Abstract
Williams Syndrome Transcription Factor (WSTF) has emerged as an incredibly versatile nuclear protein. WSTF and the ATP-dependent chromatin remodeling complexes in which it exists, WINAC, WICH and B-WICH have been studied in a variety of organisms. This research has revealed roles for WSTF in a number of diverse molecular events. WSTF function includes chromatin assembly, Pol I and III gene regulation, vitamin D metabolism and DNA repair. In addition to functioning as a subunit of several ATP-dependent chromatin remodeling complexes, WSTF binds specifically to acetylated histones and is itself a histone kinase, as well as a target of phosphorylation. This review will describe the three known WSTF-containing complexes and discuss their various roles as well as mechanisms of regulating WSTF activity.
Keywords: WSTF, WICH, WINAC, B-WICH, ATP-dependent Chromatin Remodeling Complex, Vitamin D, DNA Repair, H2A.X, Tyrosine 142
ATP-dependent Chromatin Remodeling Complexes
The eukaryotic genome exists in a highly compact form inside the nucleus of each cell. The nucleosome is the foundation of this condensed DNA structure within the cell. The nucleosome consists of 147 base pairs of DNA wrapped about 1.7 times around a histone octamer. Each histone octamer is comprised of two copies of histone H2A, H2B, H3 and H4 (Luger et al. 1997). The compaction state of DNA is dynamic, and regions of chromatin can exhibit vastly different folding levels during the cell cycle and during transcription and repair. Heterochromatin displays far less transcriptional activity than euchromatin and formation is closely associated with long term gene silencing (Rizzi et al. 2004). Heterochromatin formation creates a physical barrier between DNA and transcription machinery that acts to suppress gene expression (Grewal and Jia 2007). Nucleosomes within euchromatic regions also play an important role in gene regulation (Wyrick et al. 1999). Researchers performing high-density tiling arrays discovered that promoters typically reside in nucleosome-free regions, providing evidence that nucleosomes are not assembled onto DNA at random, or even in the most energy favorable conformation (Yuan et al. 2005). The current understanding of chromatin formation is that nuclesomes not only exist to organize and package DNA, but are also specifically positioned along the genome to help regulate gene expression (reviewed in Venters and Pugh 2009).
For molecular machinery involved in transcription, replication and repair to gain access to DNA, nucleosomes must first be displaced to expose specific segments of DNA. Nucleosomal movement and nucleosomal eviction facilitate this exposure, allowing for molecular machinery to directly associate with the DNA (Vignali et al. 2000). The molecular motors responsible for nucleosome positioning and displacement are known as ATP-dependent chromatin remodelers. These nuclear complexes utilize the energy of ATP hydrolysis for assembly, repositioning, or eviction of nucleosomes, therefore exposing (or covering) specific sections of DNA as necessary for transcription, replication, or repair (reviewed in Venters and Pugh 2009). All ATP-dependent chromatin enzymes are a part of the helicase super family 2 (SF2) because of structural similarities between the ATPase and the helicase domains (Bouazoune and Brehm 2006), though these complexes do not exhibit conventional helicase activity. Chromatin remodeling complexes are classified into subfamilies based on the specific ATPase the complex utilizes. SWI/SNF and ISWI are two of the most extensively studied subfamilies of ATP-dependent remodelers. SWI/SNF and ISWI complexes are thought to achieve nucleosome displacement in different ways. SWI/SNF remodelers expose gene segments by creating DNA loops without transferring the nucleosomes to another region (Fan, et al. 2003). ISWI complexes widen inter-nucleosomal regions by sliding nucleosomes, creating longer segments of linker DNA (reviewed in Gangaraju and Bartholomew 2007). SWI/SNF complexes have both activation and repressive functions and tend to be larger complexes, containing up to 15 subunits. ISWI complexes generally contain fewer subunits, often 2–4 and are commonly involved in remodeling chromatin to repress gene activity (reviewed in Dirscherl and Krebs 2004; Gangaraju and Bartholomew 2007; Lall 2007).
Histone modifications
Chromatin remodelers typically lack the innate ability to recognize specific DNA sequences and therefore are directed to their sites of action by sequence-specific DNA binding proteins and by histone marks left by histone modifying enzyme complexes (Heinzel et al. 1997; Yanagisawa et al. 2002). These enzyme complexes possess the ability to add or remove post-translational modifications primarily to the flexible terminal tails of histone proteins, though modifications within the histone core domains also occur. The vast array of modifications that have been identified include phosphorylation, acetylation, methylation, ubiquitination, sumoylation, ADP-ribosylation, deimination and proline isomerization (the last being a unique case of a non-covalent modification; (reviewed in Kouzarides 2007)).
These modifications can function by altering the chromatin structure itself, or by the recruitment of other nuclear proteins and complexes to specific sites of the genome. A classic example of a histone modification resulting in the recruitment of nuclear machinery is γ-H2A.X. H2A.X is a variant of histone H2A shown to play a crucial role in DNA damage response (Xiao et al. 2009). γ-H2A.X refers to the phosphorylated state of a specific serine residue, serine 139 in most multicellular organisms (S129 in yeast). This specific modification of H2A.X is essential for damage focus formation at sites of DNA double strand breaks (Rogakou et al. 1998; Rogakou et al. 1999; Paull et al. 2000). γ-H2A.X is recognized by MDC1 (Mediator of DNA damage checkpoint protein 1), a mediator protein that recruits a number of other proteins that help coordinate the assembly of DNA damage response (DDR) proteins (Stucki 2009). A number of remodeling complexes also appear to be recruited to sites of DNA damage. INO80 is remodeling complex that binds γ-H2A.X in yeast and is believed to be responsible for the nucleosomal displacement at sites of double stand breaks (Morrison et al. 2004; Wong et al. 2006). The interaction between modified histones and chromatin remodelers allows for the manipulation of nucleosomes in a targeted region. This interplay allows remodelers to establish widespread chromatin conformations (euchromatin and heterochromatin) as well as coordinate the regulation of specific gene availability (reviewed in Kouzarides 2007).
