Abstract
St. Louis encephalitis virus (SLEV; Flaviviridae; Flavivirus) is a member of the Japanese encephalitis serocomplex and a close relative of West Nile virus (WNV). Although SLEV remains endemic to the US, both levels of activity and geographical dispersal are relatively constrained when compared to the widespread distribution of WNV. In recent years, WNV appears to have displaced SLEV in California, yet both viruses currently coexist in Texas and several other states. It has become clear that viral swarm characterization is required if we are to fully evaluate the relationship between viral genomes, viral evolution, and epidemiology. Mutant swarm size and composition may be particularly important for arboviruses, which require replication not only in diverse tissues but also divergent hosts. In order to evaluate temporal, spatial, and host-specific patterns in the SLEV mutant swarm, we determined the size, composition, and phylogeny of the intrahost swarm within primary mosquito isolates from both Texas and California. Results indicate a general trend of decreasing intrahost diversity over time in both locations, with recent isolates being highly genetically homogeneous. Additionally, phylogenic analyses provide detailed information on the relatedness of minority variants both within and among strains and demonstrate how both geographic isolation and seasonal maintenance have shaped the viral swarm. Overall, these data generally provide insight into how time, space, and unique transmission cycles influence the SLEV mutant swarm and how understanding these processes can ultimately lead to a better understanding of arbovirus evolution and epidemiology.
Keywords: Arbovirus evolution, Flavivirus, St. Louis encephalitis virus, Intrahost diversity, Phylogeny
1. Introduction
St. Louis encephalitis virus (SLEV; Flaviviridae; Flavivirus) is a member of the Japanese encephalitis serocomplex and a close relative of West Nile virus (WNV). The initial isolation of SLEV occurred in St. Louis, MS in 1933, when over 1000 cases of encephalitis were recorded (Lumsden, 1958; Reisen, 2003). SLEV is ecologically similar to WNV in that it is maintained in transmission cycles between generally ornithophilic Culex mosquitoes and birds. Humans represent dead-end hosts due to generally insufficient viremia levels for successful transmission (Bernard and Kramer, 2001). The main vectors of SLEV in the western US are Culex quinqufasciatus, which is generally restricted to southern urban areas, and Culex tarsalis, which is present in agricultural and rural areas (Reisen, 2003; Darsie and Ward, 1981). Although SLEV remains endemic to the US, approximately 55 cases on average are reported per year since the last major outbreak occurred in 1975–1977 during which time more than 2500 cases were reported in 35 states. Both levels of activity and geographical dispersal of SLEV are relatively constrained when compared to the widespread distribution of WNV (http://www.cdc.gov/ncidod/dvbid/arbor). In addition, results from phylogenetic studies indicate SLEV, more so than WNV, is subject to geographic isolation (Kramer and Chandler, 2001; Baillie et al., 2008; May et al., 2008). These differing genetic and epidemiological patterns could be attributed to generally lower SLEV viremia levels in birds, resulting in a narrower competent host range and therefore, more specific transmission cycles (McLean et al., 1983; Komar et al., 2003; Marra et al., 2003). WNV is unique in that it can maintain high levels of viremia in a range of vertebrate hosts despite its need to cycle in mosquito vectors (Brault et al., 2007). In addition, over 75 mosquito species have been demonstrated to become infected with WNV (Higgs et al., 2004), although Culex mosquitoes are implicated as the predominant vectors of both viruses (Bernard and Kramer, 2001; Reisen, 2003). In recent years, WNV has appeared to displace SLEV in California (Reisen et al., 2008), yet both viruses currently coexist in Texas (Lillibridge et al., 2004; Bradford et al., 2005; Rios et al., 2006). Previous studies demonstrate that WNV strains exist as highly diverse mutant swarms in nature and that the source of this genetic diversity is likely the mosquito (Jerzak et al., 2005). The maintenance of diversity is one mechanism by which arboviruses could overcome significant fitness costs as a result of their requirement for host cycling. In vitro passage studies in mosquito cells suggest that SLEV may have a relatively decreased capacity for producing and maintaining intrahost diversity, and that this reduced diversity may have direct consequences for host range (Ciota et al., 2007). Besides being relevant for adaptability, minority variants in many viruses can play significant roles in viral fitness and pathogenesis (Martinez et al., 1991; Novella and Ebendick-Corp, 2004; Jerzak et al., 2007; Vignuzzi et al., 2006). It has become clear that viral swarm characterization is required if we are to fully evaluate the relationship between viral genomes and both viral evolution and epidemiology. In this study we determined the size of the SLEV mutant swarm in mosquitoes using molecular cloning of primary isolates from both Texas and California in order to (i) generally assess mutant swarm breadth, (ii) determine if geographical and/or temporal differences in mutant swarm size and composition exist, and (iii) elucidate the role of variable transmission cycles in shaping the mutant swarm. Advancing our understanding of these relationships will provide insight into factors important in influencing arbovirus evolution and epidemiology.
2. Materials and methods
2.1. Virus isolates
The main criterion used for the selection of SLEV isolates for this study was the availability of material from the initial amplification, i.e. single passage only (Table 1). SLEV mosquito isolates from Texas were kindly provided by R. Tesh (UTMB; Galveston, TX) as lyophilized material following a single round of amplification on C6/36 mosquito cells. Material was reconstituted in 500ul BA-1 and used directly for experimental testing. Original California mosquito pools previously determined to be SLEV positive were obtained from A. Brault (UC Davis; Davis, CA) and amplified once on C6/36 mosquito cells at the Arbovirus laboratory, Wadsworth Center. Original mosquito pools from both TX and CA were of variable size (5–50 mosquitoes), yet minimal infection rates within each state have never exceeded a level which would suggest that each pool contained more than a single, positive mosquito (http://www.cdc.gov/ncidod/dvbid/arbor). All Texas isolates originated from Cx. quinquefasciatus mosquitoes trapped in the Houston area (Harris County) whereas CA isolates originated from Cx. tarsalis mosquitoes trapped in various counties including Coachella Valley (COAV), San Bernardino (SAND), Kern (BFS), Los Angeles (SOUE), and Imperial (IMPR) counties.
Table 1.
