly T, MacLeod V, Dai Y, Theus A. Enzymatic remodeling of heparan sulfate proteoglycans within the tumor microenvironment: growth regulation and the prospect of new cancer therapies. Journal of cellular biochemistry. 2005;96:897–905. doi: 10.1002/jcb.20602. [DOI] [PubMed] [Google Scholar]
- 58.Kato M, Wang H, Bernfield M, Gallagher JT, Turnbull JE. Cell surface syndecan-1 on distinct cell types differs in fine structure and ligand binding of its heparan sulfate chains. The Journal of biological chemistry. 1994;269:18881–18890. [PubMed] [Google Scholar]
- 59.Jayson GC, Lyon M, Paraskeva C, Turnbull JE, Deakin JA, Gallagher JT. Heparan sulfate undergoes specific structural changes during the progression from human colon adenoma to carcinoma in vitro. The Journal of biological chemistry. 1998;273:51–57. doi: 10.1074/jbc.273.1.51. [DOI] [PubMed] [Google Scholar]
- 60.Shi X, Zaia J. Organ-specific heparan sulfate structural phenotypes. The Journal of biological chemistry. 2009;284:11806–11814. doi: 10.1074/jbc.M809637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brickman YG, Ford MD, Gallagher JT, Nurcombe V, Bartlett PF, Turnbull JE. Structural modification of fibroblast growth factor-binding heparan sulfate at a determinative stage of neural development. The Journal of biological chemistry. 1998;273:4350–4359. doi: 10.1074/jbc.273.8.4350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table with m/z of precursor ions isolated and their mass accuracies.
Supplementary Figure 1 showing the size exclusion chromatograms for heparin and HS digestions.
Supplementary Figure 2 comparing the MS and MS2 isotopic distributions of isolated precursor ions.