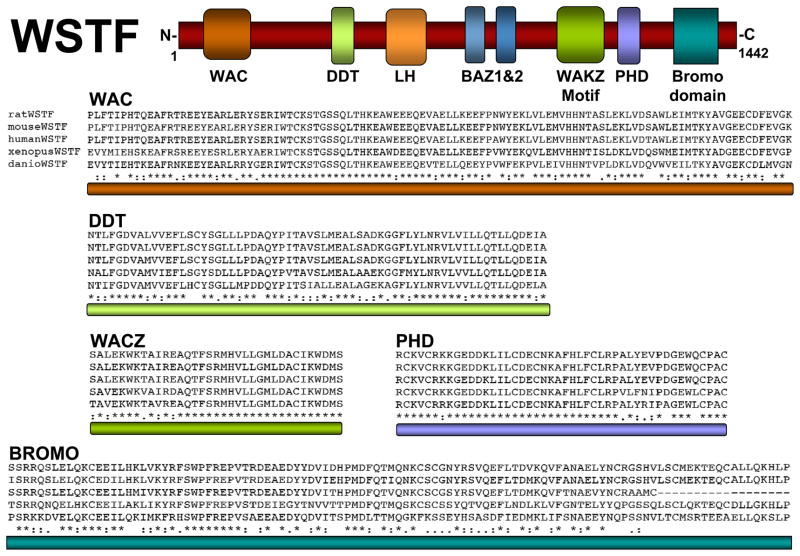
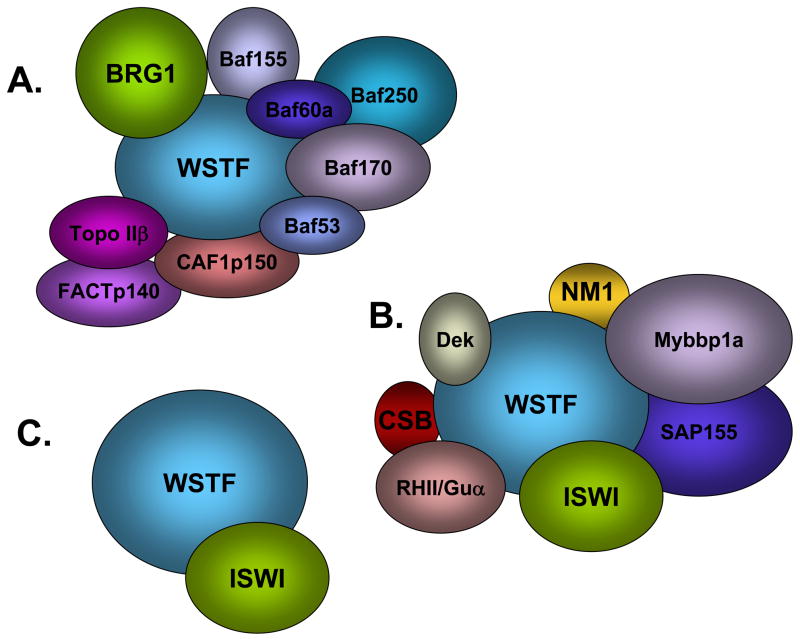
This review will focus on WSTF (Williams Syndrome Transcription Factor; Figure 1) and the functions of the three chromatin remodeling complexes in which it exists: WINAC, B-WICH and WICH (shown schematically in Figure 2). WSTF is a versatile nuclear protein that plays roles in both chromatin remodeling and covalent histone modification.
Figure 1.
Conserved motifs in WSTF. Top: schematic diagram of WSTF depicting the relative location of conserved protein domains LH, WAC, DDT, BAZ1, BAZ2, WAKZ, PHD and bromodomain. Bottom: sequence alignments of the specified domains illustrate the high degree of conservation among multiple species. “*” the residues or nucleotides in that column are identical in all sequences in the alignment; “: “ conserved substitutions have been observed; “.” semi-conserved substitutions are observed.
Figure 2.
WSTF-containing chromatin remodeling complexes. (A) WINAC. WINAC contains SWI/SNF ATPases Brg1 and Brm, 5 BAF (Brg-1 associated factor) subunits (BAF53, 60, 70, 155 and 250), TopoIIβ, FACT p140, CAF1 p150 and WSTF. (B) B-WICH. The B-WICH complex includes Myb-binding protein-1a (Myb-bp1a), Sf3b155/SAP155, Gu alpha, CSB, the proto-oncogene Dek, and nuclear myosin 1. B-WICH also includes several ribosomal RNAs 5 S rRNA, 7SL RNA, and 45 S rRNA. (C) WICH. WICH complex contains WSTF (Williams Syndrome Transcription Factor) and ISWI (Imitation Switch).
WSTF and Williams Syndrome (WS)
WSTF is a known subunit of three ATP-dependent chromatin remodelers: WINAC (WSTF including the nucleosome assembly complex), WICH (WSTF-ISWI chromatin remodeling complex) and B-WICH. The gene that codes for WSTF (WBSCR) was first identified by researchers studying a 1.5 megabase heterozygous deletion of roughly twenty genes on chromosome seven, seen in all patients with Williams Syndrome (Ewart et al. 1994). WS is a rare autosomal dominant hereditary disorder that occurs in about 1 of every 20,000 live births (Lu et al. 1998). Patients suffering from Williams Syndrome may present a number of systemic defects including characteristic personality and facial appearance, growth deficiency, mental retardation, aortic stenosis, heart disease and infantile hypercalcaemia (Bellugi et al. 1990; Morris et al. 1988; Pober 2010). Although over 20 other genes reside within the chromosome 7q11.23 deletion, studies in Xenopus, mouse and tissue culture have lead researchers to believe that WSTF heterozygosity plays an important role in a number of the systemic defects seen in WS patients (Bozhenok et al. 2002; Cus et al. 2006; Yoshimura et al. 2009).
WSTF Expression and Role in Development
The first examination of WSTF expression was done by northern blot analysis of human tissue. This revealed a 7.5-kb transcript expressed in all adult tissues (heart, brain, placental, skeletal, muscle and ovary) and four fetal tissues (brain, lung, kidney and liver) examined in non-WS individuals (Lu et al. 1998). Due to the diversity of defects seen in patients with WS, researchers have investigated WSTF expression during several stages of vertebrate development. In situ hybridization assays of developing Xenopus laevis embryos reveal that WSTF mRNA is detected in Xenopus oocytes and is ubiquitous until stage 16 (about 24 hours post-fertilization). WSTF expression first begins to localize to the closing neural tube. As neural tissue continues to differentiate into early tadpole stage, WSTF mRNA localizes to specific neural structures and cells including the neural tube, migrating head neural crest cells, anterior brain and optic cup ((Cus et al. 2006) and our unpublished data). WSTF expression is strongly detected in the optic cup of Xenopus embryos, but absent within the developing lens epithelium (Cus et al. 2006). In Xenopus embryos at tadpole stage (stages 32–39), WSTF expression within the optic cup becomes limited to the ciliary marginal zone, a regenerative structure absent in higher order vertebrates (Kubota et al. 2002). Also at this stage WSTF mRNA is strongly detected in the mid and hindbrain as well as the tectum and isthmus, structures shown to have organizing activity during brain development (Cus et al. 2006; Nakamura et al. 2005). Strong WSTF expression is detected within the migrating head neural crest cells and also within the branchial arches. The branchial or pharyngeal arches are a primordium for a number of organs and facial structures, such as the thyroid, aorta, the maxillary and mandibular bones, ligaments, and nerves as well as auditory structures like the auditory tube and the epithelium around the ear. WSTF mRNA is also faintly detected in the tailbud of these tadpole embryos. WSTF transcript detection persists within these defined domains until late tadpole stage (stage 39), where expression begins to fade until WSTF transcripts are no longer detectable by in situ hybridization (stage 42) (Cus et al. 2006). WSTF expression has been shown to have significant overlap with members of both WINAC and WICH complexes in Xenopus laevis (Dirscherl et al. 2005; Linder et al. 2004). However, researchers have also shown a distinct difference between the expression patterns of WSTF and WICH complex member ISWI in certain developmental stages of mice. This may reveal important differences in the tissues where the WICH and WINAC complexes assemble and highlight the potential for unique roles for these two WSTF-containing remodeling complexes during development (Kitagawa et al. 2003). Researchers have also noticed strong overlap of WSTF mRNA with Wnt pathway ligand Wnt-4 and transmembrane Wnt receptor frizzled-3 (Frz-3) during Xenopus development (Cus et al. 2006; Lyuksyutova et al. 2003). Both Wnt-4 and Frz-3 are expressed in domains similar to that of WSTF and both are neural-specific genes shown to be involved in eye development (Maurus et al. 2005; Rasmussen et al. 2001).