SLEV mosquito isolates.
| Strain | Collection date | Location | Source |
|---|---|---|---|
| U0476 | 22 July 2003 | Harris Co., TX | Cx. quinquefasciatus |
| V3916 | 29 September 1998 | Harris Co., TX | Cx. quinquefasciatus |
| V4179 | 6 October 1998 | Harris Co., TX | Cx. quinquefasciatus |
| V2679 | 8 August 1996 | Harris Co., TX | Cx. quinquefasciatus |
| V1042 | 28 June 1994 | Harris Co., TX | Cx. quinquefasciatus |
| V3248 | 29 July 1992 | Harris Co., TX | Cx. quinquefasciatus |
| V3222 | 14 August 1991 | Harris Co., TX | Cx. quinquefasciatus |
| V4209 | 22 August 1990 | Harris Co., TX | Cx. quinquefasciatus |
| COAV888 | 2 July 2003 | Coachella Co., CA | Cx. tarsalis |
| COAV1257 | 2 October 2003 | Coachella Co., CA | Cx. tarsalis |
| IMPR115 | 16 September 2003 | Imperial Co., CA | Cx. tarsalis |
| COAV382 | 11 July 2001 | Coachella Co., CA | Cx. tarsalis |
| COAV333 | 11 July 2001 | Coachella Co., CA | Cx. tarsalis |
| COAV332 | 15 July 2000 | Coachella Co., CA | Cx. tarsalis |
| COAV333 | 15 July 2000 | Coachella Co., CA | Cx. tarsalis |
| SAND29 | 27 August 1993 | San Bern. Co, CA | Cx. tarsalis |
| SAND39 | 27 August 1993 | San Bern. Co, CA | Cx. tarsalis |
| SOUE20 | 18 September 1984 | Los Ang. Co., CA | Cx. tarsalis |
| BFS2874 | 22 September 1960 | Kern Co., CA | Cx. tarsalis |
| BFS508 | 1 August 1950 | Kern Co, CA | Cx. tarsalis |
2.2. Molecular cloning
Molecular cloning of an approximately 2 kb region (nucleotides 1315–3325), including portions of the envelope (E) and non-structural 1 (NS1) genes, was performed as previously described (Ciota et al., 2009). RNA was extracted from virus isolates using the Qia-amp viral RNA extraction kit (Qiagen, Valencia, CA) and RT-PCR was completed with primers designed to amplify the 3′ 1302 nt of the SLEV genome (E gene) and the 5′ 3325 nt of the SLEV genome (NS1 gene). Reverse transcription was performed with Sensiscript RT (Qiagen) and cDNA was amplified with ‘high-fidelity’ PfuUltra (published error rate = 4.3 × 10−7; Stratagene, La Jolla, CA). PCR products were visualized on a 1.5% agarose gel and DNA was recovered by using a MinElute Gel Extraction kit (Qiagen). The recovered DNA was ligated into the cloning vector pCR-Blunt II-TOPO (Invitrogen, Carlsbad, CA) and transformed into One Shot TOP10 Electrocomp E. coli cells. Colonies were screened by direct PCR using primers specific for the desired insert, and plasmid DNA was purified by using a QIAprep Spin Miniprep kit (Qiagen). Sequencing was carried out with five pairs of overlapping SLEV primers together with T7 and SP6 primers at the Wadsworth Center Molecular Genetics Core using ABI 3700 and 3100 automated sequencers (Applied Biosystems, Foster City, CA). Seventeen to twenty-four clones per sample were sequenced.
2.3. Phylogenetic trees
Four different programs, including BEAST v1.4.8 (Drummond and Rambaut, 2007), RaxML v7.2.3 (Stamatakis, 2006), GARLI v0.951 (Zwickl, 2006), and MrBayes (Ronquist and Huelsenbeck, 2003), were used to construct phylogenentic trees from the 255 amino acid sequences of SLEV. BEAST and MrBayes use Bayesian Markov-Chain-Monte-Carlo (MCMC) analyses to infer phylogenies, while RAxML and GARLI use Maximum-Likelihood-based approaches. BEAST was run once using Texas sequences and once for the California sequences, each time with an MCMC length of 1 × 108, logging every 1 × 104. A general time-reversible substitution model and uncorrelated log normal molecular clock model were used. A starting tree was constructed using UPGMA (unweighted pair group method with arithmetic mean) and the tree prior was assumed to have a constant population size. All of the priors for the model parameters and statistics were left at the defaults with the exception of including isolate date. RAxML was used to perform rapid bootstrap analyses with 1000 iterations while also finding a Maximum-Likelihood tree; all other parameters were left at default. GARLI was run in addition to RAxML and MrBayes in addition to BEAST in order to provide additional information on the structure of the phylogeny. GARLI was run using the default parameters except for including 100 independent search replicates. MrBayes was run for 1 × 106 generations each. RAxML, GARLI, and MrBayes were only run on sequence data from which duplicates had been removed. All phylogenetic reconstructions were rooted using sequences from four other flaviviruses as outgroups. These included Japanese encephalitis (genbank accession number M55506), West Nile (M12294), Kunjin (D00246) and Murray Valley encephalitis (X03467).
2.4. Sequence and correlation analyses
Sequences were compiled, edited, and aligned using DNASTAR software package (Madison, WI). At least 2-fold coverage was used for sequence determination. The percentage of nucleotide diversity (total number of differences from consensus divided by total number of bases sequenced), amino acid diversity (total number of amino acid differences divided by total number of amino acids sequenced), and the sequence diversity (percent of clones with at least one difference from consensus) were used as basic indicators of genetic diversity. For nucleotide and amino acid diversity calculations redundant changes were counted as separate differences.
The ratio (ω) of nonsynonymous (dN) to synonymous (dS) substitutions per site was calculated in an attempt to evaluate the strength and direction of selection acting on these coding regions. Texas and California consensus sequences for all strains were broken up into their envelope (E) and non-structural (NS1) protein components and analyzed separately. Calculations were performed using HyPhy (Pond et al., 2005) on the Datamonkey webserver (Pond and Frost, 2005). The three methods used were single likelihood ancestor counting (SLAC), fixed effects likelihood (FEL), and random effects likelihood (REL) (Kosakovsky Pond and Frost, 2005).
