Abstract
We report a genome-wide association study of melanoma, conducted by GenoMEL, of 2,981 cases, of European ancestry, and 1,982 study-specific controls, plus a further 6,426 French and UK population controls, all genotyped for 317,000 or 610,000 SNPs. The analysis confirmed previously known melanoma susceptibility loci. The 7 novel regions with at least one SNP with p<10−5 and further local imputed or genotyped support were selected for replication using two other genome-wide studies (from Australia and Houston, Texas). Additional replication came from UK and Dutch case-control series. Three of the 7 regions replicated at p<10−3: an ATM missense polymorphism (rs1801516, overall p=3.4×10−9); a polymorphism within MX2 (rs45430, p=2.9×10−9) and a SNP adjacent to CASP8 (rs13016963, p=8.6×10−10). A fourth region near CCND1 remains of potential interest, showing suggestive but inconclusive evidence of replication. Unlike the previously known regions, the novel loci showed no association with nevus or pigmentation phenotypes in a large UK case-control series.
Cutaneous melanoma is predominantly a disease of fair-skinned individuals. Risk factors include a family history1, certain pigmentation phenotypes (notably the presence of fair skin, blue or green eyes, blond or red hair, sun sensitivity or an inability to tan2-5) and increased numbers of melanocytic nevi6,7. We previously reported Phase 1 of a genome-wide association (GWA) study of melanoma based on the Illumina 317k array8. This reinforced the importance of these genetically-determined melanoma-associated phenotypes, by showing the major common genetic determinants of risk in the populations considered were the MC1R locus (associated with red hair, freckling and sun sensitivity)4,5,9,10, tyrosinase (TYR) gene variants which code for skin color11 and a region near CDKN2A and MTAP which is associated with number of melanocytic nevi8,12. Furthermore we confirmed the importance of a haplotype spanning the agouti signaling protein locus (ASIP)11,13 and a second locus determining nevus count variation at 22q13 identified by a GWA study of nevus count12.
Both Phase 1 and Phase 2 of this study were carried out by the GenoMEL Consortium, a collaboration focusing on genetic susceptibility to melanoma. The study utilised samples collected by GenoMEL participants across populations of European ancestry living at different latitudes. In total, 14 GenoMEL groups contributed DNA samples from cases and controls of European (or Israeli) ethnicity (Supplementary Table 1). Phase 1 was based on 1,650 cases from Australian and European populations chosen to have a phenotype argued to “enrich” for genetic susceptibility (early onset, multiple primaries, or modest family history of melanoma). In Phase 2 a further 1,523 cases (1,211 of whom are genetically enriched: 532 with a family history, 277 with multiple primaries but no family history, and 402 with early disease onset but no multiple primaries or family history) and 1,112 controls were genotyped using the denser Illumina 610k array (see Supplementary Note). To optimise power for novel gene identification we combined the data from the two phases and performed an overall analysis. The Australian data used in Phase 1 were dropped from the combined Phase 1 + Phase 2 analysis as these samples are included in the Australian GWA study which formed one of the replication studies. After quality control was applied to SNPs and samples (see Supplementary Note), including Principal Components Analysis (PCA) to identify samples of non-European ethnicity (Supplementary Figure 1), the analysis utilised 2,804 cases (2,692 European and 112 Israeli) and 1,835 controls from GenoMEL studies and 5,783 controls from France and the UK Wellcome Trust Case-Control Consortium (WTCCC). A trend test, stratified by geographical region, was applied to each SNP (see Online Methods, also Figure 1). Little evidence was found of population stratification (λ=1.06, see Supplementary Figure 5).
Figure 1.