WSTF is an essential gene in mice as WSTF homozygous knockouts (WSTF−/−) develop with smaller body size and die just days after birth. WSTF−/− embryos show significant heart defects and 10% of WSTF+/− neonatal mice also display specific heart defects such as altered structure, expansion of the septal defects and hypertrophy of ventricles similar to those seen in WS patients (Yoshimura et al. 2009).
Whole mount in situ hybridizations reveal that WSTF mRNA is detectable in the headfolds, presumptive hindbrain and in the caudal tail bud of mice at 8.25 days postcoitum (dpc). At 9.5 dpc, WSTF mRNA is detected in the somites as well as the budding forelimb. WSTF expression is also present in the mesenchymal tissue of brachial arches 1 and 2, which contribute greatly to maxillary and mandibular structures of the face. WSTF expression can also be detected in the frontonasal process at 9.5 dpc as well as in the mesenchyme of medial and lateral prominences from 10.5 – 11.5 dpc (Ashe et al. 2008). These studies reveal robust WSTF expression in a number of structures critical for facial development throughout early embryogenesis in mice.
A WSTF mutant was obtained in a screen in mice for Modifiers of murine metastable epialleles (Mommes) (Ashe et al. 2008). The MommeD10 mutation was identified as a single amino acid change (L733R) within the WSTF protein. Even though WSTF transcript levels are normal in MommeD10 mutant mice, this mutation appears to disrupt protein stability as WSTF protein levels are severely diminished. Unlike WSTF−/− null animals, MommeD10 mice are able to survive and at 4 weeks exhibit an altered skull shape that includes protruding foreheads, shorter snouts, flattened nasal bone, and upward curvature of the nasal tip (Ashe et al. 2008). These craniofacial features are stirkingly similar to those of WS patients. This similarity, as well as the detection of WSTF expression in structures known to be crucial for facial morphology in both mice and Xenopus laevis strongly suggest that reduced expression WSTF may be responsible for the characteristic “elfin” features seen in WS individuals.
WSTF Protein
The WSTF protein falls into the BAZ/WAL family, a group of proteins that include six specific motifs found in sequential order (Jones et al. 2000; Poot et al. 2000): an LH motif, a proposed transactivation domain (Quong et al. 1993), BAZ 1, which mediates WSTF association with ISWI, BAZ 2 motifs, a WAKZ motif (Ito et al. 1999; Jones et al. 2000; Quong et al. 1993), a PHD finger, and a bromodomain (Jones et al. 2000). WSTF also contains an N-terminal WAC domain that does not exist in all BAZ proteins (Figure 1).
Studies with HeLa cells reveal that WSTF shows close sequence homology with a number of known subunits of other ISWI complexes, many of which fall in the BAZ/WAL family (Bochar et al. 2000). These non-catalytic subunits are believed to facilitate specific interaction and functions with chromatin (He et al. 2008). For example, ACF1 enhances nucleosome sliding and chromatin assembly by ISWI in Xenopus and ACF1-deficient cells show a reduction in chromatin bound ISWI in Drosophila (Guschin et al. 2000; Ito et al. 1999; Yoshimura et al. 2009).
Several protein domains in WSTF are implicated in binding and/or modification of chromatin. These include the PHD (Plant homeodomain) finger, the bromodomain (Bochar et al. 2000) and a histone kinase activity composed of the WAC domain and additional N-terminal sequences (Xiao et al. 2009). Proteins that contain PHD fingers, which are common methyl-lysine binding motifs, tend to have a role in transcription and are therefore almost exclusively located in the nucleus. hWSTF contains a single PHD finger that is about 50 amino acids in length and is defined by a Cys4-HisCys3 zinc binding motif (Aasland et al. 1995; Pascual et al. 2000). The hWSTF PHD finger is a β-β-α core metal-dependent folding motif that specifically binds two zinc ions (Pascual et al. 2000). The PHD finger of BPTF (bromodomain and PHD domain transcription factor), a subunit of the ISWI containing chromatin remodeling complex NURF (nucleosome remodeling factor), preferentially binds trimethylated lysine 4 on histone 3 (H3K4me3), a histone modification that correlates with the starts sites of active genes (Li et al. 2006). The PHD finger of the Mi-2 subunit of NuRD (nucleosome remodeling deacetylase), another ISWI containing complex, reveals an interaction with histone deacetylase and a connection with gene repression (Zhang et al. 1998). The specific binding target of the WSTF PHD finger remains unknown but is likely to play a role in targeting WSTF complexes.
Adjacent to the PHD finger of WSTF is a bromodomain. The bromodomain was first described as an acetylated lysine-binding domain in the histone acetyltransferase P/CAF (p300/CBP-associated factor) (Dhalluin et al. 1999). Acetylated lysine is an essential modification of histone tails that is important in gene activation (Winston and Allis 1999). The WSTF bromodomain binds acetylated lysine 14 on histone 3 (AcH3K14) and exerts a transrepressive function on gene activation (Kato et al. 2007). The results of this transrepressive activity will be described in further detail later in this review.
BPTF, like ACF1, WSTF and WCRF180 are all members of different ISWI complexes, each containing both a PHD finger and a bromodomain, suggesting that they may have similar functional roles (Bochar et al. 2000; Ito et al. 1999; Li et al. 2006; Strohner et al. 2001). The ability of these domains to recognize and bind specific modifications on histone tails provides these proteins the capacity to distinguish between nucleosomes and decipher the epigenetic marks of the histone code (Ragvin et al. 2004). It is likely that these domains facilitate these functions of WSTF and contribute greatly to its role within a complex.