Correlation analyses between nucleotide diversity and mosquito collection date were conducted with and without corrections for phylogenetic dependence. Correlation coefficients for non-phylogenetic analyses (treating each strain as independent) were calculated using the program R v2.9.2 (R Development Core Team, 2009). For phylogenetic analyses, independent contrasts were used to correct for phylogeny (Felsenstein, 1985) using the APE package (Paradis et al., 2004) in R v2.9.2.
3. Results
3.1. Phylogeny
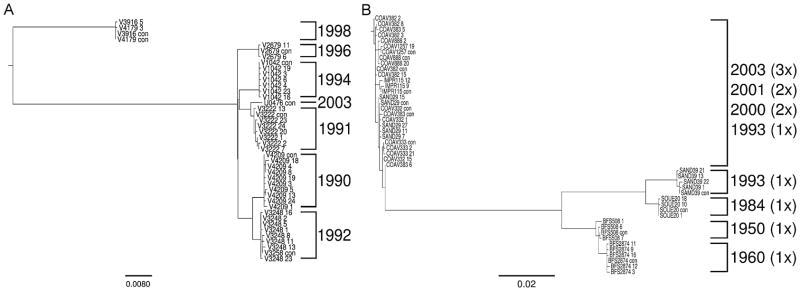
Phylogentic analysis of Texas isolates yielded relationships in which all variants within individual strains grouped together (Fig. 1A). This pattern occurred in the trees created with all four programs and is illustrated here in the phylogeny reconstructed using RAxML. Duplicate sequences from the individual strains were removed in this analysis. The lack of intersection between any single variant among years demonstrates the uniqueness of consensus sequences from year to year and the general absence of yearly maintenance of past strains within the mutant swarm. Substitution rates on the consensus level were similar from year to year from 1990 to 1996, averaging about 0.9% divergence/year, or, approximately 10 nt substitutions/genome/year. Substantially greater divergence occurred from 1996 to 1998 (Fig. 1A), with almost 10% sequence divergence occurring in the two 1998 strains (V4179 and V3916) when compared to the 1996 isolate (V2679). 87.9% of these changes were synonymous. Interestingly, the 2003 isolate (V0476) does not group with the more recent 1998 isolate but, rather, the more distant isolates. In fact, the sequence most closely related to the 2003 isolate is the 1991 isolate (V3222), in which just 0.5% sequence divergence was measured.
Fig. 1.
Phylogenetic-tree reconstruction of intrahost variants of individual isolates, using RAxML. The isolate number is followed by either the mutant number or ‘con’, indicating the consensus sequence. Duplicate sequences have been removed for this analysis. (A) Texas isolates from the same sample cluster together. Numbers on the right indicate the year of sample collection. (B) California isolates from different samples (obtained at different time points and different locations) grouped together. Multiples following the year of sample collection indicate how many independent samples are part of the same group; i.e. the top group consists of three samples taken in 2003, two taken in 2001, two in 2000, and one in 1993.
The CA data set was very different from TX both spatially and temporally, with isolates as old as 1950 (BFS 508) and from various counties. In the phylogenetic tree constructed from CA sequences just 4 of 12 strains consistently grouped together (Fig. 1B). This pattern was again consistent among all four programs and is illustrated by the RAxML-created phylogeny in which duplicate sequences from individual strains were removed. In particular, multiple minority variants for isolates after 1993 were identical between strains and this section of the phylogeny exhibited low resolution. Of particular interest, all sequences from a 1993 isolate from San Bernardino (SAND 29) were placed within this highly related clade and one mutant sequence from a 2000 isolate from Coachella valley (COAV 332) was identical to the consensus sequence from San Bernardino (SAND 29). Other than this exception all strains generally grouped both temporally and geographically. Post-1993 strains demonstrated high similarity along with a lack of phylogenetic resolution. Attempts to quantify substitution rates of SLEV in CA are clearly inaccurate when done without consideration of geographic location. For example, just 7 nt differences exist between the consensus sequences of the 1950 and 1960 isolates from BFS in the region analyzed, yet 184 substitutions were identified in this region when comparing the 1984 SOUE isolate (SOUE 20) and a 1993 San Bernardino isolate (SAND 29). When only Coachella Valley isolates are considered (2000–2003), the average substitution rates are approximately 12 substitutions/genome/year, which is similar to what was measured with the Texas isolates.
We also reconstructed phylogenetic trees based on consensus sequences for each sample (Fig. S1). The three bipartitions labeled on the Texas consensus sequence phylogeny (Fig. S1A) were present in more than 80% of bootstrapped trees using RAxML. However, a reconstruction of the phylogeny of all sequences using BEAST showed all bipartitions on the level of strains to have high posterior probabilities (>0.9). It should be noted that not all of the bipartitions in the BEAST tree were the same as those created using the other three programs with either consensus sequences or with duplicates removed. The BEAST analysis placed the 1994 strain (V1042) before the divergence of the 1991 (V3222) and 2003 (U0476) strains, while the others placed the 1994 strain in a separate clade from the 1991 and 2003 strains. However, this difference did not affect the analysis using independent contrasts (next subsection) and still support the conclusion that Texas sequences cluster by strain. The fact that Garli and MrBayes showed the same bipartions as RAxML increases confidence in the phylogeny presented.
The phylogeny reconstructed using California consensus sequences again showed very little differentiation in the area containing the eight highly related strains (Fig. S1B). In the best-scoring maximum likelihood tree reconstructed using RAxML this region only had one bipartition that appeared in more than 80% of bootstrap replicates. The rest of the bipartitions showed high bootstrap support.
3.2. Genetic diversity and correlation
Isolation of minority variants by high-fidelity molecular cloning of portions of the SLEV ENV and NS1 genes and subsequent sequencing and analysis was used to assess the level of genetic diversity within individual isolates. The purpose of these analyses was to assess both the size of the mutant swarm in recent isolates and to determine if mutant-swarm size was correlated with both temporal, geographic, and host differences. Initial analysis involved a straightforward assessment of nucleotide and amino acid diversity and overall sequence diversity (Tables 2 and 3).
Table 2.