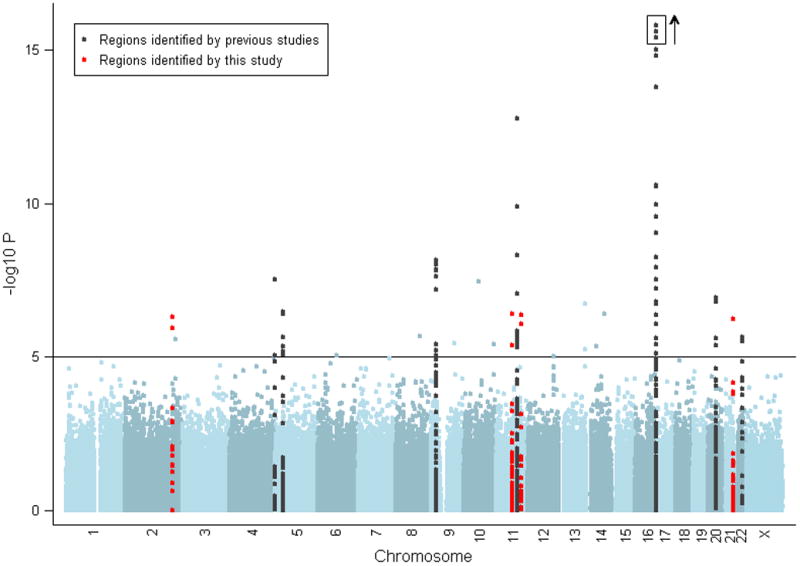
Manhattan plot of results of Cochran-Armitage (CA) trend test stratified by geographic region, with -log10 p-values shown. The solid horizontal line indicates a p-value of 10−5. Markers within 50kb of a SNP associated with melanoma are marked in black for those identified in a previous GWA and replicated here and marked in red if first identified in the current study. The y-axis is truncated at p=10−15, although three SNPs in the MC1R region have stronger p-values, up to 2.7×10−27, as signified by the box and arrow.
Strong evidence was found for the previously identified loci, (Supplementary Table 2, Supplementary Figures 2 and 3)8,11-18 and another pigmentation gene, SLC45A2, already reported to be associated with melanoma risk15. SLC45A2 is involved in melanosome maturation and pigmentation. The rs35390 SNP identified here is associated with melanoma15 and with variation in hair color15,20, in accordance with the observed pattern of known melanoma pigmentation risk factors2-5.
We also confirmed a role for SNP rs401681 in the region of TERT and CLMPT1L, which has also previously been shown to modify melanoma risk18 (Supplementary Table 2, Supplementary Figures 2 and 3)8,11-16,21. The confirmation of the SNP follows reported associations with risk of basal cell carcinomas, hematological malignancies and cancer of the bladder, cervix, lung, pancreas and prostate18,22. It was originally reported that the pattern of risk for melanoma was in the opposite direction to that for other cancers18, and we confirm this observation.
Seven further regions showed evidence of association with melanoma susceptibility (Table 1 and Supplementary Table 3). Replication was sought from two other GWA studies for the SNPs with the strongest evidence, preferentially SNPs common to all arrays. In regions with no SNP common to all platforms, we followed up both our top genotyped SNPs and the most significant imputed SNPs which had been genotyped in the replication studies. Further, these SNPs were genotyped in a replication sample set from the UK and the Netherlands (1,579 cases and 2,036 controls in total, see Supplementary Note). Table 1 contains the evidence from both the hypothesis-generating and replication datasets. Of these seven regions, three (on chromosomes 2 (rs13016963), 11 (rs1801516) and 21) showed strong evidence of replication (p<10−3), three (on chromosomes 2 (rs10932444), 12 and 13) showed no evidence of replication and one (on chromosome 11 (rs1485993)) showed marginal evidence of replication. Three of the loci showed overall combined evidence of association at p<5×10−8, based on the fixed effects meta-analysis.
Table 1.