In addition to the motifs within WSTF that may facilitate its binding to specific histone marks, WSTF can also covalently modify histone targets. A recent discovery revealed an intrinsic tyrosine kinase activity of the WAC domain (plus an additional N-terminal region) of WSTF in cultured MEF (mouse embryonic fibroblast) cells (Xiao et al. 2009). The WAC domain is not a motif that exists in all BAZ/WAL proteins (Bozhenok et al. 2002). This kinase domain has no sequence homology to any other known kinases, and has a very interesting target. WSTF phosphorylates tyrosine 142 of histone H2A.X and this modification plays a crucial role in the DNA damage response. This is described further below.
Post-translational modifications of WSTF
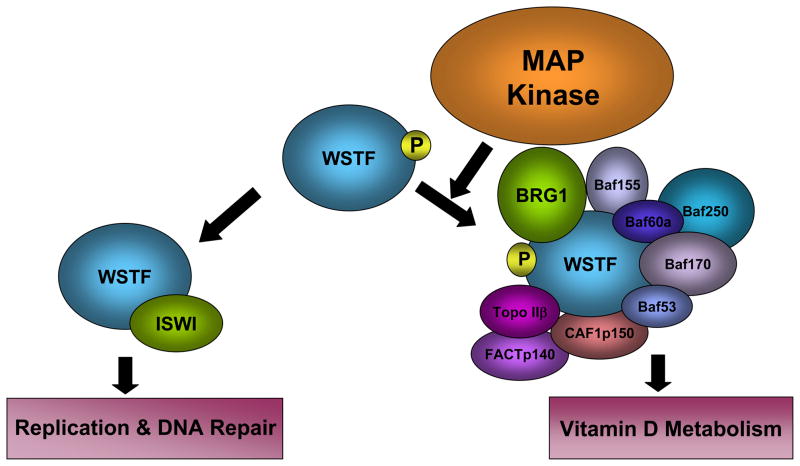
In addition to its ability to both recognize and catalyze histone modifications, WSTF itself is post-translationally modified. The WSTF protein has recently been shown to be a target of MAP kinases both in vitro and in vivo (Oya et al. 2009). MAPK phosphorylates WSTF protein within the WAC domain at serine 158 (S158). While WSTF exists in both the phosphorylated and unphosphorylated form in vivo, researchers have shown that an unphosphorylatable WSTF mutant (WSTF S158A) displays a significantly reduced affinity for other components of the WINAC chromatin remodeling complex. This mutation also results in reduced WINAC transactivation and repression activity in the breast cancer cell line MCF7 (Oya et al. 2009). Interestingly, WSTF S158A does not disrupt the association with ISWI and appears not to alter survival of cultured mouse embryonic fibroblast (MEF) cells upon treatment with MMS. This finding reveals a post-translational modification of WSTF that is important for WINAC assembly and function, but not for WICH (Figure 3). It is possible that a MAPK-dependent switch mechanism exists between WICH and WINAC assembly through the post-translational phosphorylation of the WSTF WAC domain, and that WINAC-dependent chromatin remodeling is at least partly under MAPK control (Oya et al. 2009). It is intriguing that the WAC domain is essential for WSTF’s H2A.X kinase activity; it will be interesting to see whether S158 phosphorylation impacts the kinase activity of WSTF.
Figure 3.
MAPK targets WSTF. A schematic representation of MAP kinase phosphorylation of WSTF. MAP kinase phosphorylates WSTF on Serine 158 within the WAC domain. WSTF phosphorylation is important for WSTF integration into the WINAC complex, but appears to be dispensable for assembly of the WICH complex.
WINAC
WSTF is one of 13 subunits that make up the ATP-dependent chromatin remodeling complex WINAC (WSTF including nucleosome assembly complex) (Figure 2A) (Kitagawa et al. 2003). This complex contains SWI/SNF ATPases Brg1 and Brm, a number of BAF (Brg1- associated factor) subunits, as well as TopoIIβ, FACT p140, and CAF-1 p150 (Kitagawa et al. 2003). Topoisomerase II (Topo II) is a nuclear enzyme largely responsible for the remove helical tension of DNA (Salceda et al. 2006). FACT p140 is a subunit of the FACT (Facilitates chromatin transcription) complex known to be required for Pol II transcript elongation (Belotserkovskaya et al. 2003). CAF-1p150 is a subunit of the multiprotein complex CAF-1, known for its role in chromatin assembly during DNA replication (Smith and Stillman 1989).
Knockdown experiments of WSTF and Brg1/hBrm expression via RNA interference in MCF7 cells has shown that WINAC may have a role in chromatin assembly. Depletion of WSTF or Brg1/hBrm by RNAi resulted in altered cell cycle progression, most notably lowered DNA synthesis, suggesting a role for WINAC in replication (Kitagawa et al. 2003).
Pull down assays in cultured MCF7 cells reveal an interaction between WSTF and both ligand-bound and unbound vitamin D receptor (VDR). Chromatin immuno-precipitation experiments in MCF7 cells treated with RNAi specific for WSTF reveal a significant reduction of VDR present at vitamin D-regulated promoters (Kitagawa et al. 2003). Furthermore, decreased VDR recruitment to VDR target genes observed in WS patient skin fibroblasts is rescued by WSTF overexpression. This rescue is likely to be the result of WSTF operating in the context of the WINAC complex, as experiments with MCF7 cells show WINAC-facilitated nucleosome disruption that is VDR-specific (Kitagawa et al. 2003).