Intrahost nucleotide diversity of SLEV isolates for nt 1315–3325.
| Strain | State | Year | Clones | nt changes | nt div.a (%) | nc var.b | seq div.c (%) |
|---|---|---|---|---|---|---|---|
| U0476 | TX | 2003 | 23 | 0 | 0.000 | 0 | 0.00 |
| V3916 | TX | 1998 | 24 | 1 | 0.002 | 1 | 4.17 |
| V4179 | TX | 1998 | 24 | 1 | 0.002 | 1 | 4.17 |
| V2679 | TX | 1996 | 24 | 2 | 0.004 | 2 | 8.33 |
| V1042 | TX | 1994 | 24 | 7 | 0.015 | 7 | 29.17 |
| V3248 | TX | 1992 | 24 | 18 | 0.037 | 14 | 58.33 |
| V3222 | TX | 1991 | 24 | 28 | 0.058 | 17 | 70.83 |
| V4209 | TX | 1990 | 24 | 12 | 0.025 | 9 | 37.50 |
| COAV888 | CA | 2003 | 19 | 1 | 0.008 | 1 | 5.26 |
| COAV1257 | CA | 2003 | 20 | 1 | 0.007 | 1 | 5.00 |
| IMPR115 | CA | 2003 | 19 | 5 | 0.013 | 3 | 15.79 |
| COAV382 | CA | 2001 | 19 | 6 | 0.016 | 6 | 31.58 |
| COAV383 | CA | 2001 | 19 | 6 | 0.016 | 3 | 15.79 |
| COAV332 | CA | 2000 | 18 | 3 | 0.008 | 2 | 11.11 |
| COAV333 | CA | 2000 | 19 | 2 | 0.005 | 1 | 5.26 |
| SAND29 | CA | 1993 | 19 | 7 | 0.021 | 1 | 5.88 |
| SAND39 | CA | 1993 | 17 | 11 | 0.029 | 9 | 47.37 |
| SOUE20 | CA | 1984 | 20 | 2 | 0.005 | 1 | 5.00 |
| BFS2874 | CA | 1960 | 19 | 9 | 0.024 | 6 | 31.58 |
| BFS508 | CA | 1950 | 21 | 33 | 0.078 | 7 | 33.33 |
Nucleotide diversity = total nt changes (differences from consensus)/total bases sequenced (~45,000).
Nonconsensus variants = haplotypes with at least one nt change relative to consensus.
Sequence diversity = % of clones with at least one nt change.
Table 3.
Intrahost amino acid diversity of SLEV isolates for aa 439–1109.
| Strain | State | Year | Clones | aa changes | aa div.a (%) | nc varb | seq div.c (%) |
|---|---|---|---|---|---|---|---|
| U0476 | TX | 2003 | 23 | 0 | 0.000 | 0 | 0.00 |
| V3916 | TX | 1998 | 24 | 1 | 0.006 | 1 | 4.17 |
| V4179 | TX | 1998 | 24 | 0 | 0.000 | 0 | 0.00 |
| V2679 | TX | 1996 | 24 | 1 | 0.006 | 1 | 4.17 |
| V1042 | TX | 1994 | 24 | 3 | 0.019 | 3 | 12.50 |
| V3248 | TX | 1992 | 24 | 13 | 0.081 | 11 | 45.83 |
| V3222 | TX | 1991 | 24 | 16 | 0.099 | 13 | 54.17 |
| V4209 | TX | 1990 | 24 | 10 | 0.062 | 8 | 33.33 |
| COAV888 | CA | 2003 | 19 | 1 | 0.008 | 1 | 5.26 |
| COAV1257 | CA | 2003 | 20 | 1 | 0.007 | 1 | 5.00 |
| IMPR115 | CA | 2003 | 19 | 1 | 0.007 | 1 | 5.00 |
| COAV382 | CA | 2001 | 19 | 0 | 0.000 | 0 | 0.00 |
| COAV383 | CA | 2001 | 19 | 3 | 0.024 | 2 | 10.53 |
| COAV332 | CA | 2000 | 18 | 1 | 0.008 | 1 | 5.55 |
| COAV333 | CA | 2000 | 19 | 1 | 0.007 | 1 | 5.00 |
| SAND29 | CA | 1993 | 19 | 6 | 0.052 | 5 | 26.32 |
| SAND39 | CA | 1993 | 17 | 2 | 0.015 | 1 | 5.88 |
| SOUE20 | CA | 1984 | 20 | 1 | 0.007 | 1 | 5.00 |
| BFS2874 | CA | 1960 | 19 | 5 | 0.039 | 3 | 15.79 |
| BFS508 | CA | 1950 | 21 | 1 | 0.007 | 1 | 4.76 |
Amino acid diversity = total aa changes (differences from consensus)/total sequenced (~15,000).
Nonconsensus variants = aa haplotypes with at least one aa change relative to consensus.
Sequence diversity = % of clones with at least one aa change.
The most genetically diverse isolate among the Texas strains, V3222 (1991), possessed 28 variable bases within the 24 clones analyzed from the mutant swarm. Assuming this level of genetic diversity exists genome wide, this would equate to an average of 6.4 base differences relative to the consensus sequence per genome.
Among the 58 base substitutions identified in the three most diverse isolates (1990, 1991, 1992), 39 were nonsynonomous changes (67.2%). Conversely, a total of just 2 base changes relative to consensus sequences were identified in the three most recent isolates (V0476, V3916, V4179), with not a single change identified in 23 clones of the 2003 isolate (V0476).