Summary of results from this study for the 4 regions showing evidence of replication, listing each SNP under consideration, their position (in bp) and minor allele frequency (MAF); the per-allele OR (based on the minor allele) and p-value are given for this GWA study, for the meta-analysis of the replication data sets (from the Houston GWA study, the Australian GWA study and UK/Netherlands replication samples) and for the combined genome-wide and replication analyses. The Houston GWA study and the Australian study both used a different array to the current study for at least some samples, so some of their results presented here include imputed data. Further genotyping was conducted in the UK and Netherlands replication samples for SNPs with positive support from the GWA replication data. All meta-analyses are based on a fixed effects model with the exception of those for CCND1, marked with an asterisk, where random effects analysis was used because of the observed heterogeneity. Supplementary Table 3 is a fuller version of this table.
| SNP | Chromosome | Position | Allele | MAF | GenoMEL Genome-wide | Replication samples (genotyped + imputed) | Genome-wide plus replication samples (genotyped + imputed) | Postulated gene | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||
| OR | p | OR and 95% CI | P-value | OR and 95% CI | P-value | ||||||
| rs13016963 | 2 | 201852173 | A | 0.37 | 1.18 | 5.68 × 10−7 | 1.11 (1.06, 1.18) | 9.2 × 10−5 | 1.14 (1.09, 1.19) | 8.6 × 10−10 | CASP8 |
| rs1485993 | 11 | 69071595 | A | 0.37 | 1.19 | 4.15 × 10−7 | 1.07 (1.01, 1.13) | 0.017 | 1.11 (1.04, 1.18)* | 0.0012 | CCND1 |
| rs1801516 | 11 | 107680672 | A | 0.13 | 0.79 | 4.80 × 10−7 | 0.87 (0.78, 0.90) | 3.4 × 10−4 | 0.84 (0.78, 0.90) | 3.4 × 10−9 | ATM |
| rs45430 | 21 | 41667951 | G | 0.39 | 0.85 | 5.60 × 10−7 | 0.91 (0.86, 0.96) | 4.2 × 10−4 | 0.88 (0.85, 0.92) | 2.9 × 10−9 | MX2 |
Using random effects
The CASP8 region (chromosome 2) contains a number of SNPs showing evidence for association with melanoma risk; because of the lack of overlap in the SNPs across arrays, we report multiple SNPs either genotyped or imputed across platforms, all of which show evidence of association (Table 1, Supplementary Table 3, Figure 2). The strongest evidence for a single SNP is for rs700635 (p=2.4×10−9, OR=1.15 overall). All the SNPs are in the region of CASP8, which codes for a member of a family of proteases that play a critical role in the control of cell proliferation by inducing apoptotic cell death, making them candidate cancer susceptibility genes. A recent meta-analysis23 of 3 polymorphisms in CASP8 found that individuals with one or more copies of the D302H variant have a decreased risk of multiple types of cancer. In this study, the D302H variant could be imputed, but showed only marginal evidence of association (p=0.05), suggesting this is not a variant for melanoma. The evidence for melanoma risk was consistent across populations (Figure 3).
Figure 2.
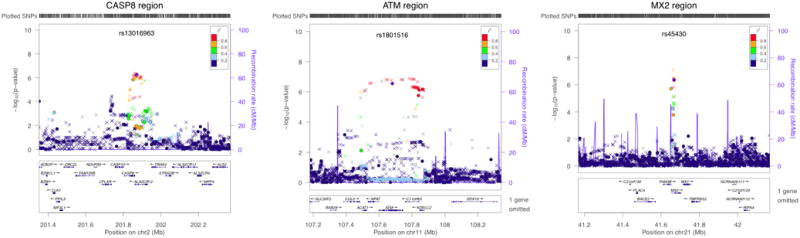
Stratified CA trend tests for the three replicated regions on chromosomes 2, 11 and 21. The log10 p-values are from the CA trend test (stratified by geographical region) for genotyped and imputed SNPs, indicated on the left-hand vertical axis. SNPs genotyped for all samples are plotted as circles, SNPs imputed for all samples as crosses and SNPs genotyped for some samples and imputed for others (due to chip differences) as squares. The most significant genotyped SNP is colored purple (with its name above) and the degree of LD between that SNP and the others is indicated by color according to the key (red being the greatest degree of LD). The estimated recombination rate is given by the blue line and indicated on the right-hand vertical axis. The genes in the region and their positions are given underneath the graph. Plots produced using LocusZoom19.