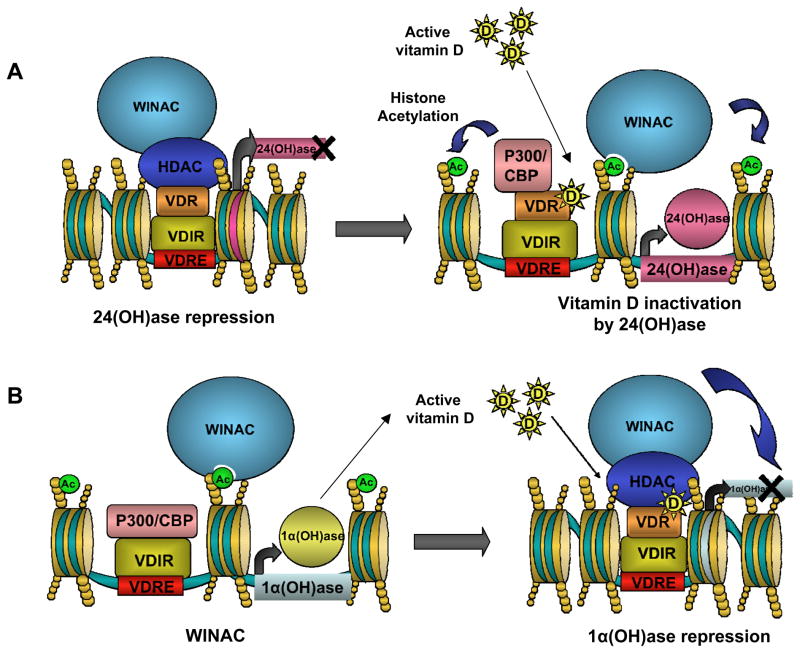
VDR binds hormonally-active vitamin D; however, association of WSTF and VDR occurs in a ligand-independent fashion. Given the infantile hypercalcaemia seen in WS patients (and the known links between vitamin D and calcium regulation), researchers have investigated the role of the WINAC complex in vitamin D homeostasis. Two genes important in vitamin D metabolism are hydroxylases 1α (OH)ase and 24(OH)ase (Bouillon et al. 1995). Vitamin D is obtained both from diet, and in the skin in a UV–dependent synthesis. However, newly synthesized vitamin D exists in an inactive hydroxylated form, 25-hydroxyvitamin D (25-OHD) (Norman 1998). The gene 25-hydroxyvitamin D-1α-hydroxylase (1α(OH)ase) is the enzyme responsible for the formation of active vitamin D (1,25-dihydroxyvitamin D). 24(OH)ase (25-hydroxyvitamin D-24-hydroxylase) is the enzyme responsible for vitamin D inactivation (Zehnder et al. 2001). WINAC activity facilitates ligand-dependent activation of 24(OH)ase and repression 1α(OH)ase by VDR in MCF7 cells (Figure 4A; Kitagawa et al. 2003). Repression of 1α(OH)ase occurs via a co-regulator swap, from histone acetyltransferase activation complex p300/CBP to a histone deacetylase. This switch occurs on the vitamin D interacting repressor (VDIR), which is constitutively bound to the 1a(OH)ase vitamin D response element (VDRE) (Figure 4B; Murayama et al. 2004).
Figure 4.
WINAC and VDR-dependent transcription regulation. (A)WINAC-VDR complex facilitates activation of 24(OH)ase. In the absence of vitamin D, unliganded VDR is bound to the VDIR along with transcriptional repressing histone deactylase. Once VDR binds active vitamin D it facilitates a co-regulator swap from histone deacetylase to histone acetyltransferase activation complex p300/CBP. The result is activation of 24(OH)ase by WINAC chromatin remodeling. (B) WINAC-VDR complex is already in position to facilitate repression of 1a(OH). WINAC and VDR are able to locate VDRE through VDR and VDIR unliganded interactions and remain at these sites through the association of the WSTF bromodomain with acetylated histones. Once VDR binds vitamin D the VDIR experiences a co-regulator swap, from histone acetyltransferase activation complex p300/CBP to a histone deacetylase. The result is repression of 1a(OH)ase by WINAC chromatin remodeling.
Researchers have shown in cultured MCF7 cells that a WSTFΔC mutant, which is missing the WAKZ, PHD and bromodomain, still possesses the ability to associate with VDR and other components of WINAC (Kato et al. 2007). However, the WSTFΔC mutant is unable to bind acetylated lysine 14 on histone 3 (AcH3K14), a bromodomain target. Therefore, the WSTFΔC mutant’s lack of chromatin interaction is thought to be due to the missing bromodomain, though a role for methyl-lysine binding has not been ruled out. WINAC is thought to facilitate the change of co-regulators on the 1a(OH)ase gene promoter via its ligand-independent binding of VDR and targeting of VDIR even in absence of active vitamin D (Kato et al. 2007). WINAC and VDR are able to locate VDRE through VDR and VDIR unligated interactions and remain at these sites through the associations of the WSTF bromodomain with acetylated histones. Once the VDR binds active vitamin D, the WINAC-VDR complex is already in position to facilitate repression of 1α(OH), which reduces the amount of active vitamin D in the cell (Kato et al. 2007). Future studies will seek to directly link the effects of loss of WSTF on VDR-dependent transcription and defects in calcium regulation in WS patients.
B-WICH
WSTF and ISWI have also found in a complex consisting of eight subunits designated B-WICH (Figure 2B). The B-WICH complex includes Myb-binding protein-1a (Myb-bp1a), Sf3b155/SAP155, Gu alpha, CSB, the proto-oncogene Dek, and nuclear myosin 1. B-WICH also includes several RNAs: 5S rRNA, 7SL RNA, and 45S rRNA (Cavellan et al. 2006). Myb-bp1a is a nucleolar protein known mainly for its role in transcriptional regulation. However, recent studies have identified Myb-bp1a as a substrate for the mitotic enzyme aurora kinase B. RNAi silencing of Myb-bp1a causes mitotic delay and aberrant spindle formation (Perrera et al.). SAP155 is a subunit known to be a part of the spliceosome complex U2 snRNP (Kramer et al. 1999). Gu-alpha is an RNA helicase involved in the processing of ribosomal RNA (Yang et al. 2005). The CSB (Cockayne syndrome protein B) is a known subunit of another ATP-dependent chromatin remodeling complex (SWI/SNF). CSB appears to bind DNA directly and is crucial for transcription-coupled DNA repair (Beerens et al. 2005). Dek is another DNA binding protein that is thought to function much like an architectural protein in chromatin (Waldmann et al. 2004). NM1 (Nuclear myosin 1) is known to be important for transcription regulation (Percipalle and Farrants 2006; Percipalle et al. 2006).
Many of the B-WICH subunits are known to play a role in gene expression and formation of B-WICH appears to correlate with active transcription (Cavellan et al. 2006). Components of B-WICH (WSTF, ISWI and NM1) are detected at RNA pol I and III genes in HeLa cells by ChIP, suggesting that the B-WICH complex might have a role in nucleosome remodeling to facilitate pol I and III transcription (Cavellan et al. 2006; Percipalle and Farrants 2006), though the specific role of B-WICH has not been elucidated. Research that addresses chromatin conformation and promoter activity are necessary to strengthen the argument for B-WICH-mediated ribosome gene activation. Also, B-WICH localization is not restricted to ribosomal gene promoters, but rather along the entire gene, suggesting that B-WICH may function in more that just activation (Percipalle and Farrants 2006).
WICH
WICH in DNA replication
WSTF is one of two subunits that comprise the Imitation Switch (ISWI)-containing remodeling complex WICH (Figure 2C). This ISWI complex was first purified from Xenopus egg extracts, and is conserved in all vertebrates. In NIH3T3 mouse cells WSTF is concentrated at the heterochromatin around the centromere during replication and prevents aberrant heterochromatin spreading (Bozhenok et al. 2002). The WICH complex also associates with the DNA clamp, PCNA (Proliferating-cell nuclear antigen) in HeLa cells. This interaction with PCNA facilitates WICH targeting to replication foci. However, DNA polymerase is still able to elongate in the absence of ISWI, suggesting a role for WICH-dependent chromatin formation after replication (MacCallum et al. 2002; Poot et al. 2004).