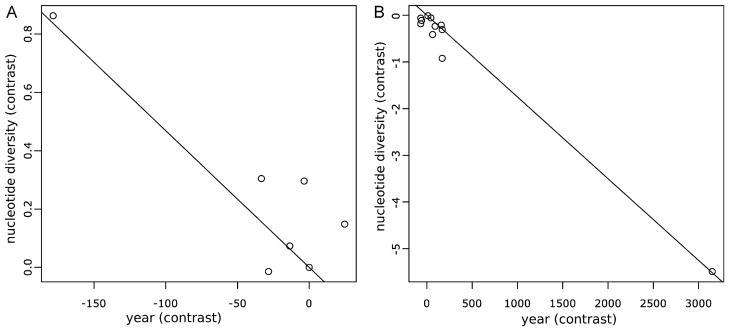
Without correcting for phylogeny, a negative correlation between isolation date and nucleotide diversity among the Texas isolates exists (R = −0.788, p = 0.020; Table 4) (Fig. 2A). This relationship, with the most highly homogeneous isolates being the most recent, also exists on the amino-acid level (R = −0.823, p = .012; Table 5). However, the isolates are not independent, and thus the correlation is confounded by the phylogenetic structure. The phylogeny that was used to correct for this dependence was created with RAxML and used only the consensus sequences from each strain. When phylogeny of the Texas isolates is corrected for by independent contrasts the correlation between time and intrahost diversity is further strengthened (R = −0.891, p = 0.003; Table 4, Fig. 3A). However, this strong correlation is driven by a single outlying contrast and disappears when this point is removed from the set (R = −0.341, p = 0.454). A Spearman rank correlation, which is less sensitive to outliers, similarly found no correlation, even when the outlying contrast was not excluded (ρ = −0.464, p = 0.302; Table 4). The outlying contrast driving the correlation is a result of a strain from 2003 (U0476) with low diversity being closely related to a strain from 1991 (V3222) with a high diversity.
Table 4.
Correlation analysis between year and intrahost nucleotide diversity.
| Data set | Analysis | R | P-value |
|---|---|---|---|
| Texas | Raw data | −0.788 | 0.020 |
| PICa | −0.891 | 0.003 | |
| PIC w/o outlier | −0.341 | 0.454 | |
| California | Raw data | −0.791 | 0.002 |
| PICa | −0.989 | <0.0001 | |
| PIC w/o outlier | −0.720 | 0.013 |
PIC refers to phylogenetic independent contrasts.
Fig. 2.
Nucleotide diversity vs. isolation year, not corrected for phylogeny. (A) Texas, (B) California.
Table 5.
Correlation analysis between year and intrahost amino-acid diversity.
| Data set | Analysis | R | P-value |
|---|---|---|---|
| Texas | Raw data | −0.822 | 0.012 |
| PICa | −0.483 | 0.226 | |
| PIC w/o outlier | −0.341 | 0.454 | |
| California | Raw data | −0.216 | 0.500 |
| PICa | −0.993 | <0.0001 | |
| PIC w/o outlier | −0.019 | 0.956 |
PIC refers to phylogenetic independent contrasts.
Fig. 3.
Phylogenetically independent contrasts (PICs) of nucleotide diversity vs. PICs of isolation year. The trees used to calculate PICs are shown in Fig. S1. (A) Texas, (B) California.
On average, recent California isolates (2000–2003) also displayed high levels of intrahost genetic homogeneity, with all of the isolates having nucleotide diversity of less than 0.016% (Table 2). The most highly homogeneous isolates were again the most recent isolates, with two of the three 2003 CA isolates (both COAV) showing just a single base change in more than 40,000 bases sequenced. In addition, the most highly diverse isolate analyzed was the oldest, BFS 508 (1950), with a nucleotide diversity of 0.078%, equating to an average of 8.6 substitutions per genome. Unlike the diverse Texas isolates, all but one of the substitutions in this isolate were synonymous. Overall, the percent of nonsynonymous changes within the CA isolates mutant swarms was significantly lower than that measured in Texas (32.0% v. 62.3%; Fisher’s exact test, p < 0.0001).
The CA isolates showed a negative correlation between nucleotide diversity and isolation date (R = −0.791, p = 0.002; Table 4, Fig. 2B), similar to the Texas isolates. When independent contrasts were performed using a phylogeny of consensus sequences the correlation strengthened greatly as with the Texas data (R = −0.989; p < 0.0001; Table 4, Fig. 3B). Unlike for the Texas data, the increased correlation after correcting for phylogenetic structure cannot easily be attributed to a single outlier although there does appear to be one (Fig. 3B). The correlations remains significant when the outlier is removed (R = −0.720, p = 0.013; Table 4) and when the non-parametric analysis is performed (ρ = −0.791, p = 0.006).
The Texas sequences show a similar relationship between diversity and time on the amino-acid level without correcting for phylogeny as with nucleotides (R = −0.822, p = 0.012; Table 5). The negative relationship seen at the nucleotide level for the California sequences is absent for amino-acids (R = −0.216, p = 0.500; Table 5). When independent contrasts were performed on the Texas data, the correlation disappeared (R = −0.483, p = 0.226; Table 5) but when this method was applied to the California amino-acid data, the strength of the correlation greatly increased (R = −0.993, p < 0.0001; Table 5). Unlike at the nucleotide level, the correlation between time and amino-acid diversity disappeared when the outlier was removed (R = −0.019, p = 0.956; Table 5) and when we calculated a Spearman correlation (ρ = −0.109, p = 0.755).
3.3. Selection
There was not enough power to infer selection at individual sites due to the relatively low number of sequences. We used three different methods provided by the DataMonkey web server (Single Likelihood Ancestor Counting, Fixed Effects Likelihood, Random Effects Likelihood), and found no site for which all methods indicated significant selection (data not shown). However, the mean ω values for the E gene were 0.017736 and 0.020650 in Texas and California, respectively, while the mean ω values for the NS1 gene were 0.065975 and 0.093021 in Texas and California, respectively (Table 6). This difference suggests that there has been a greater degree of purifying selection acting on the envelope protein than on the non-structural protein in both geographic areas.
Table 6.
dN/dS analysis.
| Data set | Protein | Length (aa) | Mean ω |
|---|---|---|---|
| Texas | E | 382 | 0.017736 |
| NS1 | 285 | 0.065975 | |
| California | E | 380 | 0.020650 |
| NS1 | 284 | 0.093021 |
4. Discussion
To some extent, all RNA viruses exist as a compilation of closely related variants, i.e. a mutant swarm (Biebricher and Eigen, 2006). Despite the fact that this swarm has been clearly implicated in viral fitness, pathogenesis, and adaptability, evolutionary studies of natural populations to date rely almost exclusively on consensus sequence analyses. Mutant swarm size and composition may be particularly important for arboviruses, which require replication not only in diverse tissues but also divergent hosts. Here, we evaluated the size and composition of SLEV swarms in single-passage mosquito isolates from Texas and California. Although our data are somewhat limited by the availability of low passage isolates, significant insight into how time, space, and transmission cycles contribute to shaping the SLEV mutant swarm was gained.