Figure 3.
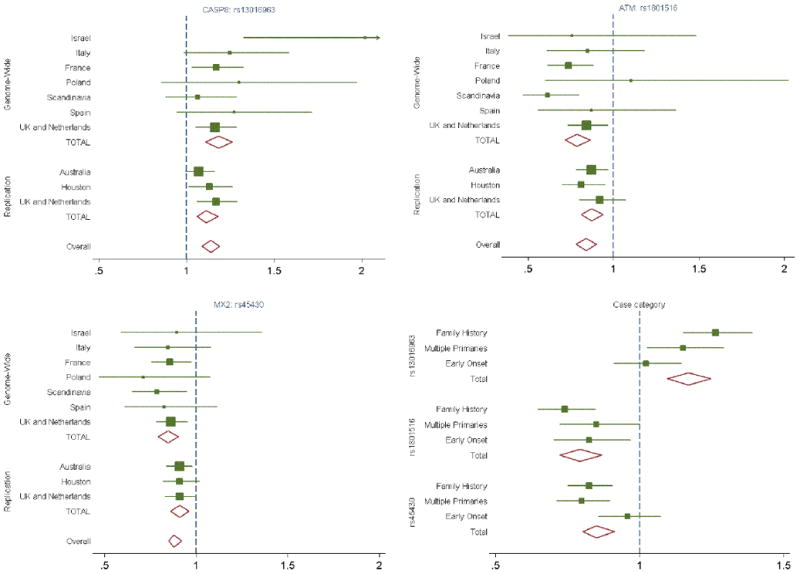
Forest plot of the per-allele OR for melanoma for SNPs in the 3 regions first identified in this study. Plots show the current evidence for effects by geography, in the genome-wide and replication samples, and by case type (family history, multiple primaries or early onset).
The ATM SNP rs1801516 (chromosome 11) (Table 1, Supplementary Table 3, Figure 2) is a missense mutation (D1853N, G > A) in a gene that codes for a protein which repairs double strand DNA breaks; association has been postulated between ATM and a number of cancer types24. For melanoma, the A allele is protective (p=3.4×10−9, OR=0.84 overall). Overall the evidence for melanoma is consistent across populations and case type, with no evidence of heterogeneity (Figure 3).
The third replicated region, around MX2 (chromosome 21), showed consistent effect sizes across the replication datasets (Table 1, Supplementary Table 3, Figure 2) and across populations (Figure 3). The SNP followed up in the replication study is rs45430 (p=2.9×10−9, OR=0.88 overall), which is intronic to MX2 and has not previously been associated with cancer susceptibility.
A fourth region, adjacent to CCND1 (chromosome 11), a proto-oncogene which is a key regulator of cell cycle progression, showed consistent effect sizes across all the replication sets (Table 1, Supplementary Table 3, Supplementary Figure 4), with best overall replication p-value of 0.011. However the replication sets produced a notably smaller OR (1.08) than the discovery set (1.19) (I2=0.507). This is potentially due to the well known Winner's Curse effect25 that causes the initial discovery set to overestimate the OR, leading in turn to a discrepancy between the overall p-value based on fixed effects and random effects meta-analysis (p=1.7×10−7 and p=0.00046 respectively). Thus, while we have strong support from this region, the evidence cannot be considered conclusive (see Supplementary Note for further details). However, further support comes from the interim analysis of a recently completed melanoma GWA study in494 melanoma cases and 5,628 controls from the Nurses' Health Study and Health Professionals Follow-up Study (OR=1.18 for rs1485993, p=0.014, unpublished data). This gene therefore remains a strong candidate, being well known in melanoma carcinogenesis26.