WICH and DNA repair
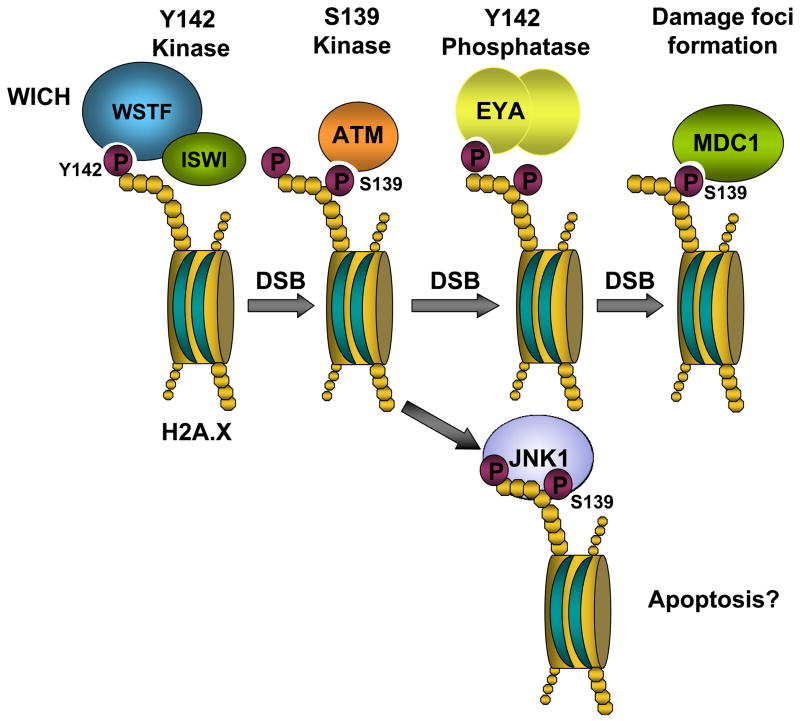
As noted above, recent work has identified an unconventional tyrosine kinase activity present in a region of WSTF encompassing the WAC domain and an additional N-terminal domain (Xiao et al. 2009). WSTF phosphorylates tyrosine 142 of histone H2A.X, and is involved in the DNA damage response. Interestingly, Y142 phosphorylation appears to exist before DNA damage and decreases in response to damage. Y142 phosphorylation is diminished when WSTF is knocked down via RNAi in MEF cells, but S139 phosphorylation (i.e. γ-H2A.X formation) still occurs in response to damage in the WSTF knockdown. However, although S139 phosphorylation occurs normally, phosphorylation and γ-H2A.X focus formation are not maintained in these WSTF-deficient MEF cells. Furthermore, the presence of Mdc1, a nuclear protein with the critical role of sustaining ATM recruitment and thus maintaining γ–H2A.X phosphorylation is diminished in WSTF RNAi-treated cells. It appears that WSTF plays an important role in maintaining S139 phosphorylation and therefore γ–H2A.X focus formation. S139 and Y142 are found in the conserved H2A.X C-terminal motif SQEY, in which S139 is phosphorylated by ATM/ATR. The corresponding motif in budding yeast is SQEL, in which the serine is phosphorylated by the yeast ATM/ATR homologs Tel1 and Mec1. Surprisingly, when the leucine residue in yeast H2A is replaced by a tyrosine, the yeast cells are able to phosphorylate the introduced tyrosine (Xiao et al. 2009), though the responsible kinase has not been identified.
Soon after the novel kinase activity of WSTF was discovered, EYA3 (Eyes Absent 3) was shown to dephosphoryate Y142 on H2A.X in human bone osteosarcoma cells (U2OS) in response to DNA damage (Krishnan et al. 2009). Similarly, EYA1 and 3 were shown to dephosphorylate Y142 in 293T human embryonic kidney cells (Cook et al. 2009). EYA proteins are transcription factors known to play developmental roles in a number of organisms (four, EYA1–4, are present in mammals). EYA proteins are the first transcription factors to also display phosphatase activity (Tootle et al. 2003). Both EYA2 and EYA3 show H2A.X Y142 specific binding and dephosphorylation in vitro. Reduction of EYA3 levels by RNAi results in both an increased level of initial tyrosine phosphorylation as well as reduced active dephosphorylation of Y142 following induction of DNA damage in vivo (Krishnan et al. 2009). Both EYA1 and EYA3 interact with γ-H2A.X following DNA damage in 293T cells, and are localized to γ-H2A.X foci after damage (Cook et al. 2009). Eya1−/− knockout mice exhibit increased apoptosis in the developing kidney at embryonic day 11.5, and EYA1 or EYA3 knockdowns in 293T cells also result in increased apoptosis in response to hypoxia. Interestingly, EYA3 is phosphorylated by ATM/ATR, and this appears to be required for EYA3-H2A.X interaction. These authors also showed that the pro-apoptotic kinase JNK1 interacts with S-Y biphosphorylated peptides, but not with singly phosphorylated peptides. These studies suggest a possible role for Y142 de/phosphorylation in the balance between repair and apoptosis.
Taken together, these discoveries reveal a complex interaction in which WSTF kinase activity targets H2A.X, providing a novel role for the WICH complex in the DNA damage response, while identifying the phosphoY142 phosphatase, EYA implicated in apoptotic pathways (the major interactions are depicted in Figure 5) (Cook et al. 2009; Krishnan et al. 2009; Xiao et al. 2009). H2A.X phosphoY142 appears to be globally dephosphorylated in response to DNA damage as antibodies are unable to detect phosphoY142 8 hours after cell irradiation (Xiao et al. 2009). This result seems contradictory to studies that show EYA acting locally, dephosphorylating H2A.X phosphoY142 at sites of DNA damage. It will be important for future studies to investigate whether H2A.X phosphoY142 exists globally or in particular areas of chromatin and to confirm that H2A.X phosphoY142 is dephosphorylated throughout the genome by EYA.
Figure 5.
The WICH complex has a role in DNA repair. The WICH complex is a kinase, targeting tyrosine 142 on histone H2A.X. Y142 is phosphorylated before damage occurs and becomes gradually dephosphorylated after DNA damage by EYA. MDC1 binds phosphorylated S139 and facilitates foci formation.
Conclusion
WSTF is a member of three known chromatin remodeling complexes: WINAC, WICH and B-WICH. The gene WBSCR that codes for WSTF is one of a number of genes within a heterozygous deletion that exists in individuals with Williams Syndrome. WSTF functions in a surprising number of different roles in a diverse set of chromatin remodeling complexes. A great deal of new information has been obtained since WSTF was first identified, yet many questions about the role of WSTF in replication, gene regulation and DNA repair—and in the clinical presentations of WS patients—still remain. However, a narrative is beginning to build as more research continues to address the questions surrounding this versatile chromatin modifying factor.