The Texas data set provided an opportunity to monitor genetic changes in a single location over time (Table 1). The Texas phylogeny clearly demonstrates that all variants within each isolate group together (Fig. 1A). The fact that variants within the same strain are more closely related to each other than other strains is not surprising, yet it is interesting that no two isolates from different years share a single variant. This, together with the relationships depicted in both the consensus and variant phylogenetic trees, demonstrates that a unique strain may take hold each successive year in a time-independent fashion (Fig. 1A and Fig. S1A). For example, the 2003 strain is more closely related to the 1991 strain than it is to any of the more recent strains, while the 1998 strains are relatively distant from all other sequences (Fig. 1A and Fig. S1A). This suggests that either a new strain was introduced each successive year or that multiple strains coexist locally. Although reintroduction does occasionally occur (Kramer and Chandler, 2001), previous studies demonstrate that the latter (i.e. local maintenance) is more likely (May et al., 2008). Without analysis of multiple strains from each year, it is not entirely clear if significant inter-host variation exists, but comparison of the sequences from the two 1998 strains suggest that this is not the case, and that a single strain likely dominates annually. The variable levels of activity in each successive year in Texas do not necessarily suggest an adaptive process (http://www.cdc.gov/ncidod/dvbid/arbor), yet individual strains may be better suited for specific environmental conditions present in different years. In addition, the percent of nonsynonomous change both on the consensus level from year to year and within the mutant swarm is not substantially different from what one would predict given a general lack of selection (Holmes, 2003). It is reasonable to assume that genetic diversity could decrease over time as a virus becomes more specialized to the hosts and ecology unique to a particular region, yet one would expect a dominance of positive selection in this case. The lack of strong statistical support for a negative correlation between intra-host heterogeneity and time when controlling for phylogeny and the outlier (Table 4) agrees with the results of the dN/dS which support a dominance of purifying selection (Table 6). Since the initially observed correlation was confounded by the phylogenetic structure of the sequences, the currently low swarm sizes might possibly be explained by other factors independent of phylogeny.
The source of all isolates from the Houston area is Cx. quinquefasciutus mosquitoes, which is the primary vector responsible for urban transmission in the southwest (Reisen, 2003). These mosquitoes are unique among primary vectors of SLEV in that they do not enter diapause (Hayes, 1975). As a result, seasonal maintenance may be achieved through a continuous low level of horizontal transmission, which could equate to significant seasonal bottlenecking during periods of decreased activity (Bellamy et al., 1968). In addition, Cx. quinquefasciutus have been shown to be less susceptible to SLEV infection than Cx. tarsalis (Meyer et al., 1983; Hardy and Reeves, 1990). Little is known about the specifics of within-host bottlenecks in mosquito vectors, but the potential for bottlenecks exists at well-documented barriers including midgut infection and egresss, and salivary gland infection and exit (Kramer et al., 1981; Girard et al., 2004). For these reasons, changes in vector competence and even more, in vectorial capacity, could play a significant role in determining the effective viral population size within specific vector populations.
The general phylogenetic grouping of CA strains from the same counties argues against drawing conclusions on the effect of time without consideration of geographic location (Fig. 1B). However, when COAV isolates are considered independently, a negative correlation between time and intrahost diversity is present although it is far less convincing on the amino-acid level (Tables 4 and 5). A larger data set would help to clarify if there is any clear temporal pattern with intrahost diversity within individual regions. The other unique characteristic of the CA strains is that, unlike Texas strains, some variants do in fact group within other isolates (Fig. 1B). This is particularly the case with COAV isolates, where there is significant intersection of sequences in all the represented strains. This degree of relatedness agrees with previous finding that yearly strain maintenance is likely common in California (Reisen, 2003). In addition to the specific role of the vector in shaping the viral swarm, a difference on the avian host side may exist. Experimental passage of SLEV in both chickens and Cx. pipiens mosquitoes has demonstrated that the size of the mutant swarm is host-dependent (Ciota et al., 2009). It is reasonable to assume that the size of the mutant swarm could also be species-dependent. Variation in host preference and host abundance has been demonstrated at the sites from which these strains were isolated and could play a major role in shaping individual populations (Reisen, 2003; Lothrop and Reisen, 2001). Studies assessing viremia levels in various bird species following infection with CA strains of SLEV found that very few species produce viremia levels sufficient for infection of mosquitoes (Reisen et al., 2001, 2003), demonstrating that the availability of competent hosts could be a significant limiting factor on both strain diversity and overall activity.
Despite the difference in the relationship between time and intrahost diversity seen in California and Texas, all of the recent isolates from both locations are highly genetically homogeneous (Tables 2 and 3). Although the requirement of all arboviruses to cycle between divergent hosts may restrict genetic change (Holmes, 2003), the extent to which these isolates lack genetic variability is somewhat surprising given the high mutation rate of RNA viruses (Drake and Holland, 1999). A previous study assessing the level of intrahost diversity within the same genomic region of WNV demonstrated that significant genetic heterogenity exists in isolates from Cx. pipiens mosquitoes from New York (Jerzak et al., 2005). Subsequent studies with adapted populations of WNV and SLEV demonstrated that differences in mutant swarm size may have significant implications for host range (Ciota et al., 2007). As with all organisms, a general lack of genetic diversity limits the ability to exploit new and changing ecology, and could therefore be an important factor in predicting exploit of new habitats and epidemiological patterns in changing landscapes. West Nile virus invaded Texas and California in 2002 and 2003, respectively (Reisen et al., 2004; Lillibridge et al., 2004). Interestingly, WNV has appeared to displace SLEV in California yet the two continue to coexist in Texas (Rios et al., 2006; Reisen et al., 2008). The combination of the close antigenic relationship and the sharing of transmission cycles by WNV and SLEV make their interaction inevitable. WNV antibody-positive birds will possess at least partial protection from SLEV and, given the lower viremia and subsequently more limited effective vector range, it is no surprise that WNV could displace SLEV in areas where they coexist. The Alphavirus Western equine encephalomyelitis virus reemerged in southeastern California in 2005, demonstrating that an antigenically distinct virus which utilizes the same transmission cycle is capable of coexisting in the region (Hom et al., 2006). Genetic diversity within a viral swarm could also correspond to antigenic diversity with variable levels of cross-reactivity to closely related viruses, which in turn could have direct affects on the capacity to evade competition. In addition, the existence of WNV could itself drive the purification of the SLEV population, which could potentially contribute to the current lack of intrahost diversity in both California and Texas.