In Phase 1 of the study, all melanoma susceptibility loci identified were associated with either skin pigmentation or nevus count variation8. For the case-control samples from Leeds, UK, detailed nevus count and pigmentation information has been obtained for cases and controls27. Table 2 shows the association between nevus count, pigmentation and all SNPs associated with melanoma. (Note that not all SNPs show convincing evidence of melanoma association within the Leeds case-control samples, reflecting limited power). As expected, MC1R, SLC45A2, IRF4 and TYR are confirmed to be associated with pigmentation, while the rs4911442 SNP on chromosome 20 shows strong association with pigmentation, increasing the evidence that ASIP truly is the focus of this hit and implicating probable linkage disequilibrium (LD) with variants within an ASIP regulatory region. SNPs in the region of CDKN2A/MTAP and PLA2G6 are associated with nevus variation. The TERT/CLMPT1L SNP is also associated with nevus count variation, suggesting its effect on melanoma risk modification may be via this mechanism. We previously showed that IRF4 had a complex relationship with nevus count and melanoma risk14, and there are suggestions for SNPs in the CASP8 region of a relationship between genotype and nevus count in controls; among cases the association is not apparent (Table 2). Finally, the SNPs in the ATM and MX2 regions show no association with either nevus count or pigmentation, suggesting alternative, unknown mechanisms, although these require evaluation in other populations (see Supplementary Note).
Table 2.
Summary of results for nevus count/pigmentation/melanoma analyses from the Leeds case-control samples examining the 11 SNPs replicated for melanoma association in this or previous studies. Results are shown for (i) the proportion and significance of log nevus count variation explained by each SNP, adjusted for age and sex among cases and controls (adjusted for case-control status), (ii) the proportion and significance of case-control adjusted pigmentation variation score explained by each SNP, where the score is calculated from factor analysis of 6 correlated pigmentation phenotypes (see Online Methods), (iii) the association with melanoma risk (both as per-allele OR with 95%CI, and by genotype (compared to a baseline of the homozygote for the common allele)).
| Chromosomal Region | Postulated gene | SNP | MAF | % of variation in log nevus count explained by SNP | % of variation in pigmentation explained by SNP | Per-allele OR (95% CI) for risk of melanoma | OR (95% CI) for risk of melanoma with one copy of Minor allele | OR (95% CI) for risk of melanoma with two copies of Minor allele | ||
|---|---|---|---|---|---|---|---|---|---|---|
| R2 | P | R2 | P | |||||||
| 2q33-q34 | CASP8 | rs13016963 | 0.33 | 0.21 | 0.083* | 0.05 | 0.33 | 1.25 (1.07, 1.46) | 1.26 (1.01, 1.56) | 1.56 (1.11, 2.18) |
| 5p15.33 | TERT/CLMPT1L | rs401681 | 0.46 | 0.50 | 0.0070 | 0.13 | 0.11 | 1.08 (0.93, 1.25) | 1.15 (0.90, 1.47) | 1.15 (0.85, 1.55) |
| 5p13.2 | SLC45A2 | rs16891982 | 0.03 | 0.02 | 0.62 | 1.33 | 1.9 × 10−6 | 0.72 (0.44, 1.18) | 0.78 (0.47, 1.30) | NA |
| 6p25-p23 | IRF4 | rs12203592 | 0.24 | 0.21 | 0.084 | 2.76 | 5.6 × 10−12 | 0.80 (0.67, 0.95) | 0.72 (0.58, 0.91) | 0.81 (0.49, 1.35) |
| 9p21 | CDKN2A/MTAP | rs7023329 | 0.49 | 0.29 | 0.047 | 0.02 | 0.55 | 0.86 (0.73, 1.00) | 0.62 (0.47, 0.82) | 0.73 (0.53, 1.01) |
| 11q14-q21 | TYR | rs1393350 | 0.27 | 0.00 | 0.95 | 1.07 | 2.0 × 10−5 | 1.34 (1.14, 1.58) | 1.19 (0.96, 1.49) | 2.12 (1.41, 3.19) |
| 11q22-q23 | ATM | rs1801516 | 0.14 | 0.07 | 0.33 | 0.00 | 0.95 | 0.88 (0.71, 1.09) | 0.93 (0.73, 1.19) | 0.59 (0.29, 1.21) |