Acknowledgments
We thank Shannon Uffenbeck and Oya Yazgan for critical reading of the manuscript and assistance with the figures. Work in our laboratory on WSTF is supported by The Whitehall Foundation #2007-08-79, NIH/NEI #EY016029-02, and Alaska INBRE, NIH/NCRR #RR016466.
References
- Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20(2):56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20(2):56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Ashe A, Morgan DK, Whitelaw NC, Bruxner TJ, Vickaryous NK, Cox LL, et al. A genome-wide screen for modifiers of transgene variegation identifies genes with critical roles in development. Genome Biol. 2008;9(12):R182. doi: 10.1186/gb-2008-9-12-r182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N, Hoeijmakers JH, Kanaar R, Vermeulen W, Wyman C. The CSB protein actively wraps DNA. J Biol Chem. 2005;280(6):4722–4729. doi: 10.1074/jbc.M409147200. [DOI] [PubMed] [Google Scholar]
- Bellugi U, Bihrle A, Jernigan T, Trauner D, Doherty S. Neuropsychological, neurological, and neuroanatomical profile of Williams syndrome. Am J Med Genet Suppl. 1990:6115–125. doi: 10.1002/ajmg.1320370621. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301(5636):1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- Bochar DA, Savard J, Wang W, Lafleur DW, Moore P, Cote J, et al. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc Natl Acad Sci U S A. 2000;97(3):1038–1043. doi: 10.1073/pnas.97.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouazoune K, Brehm A. ATP-dependent chromatin remodeling complexes in Drosophila. Chromosome Res. 2006;14(4):433–449. doi: 10.1007/s10577-006-1067-0. [DOI] [PubMed] [Google Scholar]
- Bouillon R, Okamura WH, Norman AW. Structure-function relationships in the vitamin D endocrine system. Endocr Rev. 1995;16(2):200–257. doi: 10.1210/edrv-16-2-200. [DOI] [PubMed] [Google Scholar]
- Bozhenok L, Wade PA, Varga-Weisz P. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 2002;21(9):2231–2241. doi: 10.1093/emboj/21.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavellan E, Asp P, Percipalle P, Farrants AK. The WSTF-SNF2h chromatin remodeling complex interacts with several nuclear proteins in transcription. J Biol Chem. 2006;281(24):16264–16271. doi: 10.1074/jbc.M600233200. [DOI] [PubMed] [Google Scholar]
- Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458(7238):591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cus R, Maurus D, Kuhl M. Cloning and developmental expression of WSTF during Xenopus laevis embryogenesis. Gene Expr Patterns. 2006;6(4):340–346. doi: 10.1016/j.modgep.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Dirscherl SS, Henry JJ, Krebs JE. Neural and eye-specific defects associated with loss of the imitation switch (ISWI) chromatin remodeler in Xenopus laevis. Mech Dev. 2005;122(11):1157–1170. doi: 10.1016/j.mod.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Dirscherl SS, Krebs JE. Functional diversity of ISWI complexes. Biochem Cell Biol. 2004;82(4):482–489. doi: 10.1139/o04-044. [DOI] [PubMed] [Google Scholar]
- Ewart AK, Jin W, Atkinson D, Morris CA, Keating MT. Supravalvular aortic stenosis associated with a deletion disrupting the elastin gene. J Clin Invest. 1994;93(3):1071–1077. doi: 10.1172/JCI117057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju VK, Bartholomew B. Mechanisms of ATP dependent chromatin remodeling. Mutat Res. 2007;618(1–2):3–17. doi: 10.1016/j.mrfmmm.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8(1):35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Guschin D, Geiman TM, Kikyo N, Tremethick DJ, Wolffe AP, Wade PA. Multiple ISWI ATPase complexes from xenopus laevis. Functional conservation of an ACF/CHRAC homolog. J Biol Chem. 2000;275(45):35248–35255. doi: 10.1074/jbc.M006041200. [DOI] [PubMed] [Google Scholar]
- He X, Fan HY, Garlick JD, Kingston RE. Diverse regulation of SNF2h chromatin remodeling by noncatalytic subunits. Biochemistry. 2008;47(27):7025–7033. doi: 10.1021/bi702304p. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387(6628):43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13(12):152–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, Hamana N, Nezu J, Shimane M. A novel family of bromodomain genes. Genomics. 2000;63(1):40–45. doi: 10.1006/geno.1999.6071. [DOI] [PubMed] [Google Scholar]
- Kato S, Fujiki R, Kim MS, Kitagawa H. Ligand-induced transrepressive function of VDR requires a chromatin remodeling complex, WINAC. J Steroid Biochem Mol Biol. 2007;103(3–5):372–380. doi: 10.1016/j.jsbmb.2006.12.038. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Fujiki R, Yoshimura K, Mezaki Y, Uematsu Y, Matsui D, et al. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell. 2003;113(7):905–917. doi: 10.1016/s0092-8674(03)00436-7. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kramer A, Gruter P, Groning K, Kastner B. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J Cell Biol. 1999;145(7):1355–1368. doi: 10.1083/jcb.145.7.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Jeong DG, Jung SK, Ryu SE, Xiao A, Allis CD, et al. Dephosphorylation of the C-terminal tyrosyl residue of the DNA damage-related histone H2A.X is mediated by the protein phosphatase eyes absent. J Biol Chem. 2009;284(24):16066–16070. doi: 10.1074/jbc.C900032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota R, Hokoc JN, Moshiri A, McGuire C, Reh TA. A comparative study of neurogenesis in the retinal ciliary marginal zone of homeothermic vertebrates. Brain Res Dev Brain Res. 2002;134(1–2):31–41. doi: 10.1016/s0165-3806(01)00287-5. [DOI] [PubMed] [Google Scholar]
- Lall S. Primers on chromatin. Nat Struct Mol Biol. 2007;14(11):1110–1115. doi: 10.1038/nsmb1107-1110. [DOI] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442(7098):91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Cabot RA, Schwickert T, Rupp RA. The SNF2 domain protein family in higher vertebrates displays dynamic expression patterns in Xenopus laevis embryos. Gene. 2004:32659–66. doi: 10.1016/j.gene.2003.09.053. [DOI] [PubMed] [Google Scholar]
- Lu X, Meng X, Morris CA, Keating MT. A novel human gene, WSTF, is deleted in Williams syndrome. Genomics. 1998;54(2):241–249. doi: 10.1006/geno.1998.5578. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lyuksyutova AI, Lu CC, Milanesio N, King LA, Guo N, Wang Y, et al. Anterior-posterior guidance of commissural axons by Wnt-frizzled signaling. Science. 2003;302(5652):1984–1988. doi: 10.1126/science.1089610. [DOI] [PubMed] [Google Scholar]