Overall, this study demonstrates the power of mutant spectrum analysis relative to traditional consensus sequencing. Evaluation of relationships among consensus sequences have historically provided valuable information in the understanding of virus evolution and epidemiology, yet it is now clear that our conclusions to date, particularly with regards to RNA viruses, are drawn from potentially incomplete data sets. Although the methods presented here are currently not feasible on a large scale, recent technological advances in the ability to perform high throughput deep sequencing are beginning to provide researchers with the ability to consider variation in viral swarms on a broader and more precise level. Such analyses are likely to reveal characteristics and relationships among virus variants that have broad implications for viral evolution, fitness, and epidemiology.
Supplementary Material
Acknowledgments
The authors would like to thank R. Tesh and The World Reference Center for Emerging Viruses and Arboviruses (WRCEVA), through the University of Texas Medical Branch (UTMB) in Galveston, Texas, for providing Texas mosquito isolates. Additionally, we thank A. Brault, and W. Reisen for providing original mosquito isolates from California. We also thank J. Maffei for assistance with virus amplification. The authors thank the Wads-worth Center Molecular Genetics Core for sequencing, and the Wadsworth Center Media and Tissue Culture Facility for providing cells and media for this work. This work was supported partially by federal funds from the National Institute of Allergy and Infectious Disease, National Institutes of Health (contract number NO1-AI-25490) and, National Institutes of Health (grant numbers RO1-AI-077669).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.meegid.2010.12.007.
References
- Baillie GJ, Kolokotronis SO, Waltari E, Maffei JG, Kramer LD, Perkins SL. Phylogenetic and evolutionary analyses of St. Louis encephalitis virus genomes. Molecular Phylogenetics and Evolution. 2008;47:717–728. doi: 10.1016/j.ympev.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Bellamy RE, Reeves WC, Scrivani RP. Experimental cyclic transmission of St. Louis encephalitis virus in chickens and Culex mosquitoes through a year. American Journal of Epidemiology. 1968;87:484–495. doi: 10.1093/oxfordjournals.aje.a120838. [DOI] [PubMed] [Google Scholar]
- Bernard KA, Kramer LD. West Nile virus activity in the United States, 2001. Viral Immunology. 2001;14:319–338. doi: 10.1089/08828240152716574. [DOI] [PubMed] [Google Scholar]
- Biebricher CK, Eigen M. What is a quasispecies? Current Topics in Microbiology and Immunology. 2006;299:1–31. doi: 10.1007/3-540-26397-7_1. [DOI] [PubMed] [Google Scholar]
- Bradford CM, Nascarella MA, Burns TH, Montford JR, Marsland EJ, Pepper CB, Presley SM. First report of West Nile virus in mosquitoes from Lubbock County Texas. Journal of American Mosquito Control Association. 2005;21:102–105. doi: 10.2987/8756-971X(2005)21[102:FROWNV]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Brault AC, Huang CY, Langevin SA, Kinney RM, Bowen RA, Ramey WN, Panella NA, Holmes EC, Powers AM, Miller BR. A single positively selected West Nile viral mutation confers increased virogenesis in American crows. Nature Genetics. 2007;39:1162–1166. doi: 10.1038/ng2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciota AT, Jia Y, Payne AF, Jerzak G, Davis LJ, Young DS, Ehrbar D, Kramer LD. Experimental passage of St. Louis encephalitis virus in vivo in mosquitoes and chickens reveals evolutionarily significant virus characteristics. PLoS One. 2009;4:e7876. doi: 10.1371/journal.pone.0007876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciota AT, Lovelace AO, Jones SA, Payne A, Kramer LD. Adaptation of two flaviviruses results in differences in genetic heterogeneity and virus adaptability. Journal of General Virology. 2007;88:2398–2406. doi: 10.1099/vir.0.83061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsie RF, Jr, Ward RA. Identification and geographical distribution of the mosquitoes of North America, north of Mexico. Supplement to Mosquito Systematics. 1981;1:1–313. [Google Scholar]
- Drake JW, Holland JJ. Mutation rates among RNA viruses. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. American Naturalist. 1985;125:1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- Girard YA, Klingler KA, Higgs S. West Nile virus dissemination and tissue tropisms in orally infected Culex pipiens quinquefasciatus. Vector-Borne and Zoonotic Diseases. 2004;4:109–122. doi: 10.1089/1530366041210729. [DOI] [PubMed] [Google Scholar]
- Hardy JL, Reeves WC. Experimental studies on infection in vectors. In: Reeves WC, editor. Epidemiology and control of mosquito-borne arboviruses in California, 1943–1987 Sacramento, California. California Mosquito Vector Control Association; Sacramento, CA: 1990. pp. 145–250. [Google Scholar]
- Hayes J. Seasonal changes in population structure of Culex pipiens quinque-fasciatus Say (Diptera:Culicidae): study of an isolated population. Journal of Medical Entomology. 1975;12:167–178. doi: 10.1093/jmedent/12.2.167. [DOI] [PubMed] [Google Scholar]
- Higgs S, Snow K, Gould EA. The potential for West Nile virus to establish outside of its natural range: a consideration of potential mosquito vectors in the United Kingdom. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2004;98:82–87. doi: 10.1016/s0035-9203(03)00004-x. [DOI] [PubMed] [Google Scholar]
- Holmes EC. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. Journal of Virology. 2003;77:11296–11298. doi: 10.1128/JVI.77.20.11296-11298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom A, Bonilla D, Kjemtrup A, Kramer VL, Cahoon-Young B, Barker CM, Marcus L, Glaser C, Cossen C, Baylis E, et al. Surveillance for mosquito-borne encephalitis virus activity and human disease, including West Nile virus, in California, 2005. Proceedings of the Mosquito Vector Control Association California. 2006;74:43–55. [Google Scholar]