| 16q24.3 | MC1R | rs258322 | 0.10 | 0.00 | 0.81 | 4.00 | 9.0 × 10−17 | 1.83 (1.44, 2.32) | 1.71 (1.33, 2.22) | 7.14 (1.70, 29.98) |
| 20q11.2-q12 | ASIP | rs4911442 | 0.13 | 0.07 | 0.34 | 0.93 | 8.2 × 10−5 | 1.35 (1.08, 1.68) | 1.32 (1.03, 1.69) | 2.06 (0.85, 5.00) |
| 21q22.3 | MX2 | rs45430 | 0.38 | 0.00 | 0.80 | 0.05 | 0.32 | 0.90 (0.77, 1.05) | 0.97 (0.77, 1.22) | 0.77 (0.56, 1.07) |
| 22q13.1 | PLA2G6 | rs6001027 | 0.37 | 0.39 | 0.018 | 0.12 | 0.16 | 0.78 (0.66, 0.91) | 0.79 (0.63, 0.90) | 0.60 (0.42, 0.84) |
| TOTAL | 2.33 | 9.83 | ||||||||
p = 0.004 for controls only
Overall, we report 3 novel loci associated with melanoma risk, which achieve an overall significance level of 5×10−8 based on the fixed effects meta-analysis, and a potential fourth locus. The power to detect SNPs with effect sizes similar to those estimated from the replication studies is low, and we see many more SNPs in novel regions (from across the genome) reaching p-values between 10−4 and 10−5 than expected (68 with MAF>0.05 compared with an expected 46), suggesting that there may be many other genetic regions with a similar effect on melanoma risk (see Supplementary Note). Currently 11 loci have been identified (Table 2), with a suggestion that 5 of these regions act through the pigmentation phenotype and at least 3 through the nevus phenotype, reflecting the major phenotype-associated risk factors for melanoma. Interestingly, at least two of the newly identified loci appear to influence risk through a novel mechanism, opening up potential new directions for melanoma research.
Supplementary Material
Acknowledgments
The authors are extremely grateful for the contributions of Daniela Seminara to the work of GenoMEL and also for the support of Geoff Cross (University of Leeds) for maintaining the study wiki. Overall, the GenoMEL Consortium is indebted to the organisational skills of Paul Affleck.
This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113.
The authors thank the EGEA cooperative group for having given access to data on the EGEA (Epidemiological study on the Genetics and Environment of Asthma) study. We acknowledge the biological specimens of the French Family study group were obtained from Institut Gustave Roussy and Fondation Jean Dausset-CEPH Biobanks.
Financial Support: The GenoMEL study was funded by the European Commission under the 6th Framework Programme, contract no: LSHC-CT-2006-018702; by Cancer Research UK Programme Awards, C588/A4994 and C588/A10589, and Cancer Research UK Project Grant C8216/A6129; and by US National Institutes of Health R01 ROI CA83115. This research was also supported by the Intramural Research Program of the NIH, NCI, DCEG.
Australia: Melanoma Research Alliance, the National Institutes of Health/National Cancer Institute (CA88363, CA83115, CA122838, CA87969, CA055075, CA100264, CA133996 and CA49449), the National Health and Medical Research Council of Australia (NHMRC) (107359, 200071, 241944, 339462, 380385, 389927, 389875, 389891, 389892, 389938, 402761, 443036, 442915, 442981, 496610, 496675, 496739, 552485, 552498), the Cancer Councils NSW, Victoria and Queensland, the Cancer Institute New South Wales, the Cooperative Research Centre for Discovery of Genes for Common Human Diseases (CRC), Cerylid Biosciences(Melbourne) and the Australian Cancer Research Foundation.
Cambridge: Cancer Research UK awards: C8197/A10123, C8197/A10865 & C1287/A10118
Emilia-Romagna: National Cancer Institute grants RO1 CA5558 to Maria Teresa Landi.
Genoa: IRCSS 2007 Italian Ministry of Health DGRST.4/4235-P1.9.A.B, Fondazione CARIGE, PRIN 2008 to G.B.-S.