- MacCallum DE, Losada A, Kobayashi R, Hirano T. ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP-aurora B. Mol Biol Cell. 2002;13(1):25–39. doi: 10.1091/mbc.01-09-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurus D, Heligon C, Burger-Schwarzler A, Brandli AW, Kuhl M. Noncanonical Wnt-4 signaling and EAF2 are required for eye development in Xenopus laevis. EMBO J. 2005;24(6):1181–1191. doi: 10.1038/sj.emboj.7600603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris CA, Demsey SA, Leonard CO, Dilts C, Blackburn BL. Natural history of Williams syndrome: physical characteristics. J Pediatr. 1988;113(2):318–326. doi: 10.1016/s0022-3476(88)80272-5. [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, et al. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119(6):767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- Murayama A, Kim MS, Yanagisawa J, Takeyama K, Kato S. Transrepression by a liganded nuclear receptor via a bHLH activator through co-regulator switching. EMBO J. 2004;23(7):1598–1608. doi: 10.1038/sj.emboj.7600157. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nakamura H, Katahira T, Matsunaga E, Sato T. Isthmus organizer for midbrain and hindbrain development. Brain Res Brain Res Rev. 2005;49(2):120–126. doi: 10.1016/j.brainresrev.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Norman AW. Sunlight, season, skin pigmentation, vitamin D, and 25-hydroxyvitamin D: integral components of the vitamin D endocrine system. Am J Clin Nutr. 1998;67(6):1108–1110. doi: 10.1093/ajcn/67.6.1108. [DOI] [PubMed] [Google Scholar]
- Oya H, Yokoyama A, Yamaoka I, Fujiki R, Yonezawa M, Youn MY, et al. Phosphorylation of Williams syndrome transcription factor by MAPK induces a switching between two distinct chromatin remodeling complexes. J Biol Chem. 2009;284(47):32472–32482. doi: 10.1074/jbc.M109.009738. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pascual J, Martinez-Yamout M, Dyson HJ, Wright PE. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. J Mol Biol. 2000;304(5):723–729. doi: 10.1006/jmbi.2000.4308. [DOI] [PubMed] [Google Scholar]
- Paull TT, et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10(15):886–95. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Percipalle P, Farrants AK. Chromatin remodelling and transcription: be- WICHed by nuclear myosin 1. Curr Opin Cell Biol. 2006;18(3):267–274. doi: 10.1016/j.ceb.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Percipalle P, Fomproix N, Cavellan E, Voit R, Reimer G, Kruger T, et al. The chromatin remodelling complex WSTF-SNF2h interacts with nuclear myosin 1 and has a role in RNA polymerase I transcription. EMBO Rep. 2006;7(5):525–530. doi: 10.1038/sj.embor.7400657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrera C, Colombo R, Valsasina B, Carpinelli P, Troiani S, Modugno M, et al. Identification of MYB-binding protein 1A (MYBBP1A) as a novel substrate for aurora B kinase. J Biol Chem. doi: 10.1074/jbc.M109.068312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pober BR. Williams-Beuren syndrome. N Engl J Med. 2010;362(3):239–252. doi: 10.1056/NEJMra0903074. [DOI] [PubMed] [Google Scholar]
- Poot RA, Bozhenok L, van den Berg DL, Steffensen S, Ferreira F, Grimaldi M, et al. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat Cell Biol. 2004;6(12):1236–1244. doi: 10.1038/ncb1196. [DOI] [PubMed] [Google Scholar]
- Poot RA, Dellaire G, Hulsmann BB, Grimaldi MA, Corona DF, Becker PB, et al. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 2000;19(13):3377–3387. doi: 10.1093/emboj/19.13.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quong MW, Massari ME, Zwart R, Murre C. A new transcriptional-activation motif restricted to a class of helix-loop-helix proteins is functionally conserved in both yeast and mammalian cells. Mol Cell Biol. 1993;13(2):792–800. doi: 10.1128/mcb.13.2.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragvin A, Valvatne H, Erdal S, Arskog V, Tufteland KR, Breen K, et al. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J Mol Biol. 2004;337(4):773–788. doi: 10.1016/j.jmb.2004.01.051. [DOI] [PubMed] [Google Scholar]
- Rasmussen JT, Deardorff MA, Tan C, Rao MS, Klein PS, Vetter ML. Regulation of eye development by frizzled signaling in Xenopus. Proc Natl Acad Sci U S A. 2001;98(7):3861–3866. doi: 10.1073/pnas.071586298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, et al. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell. 2004;15(2):543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, et al. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146(5):905–16. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salceda J, Fernandez X, Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25(11):2575–2583. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58(1):15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Langst G, et al. NoRC--a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001;20(17):4892–4900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M. Histone H2A.X Tyr142 phosphorylation: a novel sWItCH for apoptosis? DNA Repair (Amst) 2009;8(7):873–876. doi: 10.1016/j.dnarep.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, Mills IA, et al. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426(6964):299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit Rev Biochem Mol Biol. 2009;44(2–3):117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20(6):1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann T, Scholten I, Kappes F, Hu HG, Knippers R. The DEK protein--an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343(1):1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- Winston F, Allis CD. The bromodomain: a chromatin-targeting module? Nat Struct Biol. 1999;6(7):601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- Wong LY, Recht J, Laurent BC. Chromatin remodeling and repair of DNA double-strand breaks. J Mol Histol. 2006;37(5–7):261–269. doi: 10.1007/s10735-006-9047-4. [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, et al. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402(6760):418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457(7225):57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa J, Kitagawa H, Yanagida M, Wada O, Ogawa S, Nakagomi M, et al. Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Mol Cell. 2002;9(3):553–562. doi: 10.1016/s1097-2765(02)00478-1. [DOI] [PubMed] [Google Scholar]
- Yang H, Henning D, Valdez BC. Functional interaction between RNA helicase II/Gu(alpha) and ribosomal protein L4. FEBS J. 2005;272(15):3788–3802. doi: 10.1111/j.1742-4658.2005.04811.x. [DOI] [PubMed] [Google Scholar]
- Yoshimura K, Kitagawa H, Fujiki R, Tanabe M, Takezawa S, Takada I, et al. Distinct function of 2 chromatin remodeling complexes that share a common subunit, Williams syndrome transcription factor (WSTF) Proc Natl Acad Sci U S A. 2009;106(23):9280–9285. doi: 10.1073/pnas.0901184106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309(5734):626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zehnder D, Bland R, Williams MC, McNinch RW, Howie AJ, Stewart PM, et al. Extrarenal expression of 25-hydroxyvitamin d(3)-1 alpha-hydroxylase. J Clin Endocrinol Metab. 2001;86(2):888–894. doi: 10.1210/jcem.86.2.7220. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95(2):279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]