- Jerzak G, Bernard KA, Kramer LD, Ebel GD. Genetic variation in West Nile virus from naturally infected mosquitoes and birds suggests quasispecies structure and strong purifying selection. Journal of General Virology. 2005;86:2175–2183. doi: 10.1099/vir.0.81015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerzak GV, Bernard K, Kramer LD, Shi PY, Ebel GD. The West Nile virus mutant spectrum is host-dependant and a determinant of mortality in mice. Virology. 2007;360:469–476. doi: 10.1016/j.virol.2006.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar N, Langevin S, Hinten S, Nemeth N, Edwards E, Hettler D, Davis B, Bowen R, Bunning M. Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerging Infectious Diseases. 2003;9:311–322. doi: 10.3201/eid0903.020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Molecular Biology and Evolution. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Kramer LD, Chandler LJ. Phylogenetic analysis of the envelope gene of St. Louis encephalitis virus. Archives of Virology. 2001;146:2341–2355. doi: 10.1007/s007050170007. [DOI] [PubMed] [Google Scholar]
- Kramer LD, Hardy JL, Presser SB, Houk EJ. Dissemination barriers for western equine encephalomyelitis virus in Culex tarsalis infected after ingestion of low viral doses. American Journal of Tropical Medicine and Hygiene. 1981;30:190–197. doi: 10.4269/ajtmh.1981.30.190. [DOI] [PubMed] [Google Scholar]
- Lillibridge KM, Parsons R, Randle Y, Da Rosa APAT, Guzman H, Siirin M, Wuithiranyagool T, Hailey C, Higgs S, Bala AA, Pascua R, Meyer T, Vanlandingham DL, Tesh RB. The 2002 introduction of West Nile virus into Harris County, Texas, an area historically endemic for St. Louis encephalitis. American Journal of Tropical Medicine and Hygiene. 2004;70:676–681. [PubMed] [Google Scholar]
- Lothrop HD, Reisen WK. Landscape affects the host-seeking patterns of Culex tarsalis (Diptera: Culicidae) in the Coachella Valley of California. Journal of Medical Entomology. 2001;38:325–332. doi: 10.1603/0022-2585-38.2.325. [DOI] [PubMed] [Google Scholar]
- Lumsden LL. St. Louis encephalitis in 1933. Observations on epidemiological features. Public Health Reports. 1958;73:340. [PMC free article] [PubMed] [Google Scholar]
- Marra PP, Griffing SM, McLean RG. West Nile virus and wildlife health. Emerging Infectious Diseases. 2003;9:898–899. doi: 10.3201/eid0907.030277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MA, Carrillo C, Gonzalez-Candelas F, Moya A, Domingo E, Sobrino F. Fitness alteration of foot-and-mouth disease virus mutants: measurement of adaptability of viral quasispecies. Journal of Virology. 1991;65:3954–3957. doi: 10.1128/jvi.65.7.3954-3957.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May FJ, Li L, Zhang S, Guzman H, Beasley DW, Tesh RB, Higgs S, Raj P, Bueno R, Jr, Randle Y, Chandler L, Barrett AD. Genetic variation of St. Louis encephalitis virus. Journal of General Virology. 2008;89:1901–1910. doi: 10.1099/vir.0.2008/000190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean RG, Mullenix J, Kerschner J, Hamm J. The house sparrow (Passer domesticus) as a sentinel for St. Louis encephalitis virus. American Journal of Tropical Medicine and Hygiene. 1983;32:1120–1129. doi: 10.4269/ajtmh.1983.32.1120. [DOI] [PubMed] [Google Scholar]
- Meyer RP, Hardy JL, Presser SB, Bruen JP. Comparative arboviral susceptibility of female Culex tarsalis (Diptera: Culicidae) collected in CO2-baited traps and reared from field-collected pupae. Journal of Medical Entomology. 1983;20:56–61. doi: 10.1093/jmedent/20.1.56. [DOI] [PubMed] [Google Scholar]
- Novella IS Ebendick-Corp. Molecular basis of fitness loss and fitness recovery in vesicular stomatitis virus. Journal of Molecular Biology. 2004;342:1423–1430. doi: 10.1016/j.jmb.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2009. URL http://www.R-project.org. [Google Scholar]
- Reisen W, Lothrop H, Chiles R, Madon M, Cossen C, Woods L, Husted S, Kramer V, Edman J. West Nile virus in California. Emerging Infectious Diseases. 2004;10:1369–1378. doi: 10.3201/eid1008.040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisen WK. Epidemiology of St. Louis encephalitis virus. Advances in Virus Research. 2003;61:139–183. doi: 10.1016/s0065-3527(03)61004-3. [DOI] [PubMed] [Google Scholar]
- Reisen WK, Chiles RE, Martinez VM, Fang Y, Green EN. Experimental infection of California birds with western equine encephalomyelitis and St. Louis encephalitis viruses. Journal of Medical Entomology. 2003;40:968–982. doi: 10.1603/0022-2585-40.6.968. [DOI] [PubMed] [Google Scholar]
- Reisen WK, Kramer LD, Chiles RE, Green EGN, Martinez VM. Encephalitis virus persistence in California birds: preliminary studies with house finches (Carpodacus mexicanus) Journal of Medical Entomology. 2001;38:393–399. doi: 10.1603/0022-2585-38.3.393. [DOI] [PubMed] [Google Scholar]
- Reisen WK, Lothrop HD, Wheeler SS, Kennsington M, Gutierrez A, Fang Y, Garcia S, Lothrop B. Persistent West Nile virus transmission and the apparent displacement St. Louis encephalitis virus in southeastern California, 2003–2006. Journal of Medical Entomology. 2008;45:494–508. doi: 10.1603/0022-2585(2008)45[494:pwnvta]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios J, Hacker CS, Hailey CA, Parsons RE. Demographic and spatial analysis of West Nile virus and St. Louis encephalitis in Houston, Texas. Journal of the American Mosquito Control Association. 2006;22:254–263. doi: 10.2987/8756-971X(2006)22[254:DASAOW]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ. PhD Dissertation. The University of Texas; Austin: 2006. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets Under the Maximum Likelihood Criterion. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.