Houston: NIH awards to MD Anderson RO1CA100264 and RO1CA33996.
Leeds: Cancer Research UK Programme grants Genetic epidemiology of cancer C588/A10589, C588/A4994 and Cancer Research UK Project grant C8216/A6129.
Leiden: Grant BBMRI CO18
Lund: Swedish Cancer Society, Swedish Research Council, Region Skåne funds, Kamprad Foundation.
Norway: Grants from the Comprehensive Cancer Center, Oslo University Hospital and the University of Bergen.
Paris: Grants from Institut National du Cancer (INCa-PL016) and Ligue Nationale Contre Le Cancer (PRE05/FD and PRE 09/FD) to Florence Demenais, Programme Hospitalier de Recherche Clinique (PHRC 2007 / AOM-07-195) to Marie-Françoise Avriland Florence Demenais, Institut National du Cancer (Melanoma Network RS Number 13), Association pour la Recherche sur le Cancer (ARC N°A09/5/5003), and Société Française de Dermatologie (SFD2009) to Brigitte Bressac-de Paillerets. Brigitte Bressac-de Paillerets has been awarded an Inserm Research Fellowship for hospital-based scientists.
Utah: NIH award to L. A. Cannon Albright RO1CA102422.
Footnotes
Author Contributions: J.H.B. and M.M.I. led and carried out the statistical analysis, contributed to the design of the study and were members of the writing team; M.H. contributed to the design of the study and contributed genotyping information; J.C.T. carried out statistical analyses and was a member of the writing team; J.F.A., P.A.A., L.A.A., B.K.A., M.-F.A., E.A., W.B., D.C., A.E.C., D.D., A.D., D.F.E., E.F., P. Ghiorzo, G.G.G., M.H., V.H., C.I., M.A.J., G.J., G,L., M.T.L., J. Lang, R.M., J.M., N.G.M., A.M., G.W.M., S.N., L.P., J.A.P.-B., R.T., N. vander S. and D.C.W. contributed to the identification of suitable samples for the study; B.B. contributed to the design of the study and supervised the initial processing of samples; G.B.-S., K.M.B., B.B.-de P., L.A.C.-A., T.D., D.E.E., J.H., J.L.H., R.F.K., J. Lubiński, F.A. van N., H.O., S.P. and P.V.B. contributed to the design of the study; H.S. carried out genotyping and contributed to the interpretation of genotyping data; P. Galan, B.J., J.R.-M. and D.Z. contributed to the interpretation of genotyping data; J.H. contributed results of a confirmatory study; C.I.A., S.F., J.E.L. and Q.W. led and contributed analyses from the Houston study; N.K.H., G.J.M. and S.M. led and contributed results from the Australian study; G.M.L. contributed genotyping information and to the interpretation of genotype data; F.D., P.A.K., E.C., A.M.G. and E.M.G. advised on statistical analysis and contributed to the design of the study; N.A.G. was consortium deputy lead and contributed to the design of the study; J.A.N.B. was overall consortium lead and contributed to the design of the study; D.T.B. led the analysis group, contributed to the design of the study and was a member of the writing team.
URLs: GenoMEL www.genomel.org
EGEA (Epidemiological study on the Genetics and Environment of Asthma) study, http://cesp.vjf.inserm.fr/∼egeanet
References
- 1.Cannon-Albright LA, Bishop DT, Goldgar C, Skolnick MH. Genetic predisposition to cancer. Important Adv Oncol. 1991:39–55. [PubMed] [Google Scholar]
- 2.Naldi L, et al. Cutaneous malignant melanoma in women. Phenotypic characteristics, sun exposure, and hormonal factors: a case-control study from Italy. Ann Epidemiol. 2005;15:545–50. doi: 10.1016/j.annepidem.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Titus-Ernstoff L, et al. Pigmentary characteristics and moles in relation to melanoma risk. Int J Cancer. 2005;116:144–9. doi: 10.1002/ijc.21001. [DOI] [PubMed] [Google Scholar]
- 4.Holly EA, Aston DA, Cress RD, Ahn DK, Kristiansen JJ. Cutaneous melanoma in women. I. Exposure to sunlight, ability to tan, and other risk factors related to ultraviolet light. Am J Epidemiol. 1995;141:923–33. doi: 10.1093/oxfordjournals.aje.a117359. [DOI] [PubMed] [Google Scholar]
- 5.Holly EA, Aston DA, Cress RD, Ahn DK, Kristiansen JJ. Cutaneous melanoma in women. II. Phenotypic characteristics and other host-related factors. Am J Epidemiol. 1995;141:934–42. doi: 10.1093/oxfordjournals.aje.a117360. [DOI] [PubMed] [Google Scholar]
- 6.Bataille V, et al. Risk of cutaneous melanoma in relation to the numbers, types and sites of naevi: a case-control study. Br J Cancer. 1996;73:1605–11. doi: 10.1038/bjc.1996.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang YM, et al. A pooled analysis of melanocytic nevus phenotype and the risk of cutaneous melanoma at different latitudes. Int J Cancer. 2009;124:420–8. doi: 10.1002/ijc.23869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bishop DT, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41:920–5. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raimondi S, et al. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int J Cancer. 2008;122:2753–60. doi: 10.1002/ijc.23396. [DOI] [PubMed] [Google Scholar]
- 10.Kanetsky PA, et al. Population-based study of natural variation in the melanocortin-1 receptor gene and melanoma. Cancer Res. 2006;66:9330–7. doi: 10.1158/0008-5472.CAN-06-1634. [DOI] [PubMed] [Google Scholar]
- 11.Gudbjartsson DF, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008;40:886–91. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 12.Falchi M, et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat Genet. 2009;41:915–9. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown KM, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40:838–40. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duffy DL, et al. IRF4 variants have age-specific effects on nevus count and predispose to melanoma. Am J Hum Genet. 2010;87:6–16. doi: 10.1016/j.ajhg.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duffy DL, et al. Multiple pigmentation gene polymorphisms account for a substantial proportion of risk of cutaneous malignant melanoma. J Invest Dermatol. 2010;130:520–8. doi: 10.1038/jid.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guedj M, et al. Variants of the MATP/SLC45A2 gene are protective for melanoma in the French population. Hum Mutat. 2008;29:1154–60. doi: 10.1002/humu.20823. [DOI] [PubMed] [Google Scholar]
- 17.Nan H, Kraft P, Hunter DJ, Han J. Genetic variants in pigmentation genes, pigmentary phenotypes, and risk of skin cancer in Caucasians. Int J Cancer. 2009;125:909–17. doi: 10.1002/ijc.24327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rafnar T, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–7. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han J, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez LP, et al. SLC45A2: a novel malignant melanoma-associated gene. Hum Mutat. 2008;29:1161–7. doi: 10.1002/humu.20804. [DOI] [PubMed] [Google Scholar]
- 22.Baird DM. Variation at the TERT locus and predisposition for cancer. Expert reviews in molecular medicine. 2010;12:e16. doi: 10.1017/S146239941000147X. [DOI] [PubMed] [Google Scholar]
- 23.Yin M, Yan J, Wei S, Wei Q. CASP8 polymorphisms contribute to cancer susceptibility: evidence from a meta-analysis of 23 publications with 55 individual studies. Carcinogenesis. 2010;31:850–7. doi: 10.1093/carcin/bgq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nature reviews Molecular cell biology. 2008;9:759–69. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 25.Garner C. Upward bias in odds ratio estimates from genome-wide association studies. Genetic epidemiology. 2007;31:288–95. doi: 10.1002/gepi.20209. [DOI] [PubMed] [Google Scholar]
- 26.Curtin JA, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 27.Newton-Bishop JA, et al. Melanocytic nevi, nevus genes, and melanoma risk in a large case-control study in the United Kingdom. Cancer Epidemiol Biomarkers Prev. 2010;19:2043–54. doi: 10.1158/1055-9965.EPI-10-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


